Abstract
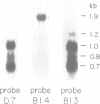
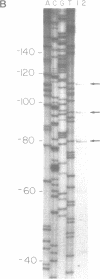
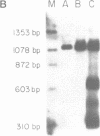
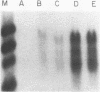
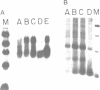
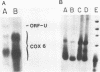
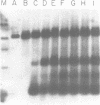
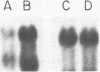
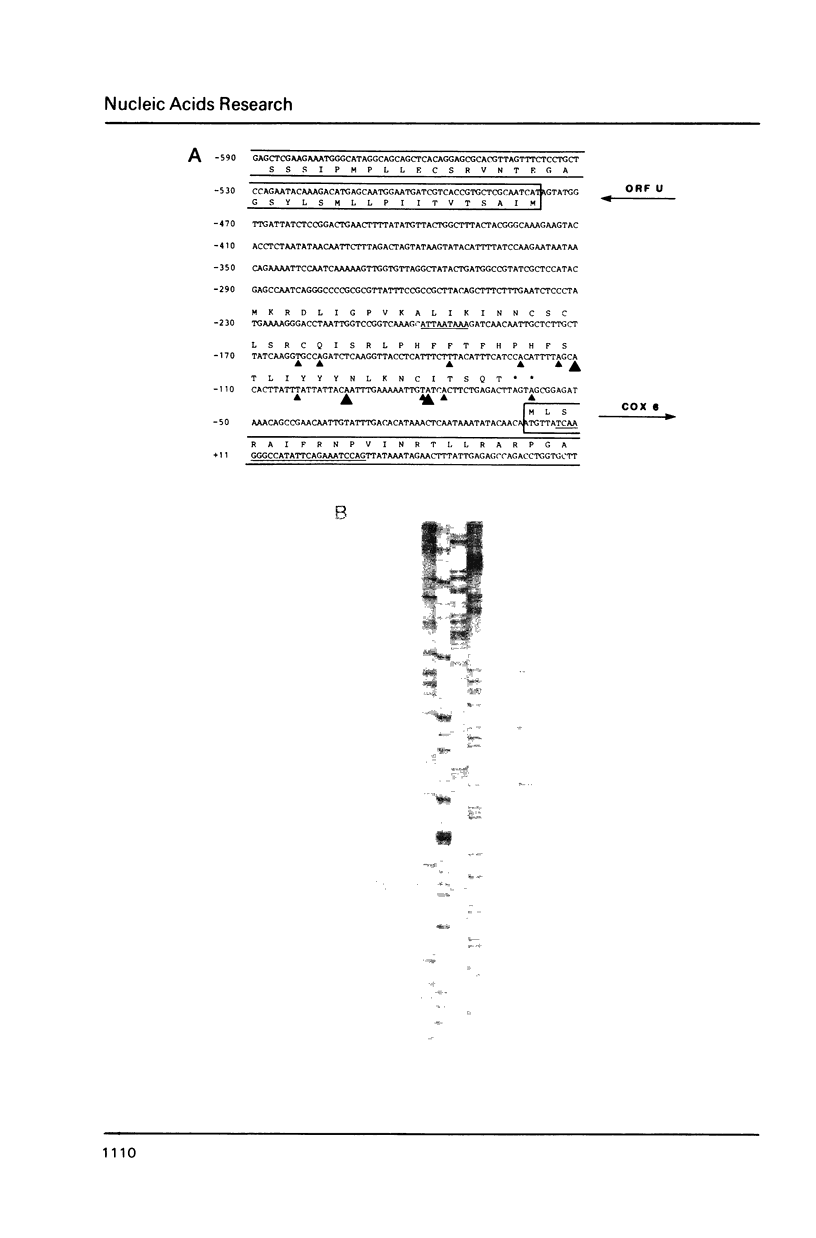
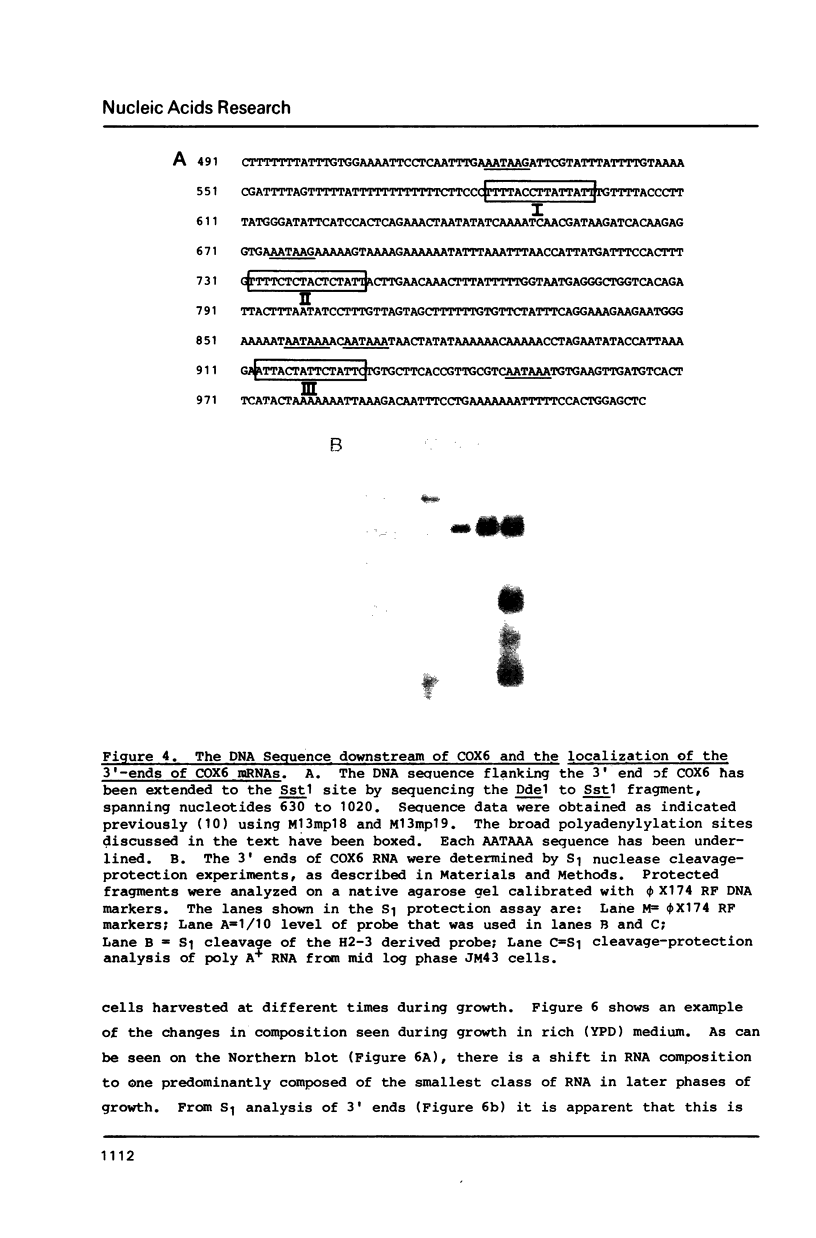
COX6 and its surrounding genetic locus have been characterized for the yeast Saccharomyces cerevisiae. Flanking genes are found closely spaced upstream and downstream of COX6. The upstream gene and COX6 are transcribed from opposite strands and are separated by no more than 300 bp. COX6 is transcribed into three different size classes of mRNA (1000b, 830b, and 700b) differing in length in their 3' untranslated regions. All three classes of mRNAs are found on polysomes and, hence, are most likely translated. The different COX6 mRNAs vary in abundance during growth in rich media and are affected differentially as cells are shifted into media containing high or low glucose concentrations. The largest mRNA is much more susceptible to glucose repression/derepression than are the two smaller mRNAs, whereas the smallest RNA is preferentially accumulated during growth in rich media. These findings demonstrate that COX6 mRNAs with different 3'-termini are either synthesized differentially or differ in stability and suggest the existence of a complex system regulating COX6 expression.
Full text
PDF















Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Bilanchone V., Duester G., Edwards Y., Smith M. Multiple mRNAs for human alcohol dehydrogenase (ADH): developmental and tissue specific differences. Nucleic Acids Res. 1986 May 12;14(9):3911–3926. doi: 10.1093/nar/14.9.3911. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Birnstiel M. L., Busslinger M., Strub K. Transcription termination and 3' processing: the end is in site! Cell. 1985 Jun;41(2):349–359. doi: 10.1016/s0092-8674(85)80007-6. [DOI] [PubMed] [Google Scholar]
- Bond B. J., Davidson N. The Drosophila melanogaster actin 5C gene uses two transcription initiation sites and three polyadenylation sites to express multiple mRNA species. Mol Cell Biol. 1986 Jun;6(6):2080–2088. doi: 10.1128/mcb.6.6.2080. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Buckholz R. G., Cooper T. G. Oxalurate induction of multiple URA3 transcripts in Saccharomyces cerevisiae. Mol Cell Biol. 1983 Nov;3(11):1889–1897. doi: 10.1128/mcb.3.11.1889. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carlson M., Osmond B. C., Botstein D. Mutants of yeast defective in sucrose utilization. Genetics. 1981 May;98(1):25–40. doi: 10.1093/genetics/98.1.25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carroll S. L., Bergsma D. J., Schwartz R. J. Structure and complete nucleotide sequence of the chicken alpha-smooth muscle (aortic) actin gene. An actin gene which produces multiple messenger RNAs. J Biol Chem. 1986 Jul 5;261(19):8965–8976. [PubMed] [Google Scholar]
- Citron B. A., Donelson J. E. Sequence of the Saccharomyces GAL region and its transcription in vivo. J Bacteriol. 1984 Apr;158(1):269–278. doi: 10.1128/jb.158.1.269-278.1984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Forsburg S. L., Guarente L. Mutational analysis of upstream activation sequence 2 of the CYC1 gene of Saccharomyces cerevisiae: a HAP2-HAP3-responsive site. Mol Cell Biol. 1988 Feb;8(2):647–654. doi: 10.1128/mcb.8.2.647. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guarente L., Mason T. Heme regulates transcription of the CYC1 gene of S. cerevisiae via an upstream activation site. Cell. 1983 Apr;32(4):1279–1286. doi: 10.1016/0092-8674(83)90309-4. [DOI] [PubMed] [Google Scholar]
- Guarente L. Regulatory proteins in yeast. Annu Rev Genet. 1987;21:425–452. doi: 10.1146/annurev.ge.21.120187.002233. [DOI] [PubMed] [Google Scholar]
- Hickok N. J., Seppänen P. J., Kontula K. K., Jänne P. A., Bardin C. W., Jänne O. A. Two ornithine decarboxylase mRNA species in mouse kidney arise from size heterogeneity at their 3' termini. Proc Natl Acad Sci U S A. 1986 Feb;83(3):594–598. doi: 10.1073/pnas.83.3.594. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johnston M. A model fungal gene regulatory mechanism: the GAL genes of Saccharomyces cerevisiae. Microbiol Rev. 1987 Dec;51(4):458–476. doi: 10.1128/mr.51.4.458-476.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kruys V., Wathelet M., Poupart P., Contreras R., Fiers W., Content J., Huez G. The 3' untranslated region of the human interferon-beta mRNA has an inhibitory effect on translation. Proc Natl Acad Sci U S A. 1987 Sep;84(17):6030–6034. doi: 10.1073/pnas.84.17.6030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kukuruzinska M. A., Robbins P. W. Protein glycosylation in yeast: transcript heterogeneity of the ALG7 gene. Proc Natl Acad Sci U S A. 1987 Apr;84(8):2145–2149. doi: 10.1073/pnas.84.8.2145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Laughon A., Gesteland R. F. Primary structure of the Saccharomyces cerevisiae GAL4 gene. Mol Cell Biol. 1984 Feb;4(2):260–267. doi: 10.1128/mcb.4.2.260. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lowry C. V., Zitomer R. S. Oxygen regulation of anaerobic and aerobic genes mediated by a common factor in yeast. Proc Natl Acad Sci U S A. 1984 Oct;81(19):6129–6133. doi: 10.1073/pnas.81.19.6129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McNeil J. B., Smith M. Transcription initiation of the Saccharomyces cerevisiae iso-1-cytochrome c gene. Multiple, independent T-A-T-A sequences. J Mol Biol. 1986 Feb 5;187(3):363–378. doi: 10.1016/0022-2836(86)90439-0. [DOI] [PubMed] [Google Scholar]
- Norrander J., Kempe T., Messing J. Construction of improved M13 vectors using oligodeoxynucleotide-directed mutagenesis. Gene. 1983 Dec;26(1):101–106. doi: 10.1016/0378-1119(83)90040-9. [DOI] [PubMed] [Google Scholar]
- Olesen J., Hahn S., Guarente L. Yeast HAP2 and HAP3 activators both bind to the CYC1 upstream activation site, UAS2, in an interdependent manner. Cell. 1987 Dec 24;51(6):953–961. doi: 10.1016/0092-8674(87)90582-4. [DOI] [PubMed] [Google Scholar]
- Parikh V. S., Morgan M. M., Scott R., Clements L. S., Butow R. A. The mitochondrial genotype can influence nuclear gene expression in yeast. Science. 1987 Jan 30;235(4788):576–580. doi: 10.1126/science.3027892. [DOI] [PubMed] [Google Scholar]
- Power S. D., Lochrie M. A., Sevarino K. A., Patterson T. E., Poyton R. O. The nuclear-coded subunits of yeast cytochrome c oxidase. I. Fractionation of the holoenzyme into chemically pure polypeptides and the identification of two new subunits using solvent extraction and reversed phase high performance liquid chromatography. J Biol Chem. 1984 May 25;259(10):6564–6570. [PubMed] [Google Scholar]
- Poyton R. O. Cooperative interaction between mitochondrial and nuclear genomes: cytochrome c oxidase assembly as a model. Curr Top Cell Regul. 1980;17:231–295. doi: 10.1016/b978-0-12-152817-1.50012-2. [DOI] [PubMed] [Google Scholar]
- Purvis I. J., Loughlin L., Bettany A. J., Brown A. J. Translation and stability of an Escherichia coli beta-galactosidase mRNA expressed under the control of pyruvate kinase sequences in Saccharomyces cerevisiae. Nucleic Acids Res. 1987 Oct 12;15(19):7963–7974. doi: 10.1093/nar/15.19.7963. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schneider J. C., Guarente L. The untranslated leader of nuclear COX4 gene of Saccharomyces cerevisiae contains an intron. Nucleic Acids Res. 1987 Apr 24;15(8):3515–3529. doi: 10.1093/nar/15.8.3515. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Struhl K. Nucleotide sequence and transcriptional mapping of the yeast pet56-his3-ded1 gene region. Nucleic Acids Res. 1985 Dec 9;13(23):8587–8601. doi: 10.1093/nar/13.23.8587. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Szekely E., Montgomery D. L. Glucose represses transcription of Saccharomyces cerevisiae nuclear genes that encode mitochondrial components. Mol Cell Biol. 1984 May;4(5):939–946. doi: 10.1128/mcb.4.5.939. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Trueblood C. E., Poyton R. O. Differential effectiveness of yeast cytochrome c oxidase subunit genes results from differences in expression not function. Mol Cell Biol. 1987 Oct;7(10):3520–3526. doi: 10.1128/mcb.7.10.3520. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tzagoloff A., Myers A. M. Genetics of mitochondrial biogenesis. Annu Rev Biochem. 1986;55:249–285. doi: 10.1146/annurev.bi.55.070186.001341. [DOI] [PubMed] [Google Scholar]
- Warner J. R., Mitra G., Schwindinger W. F., Studeny M., Fried H. M. Saccharomyces cerevisiae coordinates accumulation of yeast ribosomal proteins by modulating mRNA splicing, translational initiation, and protein turnover. Mol Cell Biol. 1985 Jun;5(6):1512–1521. doi: 10.1128/mcb.5.6.1512. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wright R. M., Ko C., Cumsky M. G., Poyton R. O. Isolation and sequence of the structural gene for cytochrome c oxidase subunit VI from Saccharomyces cerevisiae. J Biol Chem. 1984 Dec 25;259(24):15401–15407. [PubMed] [Google Scholar]