Abstract
TRAIL (tumor necrosis factor-related apoptosis-inducing ligand) is a recently identified member of the tumor necrosis factor cytokine superfamily. TRAIL has been shown to induce apoptosis in various tumor cell lines, whereas most primary cells seem to be resistant. These observations have raised considerable interest in the use of TRAIL in tumor therapy. Yet little is known about the physiological function of TRAIL. This is particularly the case in the immune system, where TRAIL has been suggested by some to be involved in target cell killing and lymphocyte death. We have developed a panel of mAbs and soluble proteins to address the role of TRAIL in lymphocyte development. These studies demonstrate activation-induced sensitization of thymocytes to TRAIL-mediated apoptosis and expression of the apoptosis-inducing TRAIL receptors. However, with the use of several model systems, our subsequent experiments rule out the possibility that TRAIL plays a major role in antigen-induced deletion of thymocytes. In contrast to thymocytes, there is no up-regulation of TRAIL receptors in peripheral T cells on activation, which remain resistant to TRAIL. Thus, susceptibility to TRAIL-induced apoptosis is controlled differently by central and peripheral T cells.
TRAIL [Apo2L or tumor necrosis factor (TNF)-related apoptosis-inducing ligand] is a recently identified member of the TNF gene superfamily (1, 2). TRAIL interacts with four cellular receptors that form a distinct subgroup within the TNF receptor superfamily. TRAIL receptor 1 (TRAIL-R1 or DR4) (3) and TRAIL receptor 2 (also called TRAIL-R2, DR5, TRICK2, or KILLER) (4–7) have cytoplasmic death domains and signal for apoptosis and for NF-κB (8). Two additional receptors highly homologous to TRAIL-R1 and -R2 were subsequently cloned that lack the death domain and have been proposed to act as functional blockers or decoys at the cell membrane. TRAIL-R3 (also called DcR1, TRAIL-R3, TRID, or LIT) (9–12) is a glycosylphosphatidylinositol-anchored cell surface protein that lacks a cytoplasmic tail. TRAIL-R4 (also called DcR2 or TRUNDD) has a truncated death domain that does not signal apoptosis induction but can activate NF-κB (13–15). A fifth, slightly weaker receptor for TRAIL is osteoprotegrin, which is a secreted TNF receptor homologue that also binds to osteoprotegrin ligand (OPGL)/receptor activator for NF-κB ligand (RANKL), inhibits osteoclastogenesis, and increases bone density (16).
T cell receptors are created by a stochastic gene rearrangement process during thymocyte development, generating thymocytes bearing useful, as well as unwanted, specificities. Within the latter group, autoreactive thymocytes arise, which are eliminated by negative selection. Negative selection is a consequence of T cell antigen receptor (TCR) ligation (17); it occurs through apoptosis (18) and involves caspase activation (19). However, the precise molecular mechanism of this deletion is unknown; indeed, it remains a matter of controversy whether cell surface receptors are involved. The paradigm of activation-induced cell death in the periphery where Fas (CD95/APO-1) occupies an important position has prompted searches for similar signaling pathways in the thymus. The role of Fas in negative selection is controversial (20, 21). Two other members of the TNF receptor family, CD30 and CD40 (22, 23), have also been implicated, although again the scale and relevance of these findings are disputed.
In this report we have studied the expression of TRAIL and its receptors on thymocytes and peripheral T cells and evaluated the role of TRAIL in activation-induced death in these two populations.
Materials and Methods
Human Thymus Organ Culture (HTOC).
Pediatric (children from 2 months to 2 years old, anonymously) thymus samples that are normally discarded during cardiac surgery were collected. Fresh thymus was cut into 1-mm3 pieces and floated on filters (Millipore) in RPMI medium 1640 containing 10% FCS at 37°C and 5% CO2. Human thymus organ culture was monitored over a period of 48 h, during which CD4, CD8, and CD3 expression remained stable. Cross-linked TRAIL [100 ng/ml + 5 μg/ml anti-flag (Sigma)], polyclonal antibodies against TRAIL-R1 and TRAIL-R2 (1:100 dilution of rabbit antiserum), anti-CD3 (PharMingen), or 1 μM dexamethasone (Sigma) were added to organ cultures for 18 h unless otherwise stated. Single suspensions of thymocytes were obtained by gentle disruption of the thymus organ, and triple staining with anti-CD4 phycoerythrin (Dako), anti-CD8 Tricolor (Caltag, South San Francisco, CA), or anti-CD19 FITC (Dako) and annexin V FITC (Roche Molecular Biochemicals) was performed.
For superantigen-induced deletion, HTOC was in the presence of 5 μg/ml superantigen enterotoxin B from Staphylococcus aureus (Sigma) and blocking proteins. After 24 h triple staining was performed with biotinylated Vβ 2 and 17 (Immunotech, Luminy, France, and Coulter) mAb followed by streptavidin-phycoerythrin (Sigma) and CD4 and CD8 staining. The remaining organs were left in culture for another 24 h in the presence of blocking proteins only and were subsequently stained as described above.
Antigenic Deletion in F5 Mice.
Thymus organ cultures were as described (24). Briefly fetal thymuses were isolated from day 15 embryos from F5 Rag-1−/− transgenic mice and cultured for 5 days. Then 10 μM nonamer peptide NP 68 from the nucleoprotein of influenza virus (ASNENMDAM) was added in the presence or absence of 20 μg/ml TRAIL-R2-Fc for 20 h. After 24 h thymocytes were harvested and stained with anti-CD4 phycoerythrin and anti-CD8 FITC antibodies. 7-amino actinomyocin D (7AAD) staining was performed as described (24).
For in vivo deletion experiments F5 Rag-1−/− were crossed to Tap-1−/− mice at the National Institute for Medical Research in Mill Hill. As described (25) F5 Rag-1−/− Tap-1−/− were coinjected i.p. with peptide NP68 (50 nmol/20 g body weight) and 150 μg TRAIL-R2-Fc. Endotoxin levels of TRAIL-R2-Fc were checked with a LAL-Endotoxin kit (Sigma) and found to be lower than 0.25 endotoxin units/ml. After 24 h the mice were killed and thymocytes were stained as described above.
51Cr Release Assay.
Cytotoxic activity was assessed by an 8-h chromium release assay. Target cells (Jurkat or BJAB) were radiolabeled with 100–150 μCi Na2[51Cr]O4 and washed three times, and 104 cells were used for each assay. Each experimental condition was set up in triplicate, and chromium release was determined by scintillation counting. Percentage specific lysis was determined as 100 × (cpm experimental wells − cpm spontaneous release)/(cpm maximum release − cpm spontaneous release).
Recombinant Proteins and Antibodies.
Recombinant proteins were prepared as described earlier (6, 26). Polyclonal antibodies against TRAIL-R1 and TRAIL-R2 were raised in rabbits against recombinant TRAIL-R1 and TRAIL-R2-Fc fusion protein. mAbs were raised in mice according to the standard procedure against TRAIL-R1, -R2, -R3, -R4 Fc-fusion proteins and TRAIL. They were tested for specificity in transfected 293T cells. Briefly 293T cells were transfected with TRAIL-R1, -R2, -R3, -R4, or TRAIL cDNA tagged with green fluorescent protein and stained with mAbs against the respective antigens followed by anti-mouse phycoerythrin and analyzed on a Becton Dickinson fluorescence-activated cell sorter. Antibodies were also checked for cross-reactivity.
Results
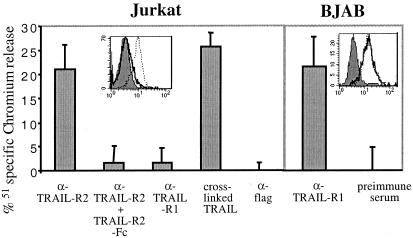
Newly generated mAbs against TRAIL-R1, -R2, -R3, -R4, and TRAIL were characterized on 293T cells transfected with the appropriate cDNA and expression analyzed by fluorescence-activated cell sorter (data not shown). Staining with the mAbs showed that Jurkat cells express TRAIL-R2 and BJAB express TRAIL-R1 and TRAIL-R2 (Fig. 1). Apoptosis was induced either by cross-linked TRAIL (TRAIL-flag + anti-flag) or by a polyclonal rabbit antiserum raised to TRAIL-R1 or TRAIL-R2. Sensitivity to the polyclonal antisera correlated with receptor expression in BJAB and Jurkat, and death was inhibited in Jurkat by TRAIL-R2-Fc (Fig. 1).
Figure 1.
Expression of TRAIL-R1 and -R2 and sensitivity to TRAIL of Jurkat and BJAB. Cells were stained with mAbs against TRAIL-R1 (solid line) or TRAIL-R2 (dashed line). Sensitivity to cross-linked TRAIL and polyclonal antisera to TRAIL-R1 and -R2 was tested with the use of a chromium release assay. Killing was shown to be specific by blocking with 20 μg/ml TRAIL-R2-Fc where indicated. Error bars were calculated from triplicates, this experiment was repeated at least three times.
When murine or human thymocytes are cultured in suspension, a high rate of nonspecific death occurs, most probably because of a lack of signal from neighboring cells, as shown in the murine system (27). We established a novel human thymus organ culture in which fresh human thymus was cut into 1-mm3 pieces and floated on filters. To determine whether HTOC had characteristics similar to those of murine thymus organ culture, we stained surface markers CD4, CD8, CD3, CD69, and annexin V over a 2-week culture period. From this analysis we chose to study events up to 48 h, after which the proportion of the different subsets changed because of increased death rather than thymic development (data not shown).
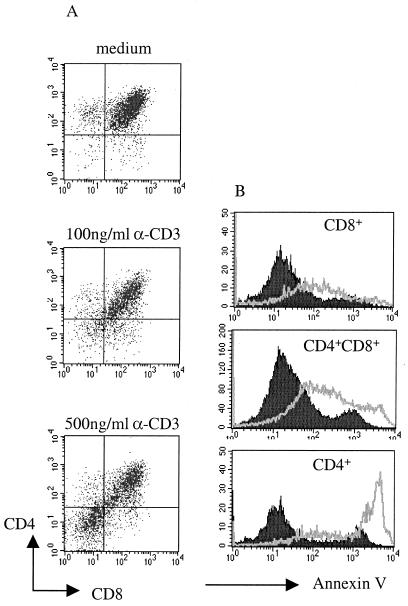
To establish a system of negative selection in this HTOC we tested the effect of CD3 stimulation, which has been used in murine models (28). Addition of anti-CD3 induced the down-modulation of the CD4 and CD8 coreceptors (Fig. 2A), which is also known to precede negative selection in murine thymocytes (17, 29). At a low dose (100 ng/ml) anti-CD3 induces coreceptor down-modulation but does not induce death, whereas high doses (500 ng/ml) mimic negative selection in the human thymus (Fig. 2). CD4+CD8+ and both single positive CD8+ and, more strikingly, CD4+ thymocytes stained positive with annexin V after addition of this high dose of anti-CD3 (Fig. 2B). The lack of requirement for cross-linking in these experiments is probably explained by capture of anti-CD3 by Fc receptors expressed on thymic epithelial cells.
Figure 2.
Human thymic organ culture. Human thymocyes were cultured in the presence or absence of anti-CD3 for 18 h. (A) Staining with anti-CD4 and anti-CD8. (B) Analysis of apoptosis by annexin V staining after electronic gating on CD4+CD8+, CD8+, or CD4+ subpopulations of anti-CD3 (500 ng/ml) stimulated thymocytes (gray line) or unstimulated thymocytes (filled histograms). These results are representative of at least five different experiments.
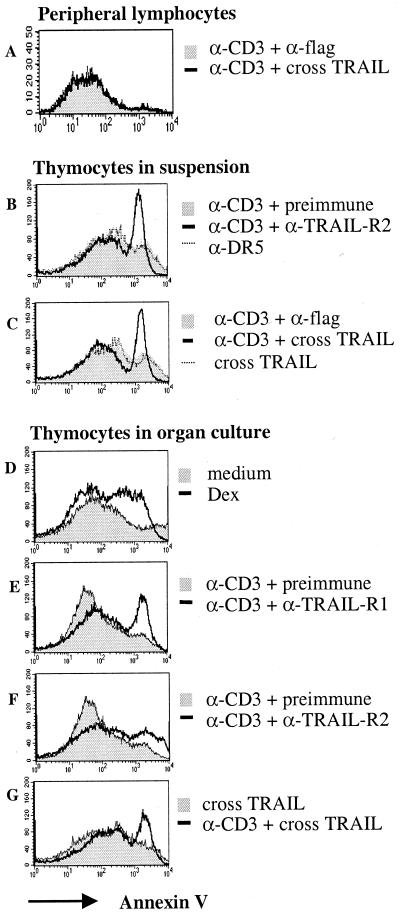
Next we tested for the sensitivity of human thymocytes to TRAIL. Human thymocytes were not sensitive to TRAIL immediately ex vivo or after overnight organ culture. However, the low/sublethal dose of anti-CD3 sensitized thymocytes to TRAIL-induced death after 6 h, 12 h, or 18 h (18 h shown in Fig. 3 D–G) of culture. Death was observed in CD4+CD8+, CD4+, and CD8+ thymocytes to the same extent (data not shown). Similar results were found for thymocytes cultured in suspension, where background death was much higher than in organ culture (Fig. 3 B–C). Trypan blue staining showed a 50% reduction in viable cells when anti-CD3-activated cells were cultured in the presence of anti-TRAIL-R2 or crosslinked TRAIL. Results were further confirmed by propidium iodide staining; subdiploid DNA-containing cells increased from 7% to 33% when TRAIL receptors were crosslinked (data not shown). Anti-CD3 (100 ng/ml), anti-TRAIL-R2, or TRAIL-flag, when used in isolation, did not induce thymocyte death (Fig. 3 B–G).
Figure 3.
Sensitivity to TRAIL. Total human thymocytes or peripheral blood mononuclear cells were cultured for 18 h with polyclonal TRAIL-R1 or -R2 antiserum, cross-linked TRAIL, dexamethasone, preimmune serum (preimmune), or anti-flag in the presence or absence of 100 ng/ml anti-CD3 and stained with annexin V. (A) Human peripheral lymphocytes. (B and C) Human thymocytes cultured in suspension. (D–G) Human thymus organ cultures.
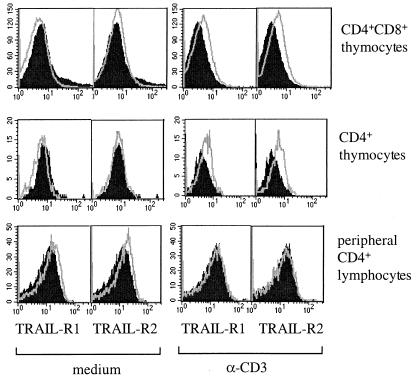
Thymocytes were tested for surface expression of TRAIL receptors and TRAIL by fluorescence-activated cell sorter. After overnight culture human thymocytes did not express any of the TRAIL receptors or TRAIL (Fig. 4). However, after overnight culture in the presence of anti-CD3 (100 ng/ml), CD4+CD8+ and CD4+ thymocytes expressed both TRAIL death receptors TRAIL-R1 and -R2 (Fig. 4). Yet we failed to detect expression of the non-apoptosis-inducing TRAIL receptors R3 and R4 (data not shown). Furthermore, we did not detect membrane expression of TRAIL by fluorescence-activated cell sorter. Finally, the supernatants of human organ cultures or lysed thymocytes stimulated with anti-CD3 were negative for TRAIL expression, according to a sensitive sandwich ELISA detecting picograms of TRAIL recombinant protein (data not shown).
Figure 4.
Human thymocytes express TRAIL-R1 and -R2 upon activation. Thymocytes from organ cultures were cultured in the absence or presence of 100 ng/ml anti-CD3 for 18 h and then stained with mAbs to TRAIL-R1 or -R2 (gray line) or with irrelevant antibody (filled histogram). This is a representative of three separate experiments.
We next looked at peripheral T cells. Human peripheral blood lymphocytes were prepared from blood and cultured in the presence of phytohemagglutinin, phorbol 12-myristate/ionomycin, or anti-CD3 alone or in combination with IL-2, IFNγ, IL-7, IL-13, Mip1α, Mip1β, regulated upon activation normal T cell expressed and selected (RANTES), IL-4, or IL-12 (results are shown only for anti-CD3 stimulation). Cells were analyzed for TRAIL and TRAIL receptor expression over a 6-day time course, and sensitivity to TRAIL was assessed in parallel. Peripheral T lymphocytes stained very weakly positive with anti-TRAIL-R1 and -R2 antibodies, this expression is not up-regulated upon anti-CD3 stimulation (CD4+ lymphocytes shown in Fig. 4). In contrast to the results obtained with thymocytes, we did not observe sensitivity to TRAIL under any of the conditions outlined above (only anti-CD3 stimulation shown in Fig. 3A).
After anti-CD3 stimulation thymocytes up-regulated both TRAIL-R1 and -R2 on their surface and, concomitantly, became susceptible to TRAIL-induced apoptosis, we therefore investigated the possibility that TRAIL is responsible for thymic negative selection. As described earlier, a lethal dose of anti-CD3 (500 ng/ml) induced the death of CD4+CD8+, CD4+, and CD8+ thymocytes (Fig. 2). To determine whether this death was due to TRAIL we performed HTOC in the presence of TRAIL-R2-Fc to block TRAIL activity. However, anti-CD3-induced death of human thymocytes was not blocked by TRAIL-R2-Fc (Fig. 5A). We repeated this experiment with a dose of 10, 20, and 40 μg/ml, and no inhibitory effect was observed at any concentration tested.
Figure 5.
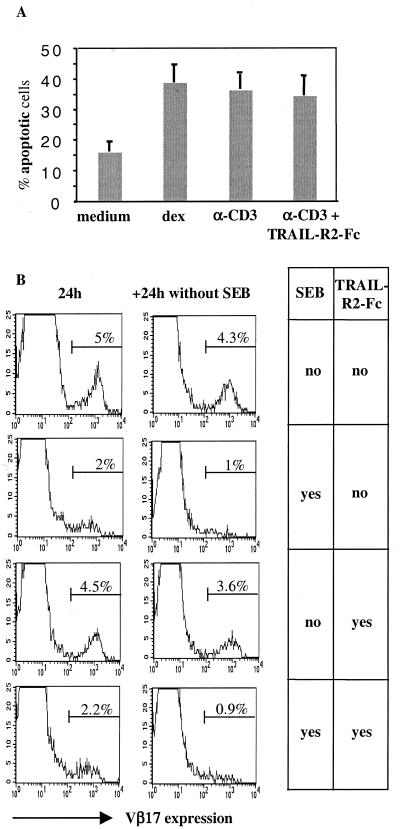
Effects of TRAIL inhibition on negative selection in HTOC. (A) Human thymocytes were cultured in organs for 18 h in the presence of dexamethasone (dex) or a lethal dose of anti-CD3, with or without 20 μg/ml of TRAIL-R2-Fc. Double-positive thymocytes stained positive with annexin V were considered apoptotic. This experiment was repeated at least five times with similar results. (B) HTOC was performed in the presence or absence of superantigen enterotoxin B and/or TRAIL-R2-Fc. After 24 h thymocytes were stained for Vβ2 (not shown) and Vβ17. In a further 24-h culture period, superantigen enterotoxin B was omitted to reveal potential TCR surface expression in the absence of antigen (Right column). Histogram plots are gated on CD4+CD8− thymocytes. This experiment was performed in triplicate with standard deviations of 0.2–0.4%.
As a second model for negative selection we tested superantigen enterotoxin B-induced deletion (30). About 5% of CD4+ thymocytes expressed Vβ17, and upon addition of superantigen the majority of these were deleted after 24 h (Fig. 5B), whereas Vβ2-expressing thymocytes (9–11%) were not deleted (data not shown). To check whether Vβ17-expressing thymocytes were indeed deleted and had not just down-regulated their TCR, culture was extended for a further 24 h without superantigen (31). Vβ17 was not reexpressed at the surface (Fig. 5B, Left), indicating that thymocytes were truly deleted. To test the effect of TRAIL we added TRAIL-R2-Fc (10, 20, and 40 μg/ml), but again there was no inhibition of negative selection, as evidenced by Vβ17 deletion (20 μg/ml shown in Fig. 5B). A further 24-h culture without superantigen but in the presence of TRAIL-R2-Fc did not rescue the expression of Vβ17 on these cells (Fig. 5B).
To test the role of TRAIL in vivo, we used a transgenic mouse system. F5 mice are transgenic for an αβ TCR pair (F5) that recognizes a nonamer peptide NP68 in the context of MHC class I H-2Db. Because allelic exclusion on the α chain is not complete, the F5 mice have been crossed onto a recombinase-deficient background to create F5 Rag-1−/−, where all mature T cells are CD8+ and express only the transgenic receptor (24). To avoid any effect of peripheral lymphocytes on thymic selection events (32), a further cross was performed. The F5 Rag-1−/− mice were crossed onto a Tap-1−/− background (lacking the peptide transporter and hence expressing very low levels of MHC class I molecules). CD4+CD8+ thymocytes from these mice do not progress to mature CD8+, as they are not positively selected. The F5 Rag−/−TAP−/− mice therefore do not contain any CD4+ or CD8+ thymocytes or any mature T lymphocytes (O.W., unpublished observation).
These mice were injected with NP68, the agonist peptide recognized by the F5 TCR. After 24 h CD4 and CD8 expression was down-modulated (Fig. 6A), and 55% of thymocytes died, as shown by 7-amino actinomycin D staining (Fig. 6B). Murine and human TRAIL have been shown to be cross-reactive, and, furthermore, human TRAIL-R2 has been shown to block murine TRAIL (1, 33). Injection of TRAIL-R2-Fc alone did not induce any death above background. Moveover, antigenic deletion of thymocytes still occurred in the presence of 150 μg per mouse TRAIL-R2-Fc (Fig. 6A). This amount was chosen according to a report by Song et al., where collagen-induced arthritis was exacerbated in the presence of 100 μg per mouse of TRAIL-R2-Fc. Furthermore, 150 μg of Fas-Fc was also sufficient to block antigenic-induced deletion of thymocytes in vivo (34).
Figure 6.
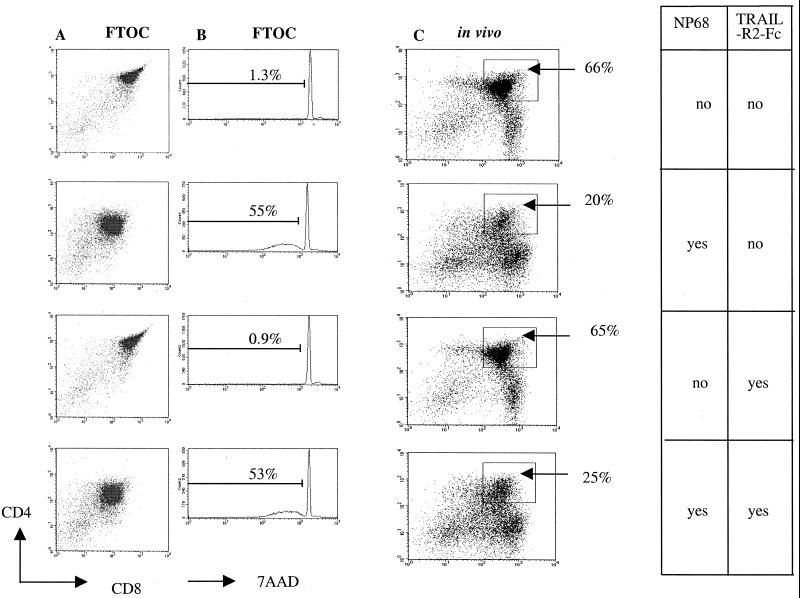
Analysis of negative selection in the murine F5 model. (C) The same experiment was performed in FTOC of F5 Rag−/−, and results were analyzed after 24 h. The loss of CD4+CD8+ thymocytes is indicated as a percentage. (B) The viability of these cells was also assessed by 7-amino antinomycin D; the percentage of dead cells is indicated. (A) F5 Rag-1−/− Tap-1−/− mice were coinjected with NP68 and TRAIL-R2-Fc where indicated and killed after 24 h, and thymocytes were analyzed for CD4 and CD8.
We cannot formally exclude the possibility that TRAIL-R2-Fc did not reach the thymus in vivo. Therefore we performed an in vitro experiment in which embryonic day 15 fetal thymi from F5 Rag-1−/− mice were cultured for 5 days in vitro for development of CD4+CD8+ thymocytes. Then agonist peptide NP68 was added for 24 h in the presence or absence of TRAIL-R2-Fc. After 5 days of culture the agonist peptide reduced the CD4+CD8+ thymocyte population from 66% to 20% (Fig. 6C). Twenty micrograms per milliliter of TRAIL-R2-Fc alone was not toxic for the thymocytes, and, as in the in vivo experiment, there was no inhibition of deletion of CD4+CD8+ thymocytes.
Discussion
Early observations with TRAIL showed that whereas most normal tissues were resistant, many tumor cell lines were extremely sensitive to its actions. One explanation for this discrepancy was the observation that normal cells expressed the decoy TRAIL receptors, whereas these are frequently absent from tumor cells. However, the “decoy” functions of decoy receptors have recently been questioned. In melanoma cells (35–37), keratinocytes (38), and dendritic cells (39, 40), TRAIL resistance did not correlate with the level of decoy receptor expression. This suggests that TRAIL sensitivity can be controlled intracellularly, perhaps by inhibitory molecules such as cFLIP, but the roles of decoy receptors in more physiological settings remain to be established. Despite the widespread interest in TRAIL as a potential tumor therapy, its biological role remains poorly understood. Such knowledge, particularly of humans, will be important in the evaluation of the potential risks of this therapy if it is used clinically. In the immune system there have been a number of reports on TRAIL, including T cell apoptosis (2, 41, 42), target killing by natural killer cells (43), macrophages (44), and dendritic cell death (39, 45).
We postulated that differential expression of TRAIL death, and decoy receptors in the thymus might control thymic selection events. Thus it is possible that negatively selected thymocytes express TRAIL receptors without decoy receptors, whereas positively selected thymocytes are rescued by the expression of decoy receptors. Interestingly, thymocytes do become sensitive to TRAIL after stimulation with anti-CD3. This sensitivity correlated with the stimulation-induced up-regulation of TRAIL-R1 and -R2; however, we found no expression of decoy receptors before or after stimulation. These initial results prompted us to test for a role in negative selection in a number of model systems. Several approaches were taken because the choice of model system for negative selection has been controversial (46). These included the establishment of a novel human thymus organ culture and the F5 TCR transgenic mice. In each of these systems blocking TRAIL with TRAIL-R2-Fc failed to inhibit negative selection. Nevertheless the possibility remains that the antigen doses we used were too high for a TRAIL-R2-Fc to block deletion. A mouse deficient for TRAIL or TRAIL-R1 and TRAIL-R2 could shed more light on the role of TRAIL in negative selection.
Our findings are reminiscent of the studies performed on other members of the TNF-R family, such as Fas. Immature CD4+CD8+ thymocytes express Fas and are susceptible to apoptosis induced by anti-Fas antibodies (47) or Fas ligand (48, 49). However, deletion of autoreactive thymocytes occurs normally in mutant lpr (50) mice and in FasL−/− (51), TNF-R1−/− (52, 53), and TNF-R2−/− (54) mice and is only partially impaired in CD30−/− mice (22), indicating that negative selection cannot be attributed to ligation of any of these receptors alone.
The identity of the downstream adaptor for TRAIL-mediated apoptosis has been debated, but it is now generally accepted that TRAIL-R1 and -R2 recruit FADD and caspase-8 (55–57). Mice expressing a dominant negative FADD under the proximal lck promoter allowing expression in the thymus do not have a defect in negative selection (58–60). Retroviral transfer of c-flip, a naturally occurring antagonist of caspase-8, similarly did not effect T cell development in the thymus (61). In another series of experiments transgenic overexpression of v-flip, the viral counterpart of c-flip under the proximal lck promoter, reduced thymic cellularity probably by inhibiting a proliferative signal emanating from FADD (62).
It has recently been demonstrated that members of the TNF receptor family, such as CD30, CD40, and TNF-R2, which lack a death domain and therefore do not interact with FADD, can use indirect pathways to trigger apoptosis (ref. 63 and reviewed in ref. 64). The partial defect in negative selection seen in CD30−/− mice may be explained by such indirect pathways. Indeed, TRAIL could be a downstream molecule of CD30. Recent mining of expressed sequence tag databases has uncovered many new members of the TNF and TNF receptor families. Many of these proteins are expressed on lymphocytes, and it may be that there is considerable functional redundancy in some of their effects. If so, then deleting or inhibiting one pathway may not be sufficient to prevent negative selection, and strains of mice lacking multiple receptors/ligands may prove more informative.
The expression of death receptors such as Fas and TRAIL-R1/R2 on activated thymocytes remains a conundrum. In all cases thymocytes expressing these molecules are exquisitely sensitive to their ligation, yet their role in thymic development is not clear. In contrast to others (42), we were unable to demonstrate sensitivity of peripheral T cells to TRAIL, even after activation or stimulation with cytokines. Indeed, TRAIL-R1 and -R2, the levels of which were low on resting cells, were not up-regulated by anti-CD3, in contrast to the receptor up-regulation seen on thymocytes. One possible explanation for this result is that signaling events downstream of anti-CD3 differ between thymocytes and mature lymphocytes, as indeed a distinct set of transcription factors is activated after antigenic stimulation of thymocytes and lymphocytes (65, 66). This also holds true for more proximal components in TCR signaling (67).
Preclinical studies in mice and nonhuman primates have shown that administration of TRAIL can induce apoptosis in human tumors, but that no cytotoxicity to normal organs or tissues is found (68, 69). This has led to the testing of TRAIL as a novel candidate drug in cancer therapy. The susceptibility of thymocytes to TRAIL reported in this study, together with the sensitivity of hepatocytes (70) and brain cells (71), necessitates a cautionary approach to the use of TRAIL in cancer therapy.
Acknowledgments
We thank Mr. Pillai, Mr. Westaby, and Nick Willcox for providing human thymus samples; Nan Chen for helping to make some of the recombinant proteins; and Raymond Daniels for characterizing the mAbs. This work was funded by the Medical Research Council.
Abbreviations
- TNF
tumor necrosis factor
- TRAIL
TNF-related apoptosis-inducing ligand
- TCR
T cell antigen receptor
- HTOC
human thymus organ culture
References
- 1.Wiley S R, Schooley K, Smolak P J, Din W S, Huang C P, Nicholl J K, Sutherland G R, Smith T D, Rauch C, Smith C A, et al. Immunity. 1995;3:673–682. doi: 10.1016/1074-7613(95)90057-8. [DOI] [PubMed] [Google Scholar]
- 2.Ashkenazi A, Dixit V M. Curr Opin Cell Biol. 1999;11:255–260. doi: 10.1016/s0955-0674(99)80034-9. [DOI] [PubMed] [Google Scholar]
- 3.Pan G, O'Rourke K, Chinnaiyan A M, Gentz R, Ebner R, Ni J, Dixit V M. Science. 1997;276:111–113. doi: 10.1126/science.276.5309.111. [DOI] [PubMed] [Google Scholar]
- 4.Walczak H, Degli Esposti M A, Johnson R S, Smolak P J, Waugh J Y, Boiani N, Timour M S, Gerhart M J, Schooley K A, Smith C A, et al. EMBO J. 1997;16:5386–5397. doi: 10.1093/emboj/16.17.5386. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Pan G, Ni J, Wei Y F, Yu G L, Gentz R, Dixit V M. Science. 1997;277:815–818. doi: 10.1126/science.277.5327.815. [DOI] [PubMed] [Google Scholar]
- 6.Screaton G R, Mongkolsapaya J, Xu X N, Cowper A E, McMichael A J, Bell J I. Curr Biol. 1997;7:693–696. doi: 10.1016/s0960-9822(06)00297-1. [DOI] [PubMed] [Google Scholar]
- 7.Wu G S, Burns T F, McDonald E R R, Jiang W, Meng R, Krantz I D, Kao G, Gan D D, Zhou J Y, Muschel R, et al. Nat Genet. 1997;17:141–143. doi: 10.1038/ng1097-141. [DOI] [PubMed] [Google Scholar]
- 8.Schneider P, Thome M, Burns K, Bodmer J L, Hofmann K, Kataoka T, Holler N, Tschopp J. Immunity. 1997;7:831–836. doi: 10.1016/s1074-7613(00)80401-x. [DOI] [PubMed] [Google Scholar]
- 9.Sheridan J P, Marsters S A, Pitti R M, Guerney A, Skubach M, Baldwin D, Ramakrishnan L, Gray C L, Baker K, Wood W I, et al. Science. 1997;277:818–821. doi: 10.1126/science.277.5327.818. [DOI] [PubMed] [Google Scholar]
- 10.Degli Esposti M A, Smolak P J, Walczak H, Waugh J, Huang C P, DuBose R F, Goodwin R G, Smith C A. J Exp Med. 1997;186:1165–1170. doi: 10.1084/jem.186.7.1165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Pan G, Ni J, Wei Y F, Yu G, Gentz R, Dixit V M. Science. 1997;277:815–818. doi: 10.1126/science.277.5327.815. [DOI] [PubMed] [Google Scholar]
- 12.Mongkolsapaya J, Cowper A E, Xu X N, Morris G, McMichael A J, Bell J I, Screaton G R. J Immunol. 1998;160:3–6. [PubMed] [Google Scholar]
- 13.Marsters S A, Sheridan J P, Pitti R M, Huang A, Skubatch M, Baldwin D, Yuan J, Gurney A, Goddard A D, Godowski P, et al. Curr Biol. 1997;7:1003–1006. doi: 10.1016/s0960-9822(06)00422-2. [DOI] [PubMed] [Google Scholar]
- 14.Pan G, Ni J, Yu G, Wei Y F, Dixit V M. FEBS Lett. 1998;424:41–45. doi: 10.1016/s0014-5793(98)00135-5. [DOI] [PubMed] [Google Scholar]
- 15.Degli Esposti M A, Dougall W C, Smolak P J, Waugh J Y, Smith C A, Goodwin R G. Immunity. 1997;7:813–820. doi: 10.1016/s1074-7613(00)80399-4. [DOI] [PubMed] [Google Scholar]
- 16.Emery J G, McDonnell P, Burke M B, Deen K C, Lyn S, Silverman C, Dul E, Appelbaum E R, Eichman C, DiPrinzio R, et al. J Biol Chem. 1998;273:14363–14367. doi: 10.1074/jbc.273.23.14363. [DOI] [PubMed] [Google Scholar]
- 17.Swat W, Ignatowicz L, von Boehmer H, Kiesilow P. Nature (London) 1991;351:150–153. doi: 10.1038/351150a0. [DOI] [PubMed] [Google Scholar]
- 18.Surh C D, Sprent J. Nature (London) 1994;372:100–103. [Google Scholar]
- 19.Clayton L K, Ghendler Y, Mizoguchi E, Patch R J, Ocain T D, Orth K, Bhan A K, Dixit V M, Reinherz E L. EMBO J. 1997;16:2282–2293. doi: 10.1093/emboj/16.9.2282. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Moulian N, Berrih-Aknin S. Semin Immunol. 1998;10:449–456. doi: 10.1006/smim.1998.0155. [DOI] [PubMed] [Google Scholar]
- 21.Kishimoto H, Sprent J. Immunol Res. 2000;21:315–323. doi: 10.1385/IR:21:2-3:315. [DOI] [PubMed] [Google Scholar]
- 22.Amakawa R, Hakem A, Kundig T M, Matsuyama T, Simard J J, Timms E, Wakeham A, Mittruecker H W, Griesser H, Takimoto H, et al. Cell. 1996;84:551–562. doi: 10.1016/s0092-8674(00)81031-4. [DOI] [PubMed] [Google Scholar]
- 23.Foy T M, Page D M, Waldschmidt T J, Schoneveld A, Laman J D, Masters S R, Tygrett L, Ledbetter J A, Aruffo A, Claassen E, et al. J Exp Med. 1995;182:1377–1388. doi: 10.1084/jem.182.5.1377. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Williams O, Tarazona R, Wack A, Harker N, Roderick K, Kioussis D. Proc Natl Acad Sci USA. 1998;95:5706–5711. doi: 10.1073/pnas.95.10.5706. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Tarazona R, Williams O, Moskophidis D, Smyth L A, Tanaka Y, Murdjeva M, Wack A, Mamalaki C, Kioussis D. J Immunol. 1998;160:5397–5403. [PubMed] [Google Scholar]
- 26.Mongkolsapaya J, Grimes J, Chen N, Xu X, Stuart D, Jones E, Screaton G. Nat Struct Biol. 1999;6:1048–1053. doi: 10.1038/14935. [DOI] [PubMed] [Google Scholar]
- 27.Anderson G, Moore N, Owen J J, Jenkinson E J. Annu Rev Immunol. 1996;14:73–99. doi: 10.1146/annurev.immunol.14.1.73. [DOI] [PubMed] [Google Scholar]
- 28.Smith C A, Williams G T, Kingston R, Jenkinson E J, Owen J J. Nature (London) 1989;337:181–184. doi: 10.1038/337181a0. [DOI] [PubMed] [Google Scholar]
- 29.Page D M, Kane L P, Allison J P, Hedrick S M. J Immunol. 1993;151:1868–1880. [PubMed] [Google Scholar]
- 30.Choi Y W, Kotzin B, L, H, Callahan J, Marrack P, Kappler J. Proc Natl Acad Sci USA. 1989;86:8941–8945. doi: 10.1073/pnas.86.22.8941. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Yonehara S, Nishimura Y, Kishil S, Yonehara M, Takazawa K, Tamatani T, Ishii A. Int Immunol. 1994;6:1849–1856. doi: 10.1093/intimm/6.12.1849. [DOI] [PubMed] [Google Scholar]
- 32.Martin S, Bevan M J. Eur J Immunol. 1997;27:2726–2736. doi: 10.1002/eji.1830271037. [DOI] [PubMed] [Google Scholar]
- 33.Song K, Chen Y, Goke R, Wilmen A, Seidel C, Goke A, Hilliard B, Chen Y. J Exp Med. 2000;191:1095–1104. doi: 10.1084/jem.191.7.1095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Castro J E, Listman J A, Jacobson B A, Wang Y, Lopez P A, Ju S, Finn P W, Perkins D L. Immunity. 1996;5:617–627. doi: 10.1016/s1074-7613(00)80275-7. [DOI] [PubMed] [Google Scholar]
- 35.Thomas W D, Hersey P. J Immunol. 1998;161:2195–2200. [PubMed] [Google Scholar]
- 36.Griffith T S, Chin W A, Jackson G C, Lynch D H, Kubin M Z. J Immunol. 1998;161:2833–2840. [PubMed] [Google Scholar]
- 37.Zhang X D, Franco A, Myers K, Gray C, Nguyen T, Hersey P. Cancer Res. 1999;59:2747–2753. [PubMed] [Google Scholar]
- 38.Leverkus M, Neumann M, Mengling T, Rauch C T, Brocker E B, Krammer P H, Walczak H. Cancer Res. 2000;60:553–559. [PubMed] [Google Scholar]
- 39.Leverkus M, Walczak H, McLellan A, Fries H W, Terbeck G, Brocker E B, Kampgen E. Blood. 2000;96:2628–2631. [PubMed] [Google Scholar]
- 40.Walczak H, Krammer P H. Exp Cell Res. 2000;256:58–66. doi: 10.1006/excr.2000.4840. [DOI] [PubMed] [Google Scholar]
- 41.Marsters S A, Pitti R M, Donahue C J, Ruppert S, Bauer K D, Ashkenazi A. Curr Biol. 1996;6:750–752. doi: 10.1016/s0960-9822(09)00456-4. [DOI] [PubMed] [Google Scholar]
- 42.Martinez-Lorenzo M J, Alava M A, S, G, Kim K J, Chuntharapai A, Pineiro A, Naval J, Anel A. Eur J Immunol. 1998;28:2714–2725. doi: 10.1002/(SICI)1521-4141(199809)28:09<2714::AID-IMMU2714>3.0.CO;2-9. [DOI] [PubMed] [Google Scholar]
- 43.Johnsen A C, Haux J, Steinkjer B, Nonstad U, Egeberg K, Sundan A, Ashkenazi A, Espevik T. Cytokine. 1999;11:664–672. doi: 10.1006/cyto.1999.0489. [DOI] [PubMed] [Google Scholar]
- 44.Griffith T S, Wiley S R, Kubin M Z, Sedger L M, Maliszewski C R, Fanger N A. J Exp Med. 1999;189:1343–1354. doi: 10.1084/jem.189.8.1343. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Wang J, Zheng L, Lobito A, Chan F K, Dale J, Sneller M, Yao X, Puck J M, Straus S E, Lenardo M J. Cell. 1999;98:47–58. doi: 10.1016/S0092-8674(00)80605-4. [DOI] [PubMed] [Google Scholar]
- 46.Page D M, Roberts E M, Peschon J J, Hedrick S M. J Immunol. 1998;160:120–133. [PubMed] [Google Scholar]
- 47.Ogasawara J, Suda T, Nagata S. J Exp Med. 1995;181:485–491. doi: 10.1084/jem.181.2.485. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Suda T, Tanaka M, Miwa K, Nagata S. J Immunol. 1996;157:3918–3924. [PubMed] [Google Scholar]
- 49.Müller K P, Mariani S M, Matiba B, Kyewski B, Krammer P H. Eur J Immunol. 1995;25:2996–2999. doi: 10.1002/eji.1830251043. [DOI] [PubMed] [Google Scholar]
- 50.Sidman C L, Marshall J D, Von Boehmer H. Eur J Immunol. 1992;22:499–504. doi: 10.1002/eji.1830220231. [DOI] [PubMed] [Google Scholar]
- 51.Adachi M, Suematsu S, Kondo T, Ogasawara J, Tanaka T, Yoshida N, Nagata S. Nat Genet. 1995;11:294–300. doi: 10.1038/ng1195-294. [DOI] [PubMed] [Google Scholar]
- 52.Pfeffer K, Matsuyama T, Kundig T M, Wakeham A, Kishihara K, Shahinian A, Wiegmann K, Ohashi P S, Kronke M, Mak T W. Cell. 1993;73:457–467. doi: 10.1016/0092-8674(93)90134-c. [DOI] [PubMed] [Google Scholar]
- 53.Rothe J, Lesslauer W, Lotscher H, Lang Y, Koebel P, Kontgen F, Althage A, Zinkernagel R, Steinmetz M, Bluethmann H. Nature (London) 1993;364:798–802. doi: 10.1038/364798a0. [DOI] [PubMed] [Google Scholar]
- 54.Erickson S L, de Sauvage F J, Kikly K, Carver-Moore K, Pitts-Meek S, Gillett N, Sheehan K C, Schreiber R D, Goeddel D V, Moore M W. Nature (London) 1994;372:560–563. doi: 10.1038/372560a0. [DOI] [PubMed] [Google Scholar]
- 55.Kischkel F C, Lawrence D A, Chuntharapai A, Schow P, Kim K J, Ashkenazi A. Immunity. 2000;12:611–620. doi: 10.1016/s1074-7613(00)80212-5. [DOI] [PubMed] [Google Scholar]
- 56.Sprick M R, Weigand M A, Rieser E, Rauch C T, Juo P, Blenis J, Krammer P H, Walczak H. Immunity. 2000;12:599–609. doi: 10.1016/s1074-7613(00)80211-3. [DOI] [PubMed] [Google Scholar]
- 57.Bodmer J L, Holler N, Reynard S, Vinciguerra P, Schneider P, Juo P, Blenis J, Tschopp J. Nat Cell Biol. 2000;2:241–243. doi: 10.1038/35008667. [DOI] [PubMed] [Google Scholar]
- 58.Zornig M, Hueber A O, Evan G. Curr Biol. 1998;8:467–470. doi: 10.1016/s0960-9822(98)70182-4. [DOI] [PubMed] [Google Scholar]
- 59.Walsh C M, Wen B G, Chinnaiyan A M, O'Rourke K, Dixit V M, Hedrick S M. Immunity. 1998;8:439–449. doi: 10.1016/s1074-7613(00)80549-x. [DOI] [PubMed] [Google Scholar]
- 60.Newton K, Harris A W, Bath M L, Smith K G C, Strasser A. EMBO J. 1998;17:706–718. doi: 10.1093/emboj/17.3.706. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Van Parijs L, Refaeli Y, Abbas A K, Baltimore D. Immunity. 1999;11:763–770. doi: 10.1016/s1074-7613(00)80150-8. [DOI] [PubMed] [Google Scholar]
- 62.OhYama T, Tsukumo S, Yajima N, Sakamaki K, Yonehara S. Microbiol Immunol. 2000;44:289–297. doi: 10.1111/j.1348-0421.2000.tb02498.x. [DOI] [PubMed] [Google Scholar]
- 63.Grell M, Zimmermann G, Gottfried E, Chen C M, Grunwald U, Huang D C, Wu Lee Y H, Durkop H, Engelmann H, Scheurich P, et al. EMBO J. 1999;18:3034–3043. doi: 10.1093/emboj/18.11.3034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Screaton G, Xu X N. Curr Opin Immunol. 2000;12:316–322. doi: 10.1016/s0952-7915(00)00093-5. [DOI] [PubMed] [Google Scholar]
- 65.Simon A K, Auphan N, Schmitt-Verhulst A M. Int Immunol. 1996;8:1421–1428. doi: 10.1093/intimm/8.9.1421. [DOI] [PubMed] [Google Scholar]
- 66.Rincón M, Flavell R A. Mol Cell Biol. 1996;16:1074–1084. doi: 10.1128/mcb.16.3.1074. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Mariathasan S, Jones R G, Ohashi P S. Semin Immunol. 1999;11:263–272. doi: 10.1006/smim.1999.0182. [DOI] [PubMed] [Google Scholar]
- 68.Walczak H, Miller R E, Ariail K, Gliniak B, Griffith T S, Kubin M, Chin W, Jones J, Woodward A, Le T, et al. Nat Med. 1999;5:157–163. doi: 10.1038/5517. [DOI] [PubMed] [Google Scholar]
- 69.Ashkenazi A, Pai R C, Fong S, Leung S, Lawrence D A, Marsters S A, Blackie C, Chang L, McMurtrey A E, Hebert A, et al. J Clin Invest. 1999;104:155–162. doi: 10.1172/JCI6926. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Jo M, Kim T H, Seol D W, Esplen J E, Dorko K, Billiar T R, Strom S C. Nat Med. 2000;6:564–567. doi: 10.1038/75045. [DOI] [PubMed] [Google Scholar]
- 71.Nitsch R, Bechmann I, Deisz R A, Haas D, Lehmann T N, Wendling U, Zipp F. Lancet. 2000;356:827–828. doi: 10.1016/S0140-6736(00)02659-3. [DOI] [PubMed] [Google Scholar]