Abstract
Salivary glands form during embryonic development by a complex process that creates compact, highly organized secretory organs with functions essential for oral health. The architecture of these glands is generated by branching morphogenesis, revealed by recent research to involve unexpectedly dynamic cell motility and novel regulatory pathways. Numerous growth factors, extracellular matrix molecules, gene regulatory pathways, and mechanical forces contribute to salivary gland morphogenesis, but local gene regulation and morphological changes appear to play particularly notable roles. Here we review these recent advances and their potential application to salivary gland tissue engineering.
Keywords: developmental biology, cell-matrix interactions, microscopy, gene expression, cell signaling, tissue engineering
Introduction
Salivary glands are created during embryonic development by complex and remarkably dynamic processes that yield the final, precisely organized architecture of normal adult glands. Efficient functioning of these salivary glands is essential to produce the approximately half-liter of saliva daily that maintains oral health. Salivary hypofunction and xerostomia can result from local surgery, therapeutic radiation for head and neck cancer, Sjögren’s disease, and side-effects of certain medications. Hypofunction can result in severe dental caries and oral ulcers, as well as difficulty in swallowing and a substantial loss of quality of life (Napenas et al., 2009; Vissink et al., 2010).
Normal salivary gland function requires a large surface area of saliva-secreting epithelium packaged efficiently into a compact gland composed of densely packed, well-organized networks of acini and ducts. A key goal of restorative tissue engineering and/or regeneration of damaged or lost salivary glands will be to replace this large expanse of secretory epithelium, ideally packaged in a similarly compact structure. One approach may be to try to mimic nature by learning how embryos generate glands and using these mechanisms to generate gland replacements. As we review below, a recurring theme in recent studies of salivary gland development is the importance of dynamic local cell and tissue re-organization regulated by novel pathways controlling cellular and extracellular matrix interactions.
Branching Morphogenesis
During embryonic salivary gland development, a simple epithelial bud is dramatically remodeled through repetitive branching (Figs. 1, 2) in a process termed ‘branching morphogenesis’. This process generates a secretory organ containing many thousands of acini connected to secretory ducts to provide sufficient secretory epithelium (Patel et al., 2006; Tucker, 2007). Branching morphogenesis also creates other branched organs, such as lungs, kidneys, and mammary and prostate glands (Lu and Werb, 2008; Affolter et al., 2009; Andrew and Ewald, 2010; Costantini and Kopan, 2010; Morrisey and Hogan, 2010). Branching morphogenesis of salivary glands has been studied extensively, providing new insights into oral biology and morphogenesis of other crucial organs (Grobstein, 1953; Melnick and Jaskoll, 2000; Kadoya and Yamashina, 2005; Patel et al., 2006; Tucker, 2007; Gresik et al., 2009; Andrew and Ewald, 2010; Hsu and Yamada, 2010; Larsen et al., 2010; Sequeira et al., 2010).
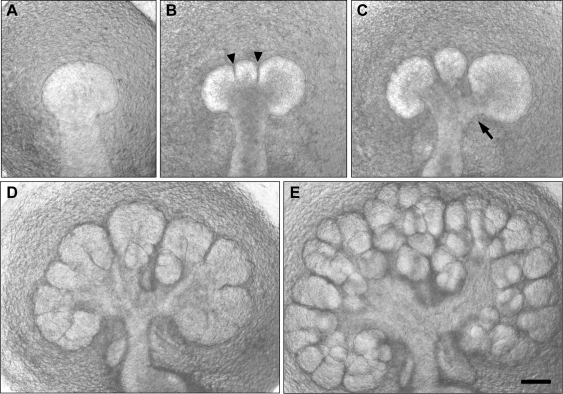
Figure 1.
Salivary gland branching morphogenesis. Transmitted light microscopy of a living submandibular salivary gland dissected out of a mouse embryo after 12 days of gestation (panel A), placed into explant culture, and photographed after being cultured for an additional 12 hrs (B), 24 hrs (C), 48 hrs (D), and 72 hrs (E). The gland starts as a single bud, which is subdivided by clefts (arrowheads), which progressively deepen. The clefts eventually widen to define secondary ducts (arrow) connected to the main duct. This process of branching morphogenesis progresses rapidly from a single bud to a complex branched structure in 3 days, and it continues to branch as the embryo develops. Scale bar = 100 µm.
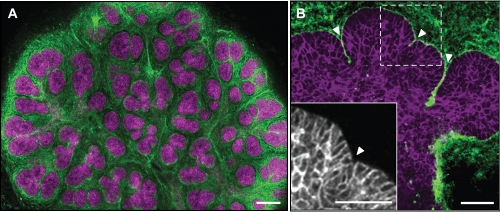
Figure 2.
Immunostaining of salivary glands. Mouse submandibular glands were fixed with paraformaldehyde and stained for the cell-to-cell adhesion molecule E-cadherin and the extracellular matrix protein fibronectin. (A) Low-magnification view of an entire salivary gland at embryonic day 14.5 showing E-cadherin (magenta) at epithelial cell junctions and the extracellular matrix protein fibronectin (green) primarily in the mesenchyme. (B) Higher-magnification view of an epithelial end bud from an embryonic day 13 salivary gland showing E-cadherin staining (magenta) and fibronectin (green) accumulating in clefts at different stages of progression (arrowheads). The inset (grayscale) shows a magnified region of the bud, highlighting the more-columnar cells by E-cadherin staining at the periphery of the bud attached to the basement membrane. Scale bars = 100 µm.
Although some morphogenetic mechanisms are shared, others are likely to be organ- or process-specific, e.g., the differences between tube and bud formation, and differing modes of branching or extension (Lu and Werb, 2008; Andrew and Ewald, 2010). For example, Drosophila salivary glands develop as relatively linear tubular structures, and major recent progress has identified key genetic regulatory pathways for their morphogenesis (Andrew and Ewald, 2010; Pirraglia and Myat, 2010). In contrast, mammalian salivary gland development involves more complex three-dimensional branching and tissue remodeling (Patel et al., 2006; Tucker, 2007; Andrew and Ewald, 2010; Hsu and Yamada, 2010). Because of space limitations, we focus this review on research published during the past three years that has provided new insights into the dynamics and mechanisms of mammalian salivary gland development.
Regulatory Gene and Growth Factor Regulation of Branching Morphogenesis
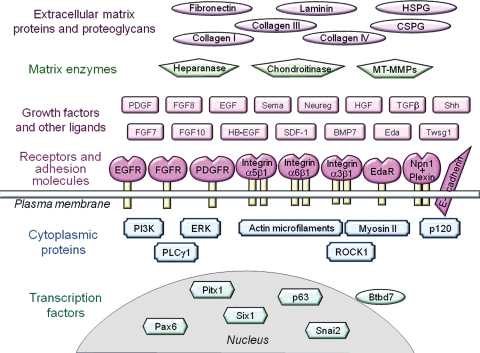
As expected for a complex developmental process, salivary gland branching requires several dozen individual genes and proteins (Fig. 3; Appendix Table), which include transcription factors, growth factors, receptors, and extracellular matrix molecules. In many cases, the precise roles of these molecules remain to be identified, but several new pathways regulating morphogenesis have been defined recently, as reviewed below.
Figure 3.
Schematic overview of proteins required for salivary gland branching morphogenesis. Multiple proteins are necessary during embryogenesis for successful branching morphogenesis, as documented in the Appendix Table. These proteins include multiple extracellular matrix proteins and proteoglycans (abbreviations: HSPG = heparan sulfate proteoglycan and CSPG = chondroitin sulfate proteoglycan); several matrix enzymes (MT-MMPs = membrane-type matrix metalloproteinases, e.g., MT2-MMP); many growth factors and ligands (Sema = soluble form of semaphorin, Neureg = neuregulin, Shh = sonic hedgehog, Eda = ectodysplasin-A, and Twsg1 = twisted gastrulation 1); and a variety of transmembrane receptors and adhesion molecules (EGFR = EGF receptor, FGFR = FGF receptor, PDGFR = PDGF receptor, EdaR = ectodysplasin-A receptor, and Npn1+Plexin = non-covalent complex of neuropilin-1 and plexin). Molecules routinely described by abbreviations, such as PDGF, are not defined here.
Salivary Gland Tissue Dynamics
A major challenge facing embryonic organs undergoing branching morphogenesis is to generate organ architecture sufficiently rapidly and reliably during fetal development. For example, the branched submandibular gland structure shown in Fig. 2A took only three days to establish after initiation at mouse embryonic day 12, with continued branching over subsequent days. Consequently, it is not surprising that these tissues undergo rapid major remodeling that involves cycles of cleft formation and bud outgrowth to generate increasing numbers of buds. A recent surprise, however, was the discovery that the cells within these tissues are themselves involved in extensive migratory movements (Appendix Movie 1, discussed below).
Cleft Formation
Cleft formation can be visualized in real time by time-lapse microscopy (Larsen et al., 2006; Kadoya and Yamashina, 2010; Onodera et al., 2010). One or more indentations appear on the outer surface of a bud, which then progress to form deepening clefts. The site of cleft formation does not appear to be precisely predetermined. Small clefts can form and regress, and sometimes two clefts can merge to form a single larger cleft (e.g., see Larsen et al., 2006) in a highly dynamic process. Because cleft formation is such a central feature of salivary gland branching morphogenesis, it is obviously important to identify the mechanisms of cleft formation and propagation. Additional mechanisms of bud extension and duct formation contribute to branching after clefting.
One approach to understanding cleft formation is to identify the key genes and proteins associated with, and especially required for, branching morphogenesis. Gene expression patterns have been determined with the use of microarrays or SAGE (serial analysis of gene expression) at early embryonic stages (Hoffman et al., 2002; Sakai et al., 2002). This approach has recently identified a daunting number of candidate genes for regulating and mediating branching morphogenesis. Conversely, genetic approaches have proven quite fruitful for identifying essential individual genes, as reviewed previously (Patel et al., 2006; Melnick et al., 2009; Larsen et al., 2010). These two approaches have been combined with functional studies involving tests of candidate growth factors and matrix molecules by localization and function-inhibition studies, which have identified numerous genes and proteins needed for successful salivary gland branching morphogenesis (Appendix Table). These findings provide a solid foundation from which to discover how these molecules interact to form clefts, buds, and ducts. To use an engineering analogy, we currently have a large parts list, and we now need to learn better how the machine works.
Over the past three years, a half-dozen different approaches have provided new insight into the mechanisms of salivary branching. These approaches include: (i) visualizing the dynamic movements of cells and tissues by fluorescence confocal time-lapse microscopy, (ii) establishing roles of mechanical forces in salivary development, (iii) identifying distinct subsets of cells in developing glands that play unique roles, (iv) determining the gene expression patterns of local subsets of cells involved in different aspects of morphogenesis, (v) identifying regulatory pathways and generating models to begin to explain how gene regulation can mediate branching morphogenesis, and (vi) taking preliminary steps toward reconstituting branching morphogenesis, with the ultimate goal of regenerating salivary gland function. In the following sections, we will review findings discovered using these complementary research approaches.
Cell Motility during Morphogenesis
Epithelial cells in salivary glands undergoing branching morphogenesis display strikingly high levels of cell motility (Larsen et al., 2006; Wei et al., 2007), as determined by confocal time-lapse microscopy of fluorescently labeled cells in developing glands in organ culture. As shown in Appendix Movie 1, these cell movements generally appear random, although peripheral bud cells can undergo a characteristic circular motion in which they relinquish contact with the basement membrane, move away, and then circle back to the basement membrane; the biological significance of these characteristic motions is not yet clear. In contrast to bud epithelial cells, the cells in duct regions tend to have lower motility. Cell motions virtually cease by the time the mice are born (Larsen et al., 2006). The signaling mechanisms that drive this motility and its function(s) remain to be identified, but this continual restless motion may provide plasticity to the embryonic organ to facilitate rapid remodeling.
A more detailed examination of these cell motility patterns reveals that the cells at the base of deepening clefts can briefly separate from each other to form gaps. These transient gaps between cells can be visualized as either dark areas in glands in which EGFP (enhanced green fluorescent protein) is expressed in the cytoplasm of all cells, or as bright red slits or gaps if the extracellular space is labeled with a fluorescent marker (Onodera et al., 2010; Appendix Movie 2). The local opening of these tiny spaces for cleft progression appears stochastic, but the progression of the main cleft by this transient gap formation proceeds relatively steadily at ~5 μm/hr. Besides these gaps between cells, local folding of the plasma membrane near the base of extending clefts can produce a “shelf” of membrane (Kadoya and Yamashina, 2010), suggesting dynamic interactions between matrix molecules filling the cleft and adjacent plasma membranes. Because efficient cleft formation involves these changes located at the base of advancing clefts, the local regulatory mechanisms that govern cell motion, cell separation, and cell-matrix interactions in this region are likely to play key roles in cleft formation and progression.
Mechanical Forces in Salivary Gland Development
The importance of the actomyosin contractile machinery of cells for cell migration and tissue morphogenesis is well known. For example, the actomyosin system is integral to cell migration (Ridley et al., 2003), and actin microfilaments were implicated in salivary gland branching more than 30 years ago (Spooner and Wessells, 1972). However, development of these concepts to understand how forces drive branching morphogenesis has accelerated only recently (Michael et al., 2005; Moore et al., 2005; Daley et al., 2009). Inhibition of Rho kinase (ROCK) and actomyosin contraction disrupts salivary gland morphogenesis; glands also begin to initiate more clefts, suggesting that ROCK/myosin may be suppressing cleft initiation by regulating tension at the periphery of the epithelial buds (Daley et al., 2009; Fig. 4A). Signaling mechanisms that could locally decrease tension at certain points in the bud to allow for cleft formation have not yet been identified. After cleft initiation, ROCK-mediated actomyosin contraction is necessary for fibronectin accumulation at the basement membrane, and for subsequent cleft progression. It is possible that myosin II contractility must first be inactivated to initiate a cleft, but must then provide the mechanical force needed to pull the fibronectin into fibrils using integrin-based adhesions for cleft elongation. These studies primarily used a ROCK inhibitor and the myosin II inhibitor blebbistatin to inhibit contractility mediated by all 3 mammalian isoforms of myosin II, which have slightly different functions, regulators, and physical properties (Vicente-Manzanares et al., 2009). It is possible that each myosin isoform plays a specific role in gland development. Moreover, Rho appears to have another function in addition to regulating myosin II-mediated contractility. ROCK I is also important for epithelial polarity (Yu et al., 2008), and it is apparently required for Par protein localization and basement membrane deposition in embryonic salivary glands (WP Daley et al., Mol Biol Cell 21:2350, 2010 [abstract]).
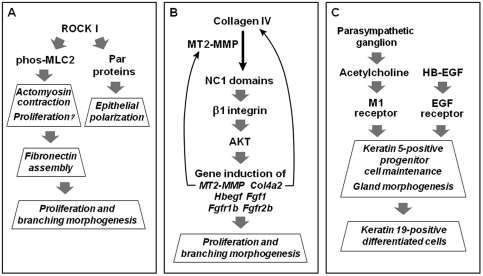
Figure 4.
New pathways regulating salivary gland morphogenesis. See text and the following citations for details: (A) Daley et al., 2009; (B) Rebustini et al., 2009; and (C) Knox et al., 2010.
A challenge for the future will be to determine the precise mechanisms of cleft initiation and progression. Current findings suggest that cleft formation is a process with at least two steps involving contractile forces: Tension inhibiting cleft initiation must be locally released to start cleft formation, but then ROCK-regulated actomyosin contraction is needed to promote fibronectin assembly and subsequent cleft elongation. It is likely that additional regulatory factors are needed to coordinate these processes.
Importance of Distinct Subsets of Epithelial Cells in Developing Glands
Even though the epithelial cells are surprisingly motile, Walker and her co-workers have established that the outer layer of bud epithelial cells adjacent to the basement membrane is morphologically distinct and different in fate from cells at the center of the buds (Walker et al., 2008). The peripheral cells are more columnar than interior cells, with more organized cell-cell junctions containing the adhesion molecule E-cadherin. Interestingly, cells at the base of forming clefts lose this organization, becoming less columnar and more variable in shape, with gaps forming between these cells (Onodera et al., 2010).
Outer bud cells express the B1 marker characteristic of more mature acini, and these cells contribute to acini rather than ducts later in development (Walker et al., 2008). Interestingly, however, as the clefts deepen and widen, they define secondary ducts (Fig. 1). That is, clefts mature three-dimensionally to form the wide spaces around the secondary ducts connecting end buds to the primary duct. An interesting puzzle is how cells adjacent to the acinar precursors of the end buds become the external cells of ducts as clefts repetitively divide up early buds into many smaller end buds. Two possibilities are that cells change fate, or that new cells are recruited from the interior of the buds to form the external cells of ducts.
Differential Gene Expression Patterns
Because epithelial cells at different sites in developing salivary glands appear morphologically distinct and have different differentiated fates, it is likely that they have distinct gene expression patterns. Laser microdissection provides a technology for identifying these differences in local salivary gland gene expression (Sakai et al., 2003). Epithelial cells in local regions of cryostat frozen sections of embryonic glands can be microdissected away from the rest of the gland tissue by means of a UV laser. Small numbers of cells (e.g., 25-50) can be collected, and the RNA from such samples can be analyzed by SAGE or whole-genome microarrays to determine gene expression patterns at different sites and stages of development (Sakai et al., 2002).
Extensive spatial-temporal gene expression data became available in March 2010 at the SGMAP (Salivary Gland Molecular Anatomy Project) Web site: http://sgmap.nidcr.nih.gov/. This public site provides information on both local spatial and temporal developmental expression patterns. Local site-specific gene expression patterns, as determined by Musselmann and his co-workers (submitted for publication), are provided. In addition, this site also provides extensive stage-specific expression data generated by Hoffman and his co-workers in a major extension of their previous developmental analyses (Hoffman et al., 2002).
This National Institute of Dental and Craniofacial Research database can be searched for gene expression patterns of individual genes by name/symbol or description, e.g., FGF or fibronectin, or by using more general gene ontology terms such as “morphogenesis” or “transcription factor”. Such searches immediately identify intriguing patterns of differential gene expression, some of which distinguish peripheral from central bud cells, developing duct from bud cells, and cells at sites of cleft progression from adjacent end bud cells. As expected, initial analyses show a considerable number of genes that are widely expressed at multiple sites (bud, cleft, and duct), comprising 79% of the genes. Interestingly, however, there was site-specific expression of 3% of the genes solely in buds, 3% only in ducts, and 6% only in cells adjacent to clefts, as well as other combinations of expression patterns (Larsen et al., 2010). These distinct patterns of expression for specific genes can then be used to identify candidate gene regulators of branching morphogenesis or local cytodifferentiation, and to develop novel hypotheses in which groups of genes regulated in parallel might guide developmental dynamics or differentiation.
An example of the value of identifying spatial differences in gene expression is seen in recent studies identifying mechanisms of cleft progression. Gene expression patterns in cells immediately adjacent to advancing clefts compared with peripheral end bud cells revealed increased expression of fibronectin, Btbd7, Snail2, and TIMP3, accompanied by decreased expression of the cell-cell adhesion protein E-cadherin (Sakai et al., 2003; Onodera et al., 2010). These findings identified a novel regulatory pathway leading from a matrix protein to a transcription factor, and then to regulation of cell motility and cleft progression (Fig. 5). Specifically, the extracellular matrix protein fibronectin induces a previously uncharacterized gene named Btbd7 (“cleftin”), e.g., within 30 minutes; Btbd7 in turn induces the well-known transcription factor Snail2 as shown by over-expression and siRNA studies, and it decreases levels of E-cadherin at the protein level. The net effect of Btbd7/cleftin function is cell separation and scattering, which appears to account for the local opening of transient gaps to promote cleft progression (Onodera et al., 2010).
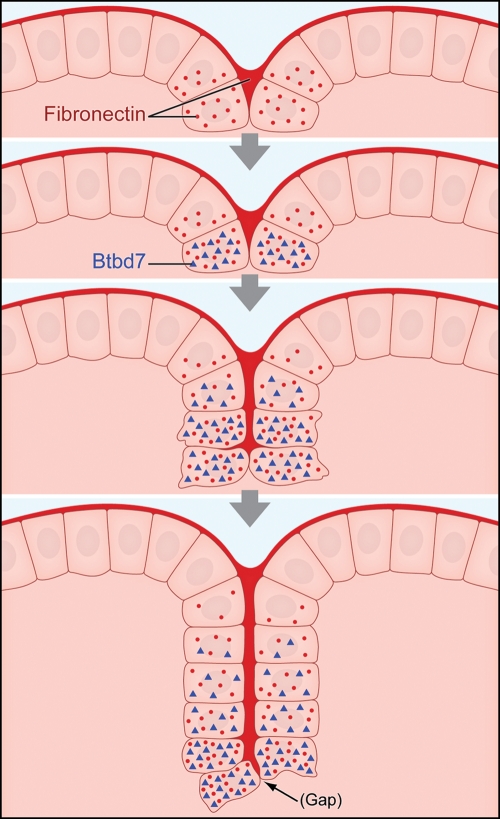
Figure 5.
Schematic representation of a new pathway regulating salivary gland cleft formation. The 4 images depict a cleft that initiates and then progresses by cleft deepening. The basement membrane at the basal surface of the peripheral epithelial cells and forming clefts both contain polymerized fibronectin (dark red), and salivary epithelial cells adjacent to the cleft synthesize new fibronectin (red dots). Fibronectin induces Btbd7. As the cleft progresses, Btbd7 decreases in cells near older (upper) parts of the cleft, as Btbd7 is mainly induced at the base of forming clefts. The cleft advances through progressive extension of the zone of fibronectin filling the cleft (Larsen et al., 2006) associated with the relatively stochastic formation of tiny gaps between cells expressing Btbd7. Average times between the second step (definitive cleft initiation) and the third step (advancing cleft) are approximately 2-4 hours, and approximately 3-5 hours between the third and the fourth steps depicted (deep cleft).
Other New Pathways Regulating Branching Morphogenesis
Classic morphological and signal transduction studies using genetic mutants, organ culture, growth factors, pharmacological inhibitors, antibodies, antisense oligonucleotides, small interfering RNAs, and enzymes have previously identified numerous pathways that are important for branching morphogenesis. These systems involve a variety of growth factors such as FGF and HB-EGF combined with various extracellular matrix molecules with a variety of downstream signaling mechanisms, including MAP kinase and phosphatidylinositol 3′-kinase pathways. These mechanisms have been the subject of excellent previous reviews (Kashimata and Gresik, 1996; Hieda and Nakanishi, 1997; Melnick and Jaskoll, 2000; Patel et al., 2006). It has become increasingly clear, however, that branching morphogenesis is extremely complex; key roles continue to be identified for more and more regulators that contribute to successful morphogenesis. For example, Fig. 4 summarizes 3 newly identified pathways. As described above, one centers on Rho kinase (ROCK I), which plays roles not only in contractility, but also apparently in epithelial cell polarity. Another novel pathway links the protease MT2-MMP to collagen IV: The release of a biologically active collagen fragment activates an integrin and AKT to induce a series of genes in a feedback mechanism, promoting cell proliferation and branching morphogenesis (Rebustini et al., 2009). A third pathway identifies novel external regulation by signaling from the nerves of the parasympathetic ganglion to maintain progenitor cell function for branching morphogenesis and differentiation (Knox et al., 2010).
An Eda/Edar pathway was known to activate NFκB analogous to the TNF/TNFR pathway (Jaskoll et al., 2003), but recent studies predict a second, more prominent pathway downstream of Eda/Edar in salivary gland morphogenesis through the candidate transcription factor C/EBPα (Melnick et al., 2009). In the Eda mutant mouse, 6 genes known to be important in gland development (Edar, Fgf8, Shh, Egf, Tgfa, and Egfr) are differentially regulated, suggesting a potentially important role for Eda/Edar as an initiation point for these major signaling molecules (Melnick et al., 2009). PDGF is now known to serve as an upstream regulator of FGF signaling (Yamamoto et al., 2008). Involvement of neuregulin and lysophosphatidic acid in morphogenesis, and a novel repulsion phenomenon between developing buds have also been discovered recently (Okamoto et al., 2010). This repulsion is promoted by FGF1, and it contributes to normal spacing between branching buds.
This tremendous regulatory complexity has precedent in other developing systems. For example, kidney development also involves a daunting number of growth factors and regulatory genes (Monte et al., 2007; Brunskill et al., 2008). This plenitude of new, essential regulatory pathways raises the obvious question of how they are all coordinated and interconnected, and how they guide morphogenetic dynamics. Systems analysis may help to clarify these complex interactions (Larsen et al., 2010).
Unanswered Mechanistic Questions
Many intriguing new questions remain unanswered. A particularly puzzling question involves the mechanisms of salivary gland morphogenetic self-organization and assembly. Unlike most developmental morphogenetic processes involving highly choreographed, spatially precise patterns of gene expression and movements of cells and tissues, salivary gland branching morphogenesis is surprisingly stochastic: Besides the relatively chaotic cell movements, initiating clefts often form and disappear, and sometimes merge. How the developing gland coordinates the formation of innumerable well-formed and perfectly connected buds and ducts remains a major challenge. Important questions include: How is a cleft initially formed prior to the fibronectin-Btbd7-Snail2 cascade? What drives the frenetic movements of the developing epithelial cells, e.g., motility or scatter factors? How are the external cells of buds specified, and how much plasticity do they normally exhibit, i.e., can they interchange with internal cells? How are the secondary ducts formed as clefts deepen to delineate new buds? Many of these questions will require increasingly sophisticated imaging and cell lineage tracking approaches. Linking the movements of single cells to local tissue organization under the regulation of gene expression, extracellular matrix, and tissue tension will require a whole new level of experimental sophistication that should have important implications for understanding the dynamic formation of other organs.
Steps toward Salivary Gland Reconstitution or Replacement
Because losses of salivary gland function can have such severe effects on oral health, restoration of function by reconstitution/regeneration or even replacement by artificial salivary glands would be a valuable therapeutic option. Although still in the distant future, there are several different approaches that currently appear promising. One strategy is to deliver stem cells to damaged glands. Although the mechanisms of such rescue remain unclear, promising restoration of saliva flow rates has been reported in a rat model system in which glands were previously damaged by irradiation (Lombaert et al., 2008). A second approach seeks to modify the remaining damaged tissue by tissue engineering, e.g., using gene transfer technology to express aquaporin to enhance water transport (Delporte et al., 1997; Shan et al., 2005). These promising approaches that use existing damaged glandular tissue as a structural framework for regenerating a functional salivary gland have been reviewed elsewhere (Baum et al., 2010; Lombaert and Hoffman, 2010; Coppes and Stokman, 2011).
A third possibility would be to try to use the dynamic cell and tissue mechanisms that underlie normal embryonic salivary gland morphogenesis to regenerate damaged tissues or to create an artificial salivary gland. Although obviously quite challenging, this approach would take advantage of the sophisticated self-assembly mechanisms used in normal development to generate the vast numbers of acini of normal glands to provide enough epithelial surface area for adequate production of saliva. Because another oral tissue, the tooth, can be regenerated in mice by transplantation of a bioengineered embryonic tooth germ (Ikeda et al., 2009), it is possible that oral tissues could be replaced by dedifferentiated adult tissue that is guided to recapitulate a normal developmental program to form a functional replacement.
The intrinsic high motility of embryonic salivary gland epithelial cells during normal morphogenesis suggests that salivary gland architecture is not fixed, but instead involves a highly dynamic, self-assembling system. Consequently, isolated dedifferentiated or progenitor/stem cells might reassemble to generate gland structures. Isolated embryonic salivary gland epithelial cells, or a mixture of epithelial and mesenchymal cells, can in fact readily self-assemble into bud-like structures that closely resemble normal developing salivary tissue and can even synthesize salivary gland differentiation markers (Wei et al., 2007). Gland branching or formation of acinar structures can be stimulated by biologically active peptides from laminin or perlecan (Pradhan et al., 2009; Kadoya and Yamashina, 2010), or by a biomaterial such as chitosan (Yang and Young, 2009). However, one uncertainty about this developmental recapitulation approach involves the question of how to establish the ductal system. One approach might be to bioengineer this system using biomaterials and epithelial cells (Baum, 2000). These challenging regenerative and tissue engineering approaches will benefit from a better understanding of the detailed regulatory and mechanical mechanisms of gland formation.
Conclusions and Perspective
Research on salivary gland morphogenesis has advanced rapidly over the past several years with the discovery of novel regulatory pathways and direct visualization of dynamic cell motility. Besides biochemical and gene regulation, mechanical forces are also likely to play important roles in determining how glands develop. In this regard, salivary gland tissue engineering approaches will have the added benefit of providing opportunities to explore the mechanisms of gland formation. That is, experimental approaches to create glands de novo will allow researchers to determine which types of local physical, biochemical, and genetic inputs can produce different types of gland morphology and differentiation.
We have learned a great deal about the roles of many individual molecules and multiple specific regulatory pathways, as detailed in this review. However, it is not entirely clear how these numerous growth factors, matrix molecules, gene regulatory pathways, and mechanical forces are integrated during salivary gland morphogenesis. Further research should provide exciting new insight into this important process at the intersection of developmental biology, matrix biology, and tissue engineering.
Supplementary Material
Acknowledgments
The authors acknowledge support from the Intramural Research Program of the National Institute of Dental and Craniofacial Research.
Footnotes
They declare no potential conflicts of interest with respect to the authorship and/or publication of this article.
A supplemental appendix to this article is published electronically only at http://jdr.sagepub.com/supplemental.
References
- Affolter M, Zeller R, Caussinus E. (2009). Tissue remodelling through branching morphogenesis. Nat Rev Mol Cell Biol 10:831-842 [DOI] [PubMed] [Google Scholar]
- Andrew DJ, Ewald AJ. (2010). Morphogenesis of epithelial tubes: insights into tube formation, elongation, and elaboration. Dev Biol 341:34-55 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baum BJ. (2000). Prospects for re-engineering salivary glands. Adv Dent Res 14:84-88 [DOI] [PubMed] [Google Scholar]
- Baum BJ, Zheng C, Alevizos I, Cotrim AP, Liu S, McCullagh L, et al. (2010). Development of a gene transfer-based treatment for radiation-induced salivary hypofunction. Oral Oncol 46:4-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brunskill EW, Aronow BJ, Georgas K, Rumballe B, Valerius MT, Aronow J, et al. (2008). Atlas of gene expression in the developing kidney at microanatomic resolution. Dev Cell 15:781-791 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Coppes RP, Stokman MA. (2011). Stem cells and the repair of radiation-induced salivary gland damage. Oral Dis 17:143-153 [DOI] [PubMed] [Google Scholar]
- Costantini F, Kopan R. (2010). Patterning a complex organ: branching morphogenesis and nephron segmentation in kidney development. Dev Cell 18:698-712 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Daley WP, Gulfo KM, Sequeira SJ, Larsen M. (2009). Identification of a mechanochemical checkpoint and negative feedback loop regulating branching morphogenesis. Dev Biol 336:169-182 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Delporte C, O’Connell BC, He X, Lancaster HE, O’Connell AC, Agre P, et al. (1997). Increased fluid secretion after adenoviral-mediated transfer of the aquaporin-1 cDNA to irradiated rat salivary glands. Proc Natl Acad Sci USA 94:3268-3273 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gresik EW, Koyama N, Hayashi T, Kashimata M. (2009). Branching morphogenesis in the fetal mouse submandibular gland is codependent on growth factors and extracellular matrix. J Med Invest 56(Suppl): 228-233 [DOI] [PubMed] [Google Scholar]
- Grobstein C. (1953). Morphogenetic interaction between embryonic mouse tissues separated by a membrane filter. Nature 172:869-870 [DOI] [PubMed] [Google Scholar]
- Hieda Y, Nakanishi Y. (1997). Epithelial morphogenesis in mouse embryonic submandibular gland: its relationships to the tissue organization of epithelium and mesenchyme. Dev Growth Differ 39:1-8 [DOI] [PubMed] [Google Scholar]
- Hoffman MP, Kidder BL, Steinberg ZL, Lakhani S, Ho S, Kleinman HK, et al. (2002). Gene expression profiles of mouse submandibular gland development: FGFR1 regulates branching morphogenesis in vitro through BMP- and FGF-dependent mechanisms. Development 129: 5767-5778 [DOI] [PubMed] [Google Scholar]
- Hsu JC, Yamada KM. (2010). Salivary gland branching morphogenesis—recent progress and future opportunities. Int J Oral Sci 2:117-126 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ikeda E, Morita R, Nakao K, Ishida K, Nakamura T, Takano-Yamamoto T, et al. (2009). Fully functional bioengineered tooth replacement as an organ replacement therapy. Proc Natl Acad Sci USA 106:13475-13480 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jaskoll T, Zhou YM, Trump G, Melnick M. (2003). Ectodysplasin receptor-mediated signaling is essential for embryonic submandibular salivary gland development. Anat Rec A Discov Mol Cell Evol Biol 271: 322-331 [DOI] [PubMed] [Google Scholar]
- Kadoya Y, Yamashina S. (2005). Salivary gland morphogenesis and basement membranes. Anat Sci Int 80:71-79 [DOI] [PubMed] [Google Scholar]
- Kadoya Y, Yamashina S. (2010). Cellular dynamics of epithelial clefting during branching morphogenesis of the mouse submandibular gland. Dev Dyn 239:1739-1747 [DOI] [PubMed] [Google Scholar]
- Kashimata M, Gresik EW. (1996). Contemporary approaches to the study of salivary gland morphogenesis. Eur J Morphol 34:143-147 [DOI] [PubMed] [Google Scholar]
- Knox SM, Lombaert IM, Reed X, Vitale-Cross L, Gutkind JS, Hoffman MP. (2010). Parasympathetic innervation maintains epithelial progenitor cells during salivary organogenesis. Science 329:1645-1647 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Larsen M, Wei C, Yamada KM. (2006). Cell and fibronectin dynamics during branching morphogenesis. J Cell Sci 119(Pt 16):3376-3384 [DOI] [PubMed] [Google Scholar]
- Larsen M, Yamada KM, Musselmann K. (2010). Systems analysis of salivary gland development and disease. Wiley Interdiscip Rev Syst Biol Med 2:670-682 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lombaert IM, Hoffman MP. (2010). Epithelial stem/progenitor cells in the embryonic mouse submandibular gland. Front Oral Biol 14:90-106 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lombaert IM, Brunsting JF, Wierenga PK, Faber H, Stokman MA, Kok T, et al. (2008). Rescue of salivary gland function after stem cell transplantation in irradiated glands. PLoS One 3:e2063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lu P, Werb Z. (2008). Patterning mechanisms of branched organs. Science 322:1506-1509 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Melnick M, Jaskoll T. (2000). Mouse submandibular gland morphogenesis: a paradigm for embryonic signal processing. Crit Rev Oral Biol Med 11:199-215 [DOI] [PubMed] [Google Scholar]
- Melnick M, Phair RD, Lapidot SA, Jaskoll T. (2009). Salivary gland branching morphogenesis: a quantitative systems analysis of the Eda/Edar/NFkappaB paradigm. BMC Dev Biol 9:32. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Michael L, Sweeney DE, Davies JA. (2005). A role for microfilament-based contraction in branching morphogenesis of the ureteric bud. Kidney Int 68:2010-2018 [DOI] [PubMed] [Google Scholar]
- Monte JC, Sakurai H, Bush KT, Nigam SK. (2007). The developmental nephrome: systems biology in the developing kidney. Curr Opin Nephrol Hypertens 16:3-9 [DOI] [PubMed] [Google Scholar]
- Moore KA, Polte T, Huang S, Shi B, Alsberg E, Sunday ME, et al. (2005). Control of basement membrane remodeling and epithelial branching morphogenesis in embryonic lung by Rho and cytoskeletal tension. Dev Dyn 232:268-281 [DOI] [PubMed] [Google Scholar]
- Morrisey EE, Hogan BL. (2010). Preparing for the first breath: genetic and cellular mechanisms in lung development. Dev Cell 18:8-23 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Napenas JJ, Brennan MT, Fox PC. (2009). Diagnosis and treatment of xerostomia (dry mouth). Odontology 97:76-83 [DOI] [PubMed] [Google Scholar]
- Okamoto K, Kikuchi-Handa T, Nogawa H. (2010). Evidence of interlobular repulsion during branching morphogenesis in mouse salivary glands. Dev Dyn 239:2208-2218 [DOI] [PubMed] [Google Scholar]
- Onodera T, Sakai T, Hsu JC, Matsumoto K, Chiorini JA, Yamada KM. (2010). Btbd7 regulates epithelial cell dynamics and branching morphogenesis. Science 329:562-565 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Patel VN, Rebustini IT, Hoffman MP. (2006). Salivary gland branching morphogenesis. Differentiation 74:349-364 [DOI] [PubMed] [Google Scholar]
- Pirraglia C, Myat MM. (2010). Genetic regulation of salivary gland development in Drosophila melanogaster. Front Oral Biol 14:32-47 [DOI] [PubMed] [Google Scholar]
- Pradhan S, Zhang C, Jia X, Carson DD, Witt R, Farach-Carson MC. (2009). Perlecan domain IV peptide stimulates salivary gland cell assembly in vitro. Tissue Eng Part A 15:3309-3320 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rebustini IT, Myers C, Lassiter KS, Surmak A, Szabova L, Holmbeck K, et al. (2009). MT2-MMP-dependent release of collagen IV NC1 domains regulates submandibular gland branching morphogenesis. Dev Cell 17:482-493 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ridley AJ, Schwartz MA, Burridge K, Firtel RA, Ginsberg MH, Borisy G, et al. (2003). Cell migration: integrating signals from front to back. Science 302:1704-1709 [DOI] [PubMed] [Google Scholar]
- Sakai T, Larsen M, Yamada KM. (2002). Microanalysis of gene expression in tissues using T7-SAGE: serial analysis of gene expression after high-fidelity T7-based RNA amplification. Curr Protoc Cell Biol Chapter 19:Unit 19.3 [DOI] [PubMed]
- Sakai T, Larsen M, Yamada KM. (2003). Fibronectin requirement in branching morphogenesis. Nature 423:876-881 [DOI] [PubMed] [Google Scholar]
- Sequeira SJ, Larsen M, DeVine T. (2010). Extracellular matrix and growth factors in salivary gland development. Front Oral Biol 14:48-77 [DOI] [PubMed] [Google Scholar]
- Shan Z, Li J, Zheng C, Liu X, Fan Z, Zhang C, et al. (2005). Increased fluid secretion after adenoviral-mediated transfer of the human aquaporin-1 cDNA to irradiated miniature pig parotid glands. Mol Ther 11: 444-451 [DOI] [PubMed] [Google Scholar]
- Spooner BS, Wessells NK. (1972). An analysis of salivary gland morphogenesis: role of cytoplasmic microfilaments and microtubules. Dev Biol 27:38-54 [DOI] [PubMed] [Google Scholar]
- Tucker AS. (2007). Salivary gland development. Semin Cell Dev Biol 18:237-244 [DOI] [PubMed] [Google Scholar]
- Vicente-Manzanares M, Ma X, Adelstein RS, Horwitz AR. (2009). Non-muscle myosin II takes centre stage in cell adhesion and migration. Nat Rev Mol Cell Biol 10:778-790 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vissink A, Mitchell JB, Baum BJ, Limesand KH, Jensen SB, Fox PC, et al. (2010). Clinical management of salivary gland hypofunction and xerostomia in head-and-neck cancer patients: successes and barriers. Int J Radiat Oncol Biol Phys 78:983-991 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Walker JL, Menko AS, Khalil S, Rebustini I, Hoffman MP, Kreidberg JA, et al. (2008). Diverse roles of E-cadherin in the morphogenesis of the submandibular gland: insights into the formation of acinar and ductal structures. Dev Dyn 237:3128-3141 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wei C, Larsen M, Hoffman MP, Yamada KM. (2007). Self-organization and branching morphogenesis of primary salivary epithelial cells. Tissue Eng 13:721-735 [DOI] [PubMed] [Google Scholar]
- Yamamoto S, Fukumoto E, Yoshizaki K, Iwamoto T, Yamada A, Tanaka K, et al. (2008). Platelet-derived growth factor receptor regulates salivary gland morphogenesis via fibroblast growth factor expression. J Biol Chem 283:23139-23149 [DOI] [PubMed] [Google Scholar]
- Yang TL, Young TH. (2009). The specificity of chitosan in promoting branching morphogenesis of progenitor salivary tissue. Biochem Biophys Res Commun 381:466-470 [DOI] [PubMed] [Google Scholar]
- Yu W, Shewan AM, Brakeman P, Eastburn DJ, Datta A, Bryant DM, et al. (2008). Involvement of RhoA, ROCK I and myosin II in inverted orientation of epithelial polarity. EMBO Rep 9:923-929 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.