LETTER
OXA-48 is an emerging class D carbapenemase originally identified in isolates from Turkey (14) and subsequently detected in several European and north African countries (10). Klebsiella pneumoniae is the most common host for OXA-48, but the enzyme has also been detected in Escherichia coli and Enterobacter cloacae (10). The blaOXA-48 gene is carried by the composite transposon Tn1999 or a variant thereof, named Tn1999.2 (1), and is plasmid mediated. A 60-kb IncL/M-related conjugative plasmid, named pOXA-48a, which carries the carbapenemase gene as the only resistance determinant, was found to mediate the horizontal dissemination of blaOXA-48 among strains of different species and from different sources (13).
In this report we describe the first case of a strain from Italy producing OXA-48, encoded by a pOXA-48a-like plasmid which carries a new variant of Tn1999.
E. coli ECBZ-1 was isolated in April 2011 from a urinary tract infection (UTI) of a 54-year-old female patient living in northern Italy. Susceptibility testing, performed by Etest (bioMérieux, Marcy l'Etoile, France), revealed that the isolate was resistant to trimethoprim-sulfamethoxazole, penicillins (including β-lactamase inhibitor combinations), narrow-spectrum cephalosporins, and ertapenem (even by disk diffusion), while it remained susceptible to other carbapenems, expanded-spectrum cephalosporins, aminoglycosides, fluoroquinolones, and fosfomycin (Table 1). Initially treated empirically with trimethoprim-sulfamethoxazole without improvement, the infection was successfully treated with oral fosfomycin (3 g per day for 2 days). A modified Hodge test (9) suggested production of carbapenemase, but neither EDTA nor boronic acid affected meropenem susceptibility in combination disk tests (16). PCR analysis (3, 4, 6, 11, 12, 14), and sequencing revealed the presence of a blaOXA-48 carbapenemase gene and of a blaTEM-1b gene. This β-lactamase profile was consistent overall with the resistance phenotype of ECBZ-1, since OXA-48 hydrolyzes carbapenems (although weakly), penicillins, and narrow-spectrum cephalosporins but not expanded-spectrum cephalosporins and is not inhibited by clavulanate and sulfones (5, 14). The peculiar resistance phenotype of ECBZ-1 underlines the importance of surveillance of isolates showing resistance only to ertapenem even in the presence of susceptibility to expanded-spectrum cephalosporins.
Table 1.
MICs for E. coli ECBZ-1, J53(pECBZ-1), and J53
| Antibiotic | MIC (μg/ml) (category)a |
||
|---|---|---|---|
| ECBZ-1 | J53 (pECBZ-1) | J53 | |
| Ampicillin | >32 (R) | >32 | 0.5 |
| Amoxicillin-clavulanate | >32 (R) | >32 | 0.5 |
| Cefotaxime | 0.75 (S) | 0.75 | 0.25 |
| Ceftazidime | 1 (S) | 0.38 | 0.25 |
| Cefepime | 0.75 (S) | 0.125 | 0.047 |
| Imipenem | 1b (S) | 0.5 | 0.064 |
| Ertapenem | 2b (R) | 1 | 0.016 |
| Meropenem | 1b (S) | 0.5 | 0.008 |
| Piperacillin-tazobactam | 256 (R) | 8 | 0.25 |
| Levofloxacin | 0.064 (S) | ND | ND |
| Ciprofloxacin | 0.023 (S) | ND | ND |
| Amikacin | 2 (S) | ND | ND |
| Gentamicin | 0.064 (S) | ND | ND |
| Tobramycin | 1 (S) | ND | ND |
| Trimethoprim-sulfamethoxazole | 256 (R) | 0.023 | 0.023 |
| Tigecycline | 0.75 (S) | ND | ND |
| Colistin | 0.19 (S) | ND | ND |
MICs were determined for E. coli ECBZ-1, E. coli transconjugant J53(pECBZ-1), and recipient strain E. coli J53. Category classification was based on updated EUCAST breakpoints (http://www.eucast.org/clinical_breakpoints/ [version 2.0, January 1 2012]). R, resistant; S, susceptible; ND, not determined.
Diameters of inhibition: imipenem, 23 mm (S); meropenem, 22 mm (S); ertapenem, 16 mm (R).
Analysis of clinical records revealed a history of recurrent UTIs and a previous vacation (in November 2010) in Egypt, a country where OXA-48-producing isolates have been reported (10), suggesting a cross-border source for the OXA-48-producing strain.
ECBZ-1 belongs in phylogenetic group D (2) and in sequence type ST2076 (http://mlst.ucc.ie/mlst/dbs/Ecoli) related to clonal complex 394, which also includes ST471, which was found to be associated with diffusion of KPC-2 carbapenemase in Israel (7).
A conjugation experiment (8) using E. coli J53 (pro met Rifr Nalr) (17) as the recipient and selection with ertapenem (0.5 μg/ml), nalidixic acid (32 μg/ml), and rifampin (250 μg/ml) yielded J53 transconjugants (frequency, 7 × 10−5 ± 1.5 × 10−5 transconjugants per recipient) containing the blaOXA-48 but not the blaTEM-1b gene, as assessed by PCR, and showing a resistance phenotype resembling that of the donor except for resistance to trimethoprim-sulfamethoxazole (Table 1). Analysis of the plasmid (15) from a randomly selected transconjugant revealed a ca. 60-kb plasmid, named pECBZ-1 (data not shown), whose backbone was confirmed to be identical or closely related to that of the pOXA-48a IncL/M type plasmid by PCR analysis of the repA, traU, and parA genes (13).
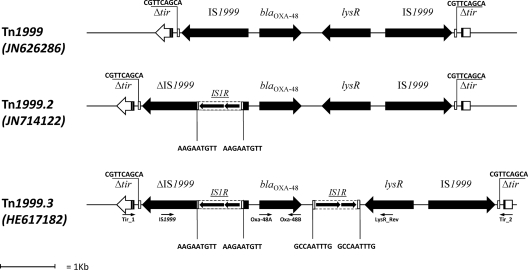
Mapping of the blaOXA-48-flanking regions and sequencing of the amplicons revealed an original genetic context that was similar overall to Tn1999.2 (1) but differed from the latter by the presence of a second copy of IS1R inserted 199 bp downstream blaOXA-48 (Fig. 1). In this new Tn1999 transposon variant, named Tn1999.3, blaOXA-48 could have the advantage of overexpression driven by the upstream IS1R copy (as in Tn1999.2 [1]), while the two copies of IS1R flanking blaOXA-48 define a new putative composite transposon that might further mobilize the carbapenemase gene (Fig. 1). Mapping of the Tn1999.3-flanking regions revealed that the transposon was inserted into a tir gene, at the same position reported for Tn1999 in plasmid pOXA-48a (13) (Fig. 1), suggesting that that Tn1999.3 evolved from Tn1999 by stepwise insertion of the IS1R elements into the latter transposon, once it had been incorporated into the plasmid backbone. This hypothesis for the evolution of Tn1999.3 is also consistent with the fact that no direct repeats are present flanking the putative composite transposon made up of the two IS1R elements.
Fig 1.
Linear map of the transposon Tn1999.3 carried by ECBZ-1, compared with the other Tn1999 variants. Small arrows show primer positions. Primers used to characterize Tn1999.3 were OXA-48A and OXA-48B (14), IS1999 (5′-TGATGTTGTGCTTGGTTCGG-3′), LysR_Rev (5′-GCTAGTGCCAATCTTACAGG-3′), Tir_1 (5′-GCCCAACAGTAAACCCAGC-3′), and Tir_2 (5′-AGTTTATGCTGGTTCTGCTGG-3′). The putative composite transposon formed by the two IS1R insertion sequences, carrying blaOXA-48, is underlined.
Nucleotide sequence accession number.
The nucleotide sequence of the blaOXA-48 gene and flanking regions has been submitted to the GenBank/EMBL database and assigned accession no. HE617182.
ACKNOWLEDGMENT
This work was partially supported by funding from the European Commission under the 7th Framework Programme (TROCAR contract HEALTH-F3-2008-223031 and TEMPOtest-QC contract HEALTH-F3-2009-241742).
Footnotes
Published ahead of print 30 January 2012
Contributor Information
Vincenzo Di Pilato, Department of Biotechnologies Section of Microbiology University of Siena Siena, Italy.
Richard Aschbacher, Laboratory of Microbiology and Virology Bolzano Central Hospital Bolzano, Italy.
Cordula Weber, Ortisei, Italy.
Clara Larcher, Laboratory of Microbiology and Virology Bolzano Central Hospital Bolzano, Italy.
Gian Maria Rossolini, Department of Biotechnologies Section of Microbiology University of Siena Siena, Italy.
REFERENCES
- 1. Carrer A, et al. 2008. Spread of OXA-48-positive carbapenem-resistant Klebsiella pneumoniae isolates in Istanbul, Turkey. Antimicrob. Agents Chemother. 52:2950–2954 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Clermont O, Bonacorsi S, Bingen E. 2000. Rapid and simple determination of the Escherichia coli phylogenetic group. Appl. Environ. Microbiol. 66:4555–4558 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. D'Andrea MM, et al. 2009. Characterization of pABVA01, a plasmid encoding the OXA-24 carbapenemase from Italian isolates of Acinetobacter baumannii. Antimicrob. Agents Chemother. 53:3528–3533 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. D'Andrea MM, et al. 2011. Persistent carriage and infection by multidrug-resistant Escherichia coli ST405 producing NDM-1 carbapenemase: report on the first Italian cases. J. Clin. Microbiol. 49:2755–2758 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Docquier JD, et al. 2009. Crystal structure of the OXA-48 β-lactamase reveals mechanistic diversity among class D carbapenemases. Chem. Biol. 16:540–547 [DOI] [PubMed] [Google Scholar]
- 6. Giani T, et al. 2009. Emergence in Italy of Klebsiella pneumoniae sequence type 258 producing KPC-3 carbapenemase. J. Clin. Microbiol. 47:3793–3794 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Goren MG, Navon-Venezia S, Chmelnitsky I, Carmeli Y. 2010. Carbapenem-resistant KPC-2-producing Escherichia coli in a Tel Aviv medical center, 2005 to 2008. Antimicrob. Agents Chemother. 54:2687–2691 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Lauretti L, et al. 1999. Cloning and characterization of blaVIM, a new integron-borne metallo-β-lactamase gene from a Pseudomonas aeruginosa clinical isolate. Antimicrob. Agents Chemother. 43:1584–1590 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Lee K, et al. 2001. Modified Hodge and EDTA-disc synergy tests to screen metallo-β-lactamase-producing strains of Pseudomonas and Acinetobacter species. Clin. Microbiol. Infect. 7:88–91 [DOI] [PubMed] [Google Scholar]
- 10. Nordmann P, Naas T, Poirel L. 2011. Global spread of carbapenemase-producing Enterobacteriaceae. Emerging Infect. Dis. 17:1791–1798 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Pallecchi L, et al. 2004. Detection of CTX-M-type β-lactamase genes in fecal Escherichia coli isolates from healthy children in Bolivia and Peru. Antimicrob. Agents Chemother. 48:4556–4561 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Perilli M, et al. 2002. Molecular characterization of extended-spectrum β-lactamases produced by nosocomial isolates of Enterobacteriaceae from an Italian nationwide survey. J. Clin. Microbiol. 40:611–614 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Poirel L, Bonnin RA, Nordmann P. 2011. Genetic features of the widespread plasmid coding for the carbapenemase OXA-48. Antimicrob. Agents Chemother. 2012. 56:559–562 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Poirel L, Héritier C, Tolün V, Nordmann P. 2004. Emergence of oxacillinase-mediated resistance to imipenem in Klebsiella pneumoniae. Antimicrob. Agents Chemother. 48:15–22 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Sambrook J, Fritsch EF, Maniatis T. 1989. Molecular cloning: a laboratory manual. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY [Google Scholar]
- 16. Tsakris A, et al. 2009. Use of boronic acid disk tests to detect extended-spectrum β-lactamases in clinical isolates of KPC carbapenemase-possessing Enterobacteriaceae. J. Clin. Microbiol. 47:3420–3426 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Wolfson JS, Hooper DC, Swartz MN, Swartz MD, McHugh GL. 1983. Rapid method for screening large numbers of Escherichia coli colonies for production of plasmid-mediated β-lactamases. Antimicrob. Agents Chemother. 23:308–312 [DOI] [PMC free article] [PubMed] [Google Scholar]