Abstract
Efflux pumps extrude a wide variety of chemically unrelated compounds conferring multidrug resistance and participating in numerous physiological processes. Mycobacterium tuberculosis possesses many efflux pumps, and their roles in drug resistance and physiology are actively investigated. In this work we found that tap mutant cells showed changes in morphology and a progressive loss of viability upon subcultivation in liquid medium. Transcriptome analysis in Mycobacterium bovis BCG revealed that disruption of the Rv1258c gene, encoding the Tap efflux pump, led to an extensive change in gene expression patterns during stationary phase, with no changes during exponential growth. In stationary phase, Tap inactivation triggered a general stress response and led to a general repression of genes involved in cell wall biosynthesis, in particular the formation of the peptidoglycan; this suggested the accumulation of an unknown Tap substrate that reaches toxic concentrations during stationary phase. We also found that both disruption and overexpression of tap altered susceptibility to many clinically approved antibiotics in M. bovis BCG. Acriflavine and tetracycline accumulation assays and carbonyl cyanide m-chlorophenylhydrazone (CCCP) potentiation experiments demonstrated that this phenotype was due to an active efflux mechanism. These findings emphasize the important role of the Tap efflux pump in bacterial physiology and intrinsic drug resistance.
INTRODUCTION
Bacterial efflux pumps are energy-dependent membrane proteins capable of exporting a wide variety of compounds from the cytoplasm. Substrates of efflux pumps include antimicrobials, synthetic compounds, lipids, toxic metabolites, host-derived antimicrobial agents, and virulence factors, among others. Such a heterogeneous substrate profile allows bacterial efflux pumps to play diverse roles in bacterial cell physiology, drug resistance, detoxification, and virulence (41). In fact, the ability of many bacterial pathogens such as Salmonella enterica serovar Typhimurium, Pseudomonas aeruginosa, Campylobacter jejuni, or Neisseria gonorrhoeae to cause disease relies on the activities of efflux pumps (42, 47). In addition, efflux-mediated drug resistance has become clinically relevant in some bacterial pathogens, such as P. aeruginosa, and attempts have been made to develop efflux pump inhibitors as a strategy to overcome drug resistance (27). Efflux pumps usually confer low levels of drug resistance but can also play critical roles in the evolution to high levels of resistance. The activities of drug transporters can facilitate the progressive acquisition of chromosomal mutations conferring higher levels of resistance to a particular antibiotic (46). In addition to their roles in drug resistance and virulence, efflux pumps have also been associated with cell division (11, 25) and have physiological roles supporting pH homeostasis and alkali tolerance (24). Interestingly, the physiological functions of these pumps may foster their persistence as potential resistance determinants (24).
In Mycobacterium, the genus including the important human pathogen Mycobacterium tuberculosis, clinically relevant drug resistance is mainly due to chromosomal mutations in genes encoding the drug target or prodrug-activating enzymes (29). As in other bacterial pathogens, drug efflux pumps could contribute to the acquisition of such mutations in M. tuberculosis and explain why mutations in the target genes were not found in many low-level resistant strains (8). In this regard, rifampin resistance in M. tuberculosis has been traditionally correlated with specific mutations in the gene encoding the β-subunit of the RNA polymerase (rpoB). It has recently been proposed that the level of rifampin resistance in these mutant strains is defined by efflux (28); moreover, it was shown that activation of efflux pump genes by rifampin led to cross-resistance, i.e., a decrease in susceptibility to ofloxacin. The loss of particular efflux pumps also results in M. tuberculosis strains having a decreased virulence phenotype in an animal model of infection (6), probably due to their inabilities to properly secrete and locate essential cell envelope components (5). In addition, efflux pumps in mycobacteria also play fundamental roles in intrinsic drug resistance, oxidative stress responses, cell wall assembly, and growth (8, 15, 36, 53). These findings highlight the relevance of efflux pumps for establishing latency, in which a subpopulation of mycobacteria that are slowly dividing, metabolically active, and drug tolerant is able to persist in tuberculosis (TB) patients. The persistent state of mycobacterium has some similarities to cultures in stationary growth phase (23).
We have previously characterized the Tap efflux pumps from Mycobacterium fortuitum and M. tuberculosis, focusing on their contributions to drug resistance. These genes were expressed in the fast-growing nonpathogenic species Mycobacterium smegmatis. Here we extend these studies, further documenting the role of Tap efflux pumps in providing intrinsic drug resistance as well as its essential roles in physiology, growth, and cell morphology in Mycobacterium bovis BCG Pasteur, a slow-growing, more closely related model system for M. tuberculosis.
MATERIALS AND METHODS
Bacterial strains, growth conditions, and chemicals.
Strains used in this study are listed in Table 1. Sequences of oligonucleotides and plasmids used are available upon request. M. bovis BCG was cultivated at 37°C and 5% CO2 in Middlebrook 7H9 broth (Difco) supplemented with 10% Middlebrook ADC (Difco) and 0.05% (vol/vol) Tween 80 or on Middlebrook 7H10 agar plates (Difco) supplemented with 10% (vol/vol) oleic acid-albumin-dextrose-catalase (OADC; Difco). Escherichia coli was grown at 37°C in LB broth or on LB 1.5% agar plates. For the selection of vectors in mycobacteria, hygromycin or kanamycin was added to cultures at final concentrations of 10 mg/liter or 20 mg/liter, respectively. Plasmids were maintained in E. coli with appropriate antibiotics for selection (ampicillin 100 mg/liter, kanamycin 20 mg/liter).
Table 1.
Strains used in this study
| Strain | Description | Reference or source |
|---|---|---|
| M. bovis BCG Pasteur 1173 | Wild type | Laboratory collection |
| M. bovis SUM36 | Derivative of wild type containing vector pSUM36 | This study |
| M. bovis KOTap | Derivative of wild type with the tap gene disrupted; tapTB::Ωhyg | This study |
| M. bovis PAZ11 | Derivative of wild type expressing the tapTB gene under its own promoter in the multicopy pSUM36 plasmid | This study |
| M. bovis AC48 | Derivative of wild type expressing the tapFR gene under its own promoter in the multicopy pSUM36 plasmid | This study |
| M. bovis KOP55 | Derivative of wild type with the p55 gene disrupted; p55::Ωhyg | 53 |
| M. bovis KO2333 | Derivative of wild type with the stp gene disrupted; stp::Ωhyg | 52 |
DNA manipulations.
DNA manipulations were carried out according to standard techniques (54). Mycobacterial genomic DNA isolation was performed as previously described (43). Southern blotting was done using the ECL direct nucleic acid labeling and detection system (Amersham Biosciences) according to the manufacturer's instructions. Both E. coli and mycobacteria were transformed by electroporation with a gene pulser (Bio-Rad Laboratories Inc., Richmond, CA) (43).
Strain construction.
The nucleotide sequence of the M. tuberculosis Rv1258c gene (http://genolist.pasteur.fr/TubercuList/) is identical to that of BCG1316c from M. bovis BCG Pasteur 1173 P2 (http://genolist.pasteur.fr/BCGList/). In this study, both Rv1258c and BCG1316c will be referred to as tapTB and the tap gene from M. fortuitum as tapFR.
(i) Overexpression.
The tapTB and tapFR genes expressed under the control of their respective promoters were cloned previously into the pSUM36 vector, yielding pPAZ11 (9) and pAC48 (2). Plasmids pPAZ11 and pAC48 were introduced to M. bovis BCG, resulting in M. bovis BCG PAZ11 and M. bovis BCG AC48, respectively.
(ii) Disruption.
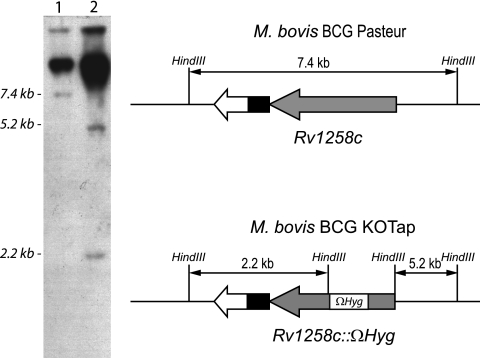
A suicide delivery plasmid was constructed containing the tapTB gene interrupted by the insertion of a hygromycin resistance cassette (Ωhyg) with flanking termination sequences (49). Briefly, a 2.3-kb PCR product from M. tuberculosis H37Rv genomic DNA containing tapTB was cloned into pUC19. The tapTB gene was then interrupted by the insertion of the Ωhyg cassette. The tap::Ωhyg fragment was isolated by PstI digestion and cloned into the PstI-digested p2NIL vector (44), yielding pVZ16. A cassette containing lacZ and sacB genes from pGOAL17 (44) was then cloned into the single PacI site of pVZ16 to generate the suicide delivery vector pVZ17. pVZ17 was used to transform M. bovis BCG. Single- and double-crossover (DXO) transformants were selected as described elsewhere (44). Candidates for DXO were analyzed by PCR with primers for the tapTB gene flanking the Ωhyg insertion point. The mutant DNA generated a large PCR fragment compared to the wild-type fragment, because it included the inserted hygromycin cassette (data not shown). Candidates with the expected PCR patterns were streaked onto plates with X-Gal (5-bromo-4-chloro-3-indolyl-β-d-galactopyranoside) and either kanamycin or hygromycin for phenotypic analysis. Finally, Southern blotting was done to confirm DXO (Fig. 1). The M. bovis BCG strain with the disrupted tapTB gene was named M. bovis BCG KOTap (Table 1).
Fig 1.
Southern blot analysis for tap gene inactivation in M. bovis BCG. Genomic DNA isolated from M. bovis BCG wild type (lane 1) and the M. bovis BCG double-crossover (DXO) strain (lane 2) was digested with HindIII and hybridized to a probe corresponding to a 0.86-kb tapTB internal region flanking the Ωhyg insertion point. HindIII digestion released the Ωhyg cassette, yielding two fragments of 5.2 kb and 2.2 kb for the DXO strain, while a single 7.4-kb fragment was observed in the wild-type strain. Loading wells and undigested DNA can be also seen on the upper part of the image. The diagram showing the expected HindIII digestion fragment is not shown to scale. In M. bovis BCG Pasteur, the RD13 deletion (4) comprises the region containing the Rv1255c, Rv1256c, and Rv1257c genes, compared to the M. tuberculosis H37Rv genome. The chimeric gene after the fusion of the Rv1255c and Rv1257c genes is shown in white and black.
Drug susceptibility assays.
MICs were determined using 2-fold serial dilutions of compounds. For determination in liquid medium, the resazurin assay was carried out essentially as previously described (51), except plates were incubated for 8 days at 37°C and for two additional days after the addition of the redox indicator (resazurin). For agar-based determinations, exponential-phase cultures were diluted to 105 cells/ml, and 10 μl was spotted onto 25-ml 7H10 agar plates containing serial 2-fold antibiotic dilutions. Additionally, 10-fold serial dilutions of a 107-cell/ml exponential-phase culture were inoculated on a fixed subinhibitory concentration of antibiotic using a Steer replicator. A conventional disc diffusion assay was used to test the redox compounds. Briefly, an exponential-phase culture dilution containing 106 cells/ml was spread on Middlebrook 7H10 agar plates, and discs containing the redox compounds were placed onto the lawn. Visible growth was scored after 21 days of incubation. Experiments were carried out in triplicate and repeated at least three times.
Accumulation experiments. (i) [3H]tetracycline accumulation.
Uptake experiments were essentially performed as previously described (10, 35). Exponential-phase cultures of M. bovis BCG wild-type and KOTap strains growing in 7H9 broth were harvested by centrifugation at room temperature, washed twice with 0.1 M potassium phosphate (pH 7.0), and resuspended in prewarmed assay phosphate buffer (0.1 M potassium phosphate [pH 7.0], 1 mM MgCl2). Aliquots of 1 ml were preincubated for 15 min at 37°C with vigorous aeration by shaking, and the assay was started by the addition of [3H]tetracycline (0.76 Ci/mmol; New England Nuclear) to a final concentration of 5 μM. At various time intervals thereafter, 50 μl of the suspension was removed, diluted in 1 ml of ice-cold 0.1 M potassium phosphate (pH 7.0) buffer containing 0.1 M LiCl, and immediately filtered through a 0.45-μm-pore-size filter (Millipore). The filter was rapidly washed twice with 4 ml of the same buffer and dried. The radioactivity was then determined in a Beckman LS 7000 liquid scintillation counter by using Ecolume scintillation cocktail (ICN Biomedicals). Uptake experiments were performed at least three times.
Acriflavine accumulation.
Exponential-phase cultures of M. bovis BCG SUM36 and PAZ11 cultures grown in 7H9 broth were washed twice and resuspended in 0.1 M potassium phosphate (pH 7.0). Cells were incubated at 37°C for 15 min to allow stabilization of the intrinsic fluorescence (F0) of the cells. The experiment was initiated by the addition of acriflavine at a final concentration of 5 μM. At every time point, fluorescence was measured for 20 s, and the highest [F1(max)] and lowest [F1(min)] values were recorded. The mean fluorescence of the two values was then calculated (F1). Acriflavine accumulation was expressed as an arbitrary unit (F1/F0). Acriflavine has an excitation wavelength (λex) of 485 nm and an emission wavelength (λex) of 501 nm. Fluorescence was determined using a Perkin Elmer LS-3 fluorimeter.
RNA extraction, cDNA labeling, microarray hybridization, and data analysis.
Two independent cultures of M. bovis BCG wild-type and KOTap strains grown in 75-cm2 tissue culture standing flasks were harvested during exponential growth phase (1 week; optical density at 600 nm [OD600] = 0.2 to 0.3) or stationary growth phase (6 weeks; OD600 = 0.8 to 1.0). Cultures were pelleted at room temperature. After removing the supernatant, pellets were frozen on dry ice and stored at −80°C. RNA extraction was performed as previously described (30). Fluorescently labeled cDNA copies of total RNA were prepared by direct incorporation of fluorescent nucleotide analogues during a first-strand reverse transcription (RT) reaction as previously described (30). M. tuberculosis oligoarrays were obtained from the Center for Applied Genomics, International Center for Public Health (Newark, NJ). These microarrays consist of 4,295 70-mer oligonucleotides representing 3,924 open reading frames (ORFs) from M. tuberculosis strain H37Rv and 371 unique ORFs from strain CDC1551 that are not present in H37Rv. Each microarray was processed for hybridization as previously described (30). Hybridizations were performed using RNA extracted from two cultures, each started from different colonies. Each sample was hybridized twice through reverse labeling of their cDNAs. Fluorescence intensity data from each array were collected with an Affymetrix 428 scanner. Fluorescence intensities of Cy3 and Cy5 dyes at each spot were quantified using the ImaGene software 5.0 (BioDiscovery, Inc.), and data obtained from qualified spots on each chip were normalized using the print-tip Lowess implementation included in GEPAS v1.1 (62). The expression ratio for the wild-type and the mutant genes was determined from the normalized fluorescence intensity and was calculated as the average fold change. Significance analysis of microarrays (SAM) identified genes whose expression was affected by the disruption of tap (60), defined by a q value (percent chance that the gene is a false positive) of ≤1% and a minimum change in expression of 2.0- or 0.5-fold.
RT-qPCR.
Reverse transcription was performed with random hexamer primers using murine leukoblastoma virus retrotranscriptase (Applied Biosystems), as previously described (30). Quantitative PCR (qPCR) was performed with SYBR green master mix (Applied Biosystems). After 10 min at 95°C to activate the enzyme, 40 amplification cycles were performed using an Applied Biosystems 7700 Prism spectrofluorometric thermal cycler (Perkin-Elmer) under the following conditions: 1 min denaturation at 95°C, 30 s annealing at 60°C, and 30 s extension at 72°C. Results were normalized to the amount of sigA mRNA, as previously described (31). RNA samples that had not been reverse transcribed were included in all experiments to exclude samples with significant DNA contamination. For each sample, melting curves were performed to confirm the purity of the products. Sequences of the primers for qRT-PCR are available upon request.
Auramine O stain.
Samples were taken to an OD600 of 0.5 and fixed on a heating block for 1 h at 70 to 80°C. A fluorescent stain kit for mycobacteria (Fluka) was used. Glass slides were flooded with phenolic auramine O solution and stained for 30 min. Samples were decolorized for 1 min with an acid-alcohol solution. Finally, counterstaining was performed for 2 min using a potassium permanganate solution. Samples were air dried and visualized by fluorescence microscopy. Images were captured with a Hamamatsu Orca camera system coupled to a Nikon Eclipse TE2000-U microscope equipped with a 100× objective and a 10× ocular. Micrograph analyses were performed using ImageJ.
Microarray data accession number.
Microarray data have been deposited in the Gene Expression Omnibus public database (http://www.ncbi.nlm.nih.gov/geo) under the accession number GSE32249.
RESULTS AND DISCUSSION
KOTap cells had a progressive growth defect and altered morphology.
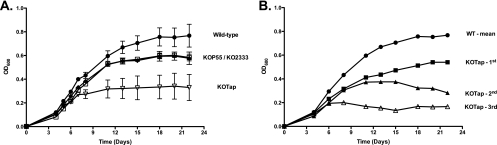
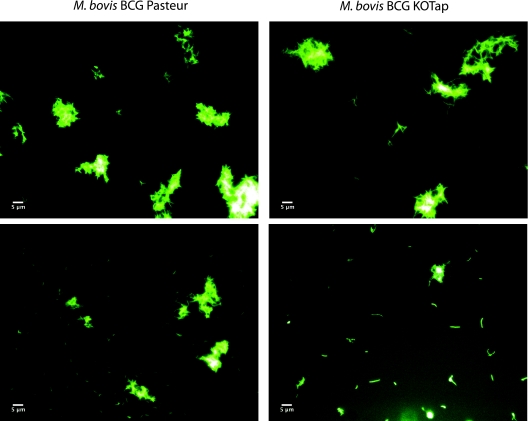
In the course of this study, we found that KOTap cells (M. bovis BCG with a disrupted homolog of the tapTB gene) grew less rapidly and arrested growth prematurely after cycles of subcultivation in liquid cultures (Fig. 2). This progressive growth defect could be reversed by cultivation on solid medium; when these new single colonies were reinoculated into liquid medium, growth kinetics were similar to those of the original transformants. Interestingly, after subcultivation in liquid, the mutant bacilli became more elongated (Fig. 3). The progressive growth defect and altered morphology phenotypes were specific for the KOTap strain; other efflux pump mutants (p55 [53] and stp [52]) were included for comparison and did not display the progressive growth defect (Fig. 2A) or elongated bacillus morphology (data not shown). Surprisingly, KOTap cells resembled the morphology observed after overexpression of ftsZ (12) or an E. coli AcrEF-TolC efflux pump mutant (25). The ftsZ gene determines molecular events involved in cell division in mycobacteria. Dziadek et al. (12) showed that overproduction of FtsZ generated filamentous cells that lacked visible septa and were not viable. Finally, while the p55 colonies were smaller than those of its parental strain (53), this phenotype was not observed in KOTap or KO2333 (data not shown), suggesting different physiological roles for these efflux pumps.
Fig 2.
Growth kinetics of M. bovis BCG wild-type and efflux pump mutant strains. Single colonies were inoculated in 10 ml of broth as preinoculum and subcultivated at 37°C, 5% CO2 in 100-ml cultures in 75-cm2 tissue culture standing flasks with occasional shaking. After 1 week, strains were subcultivated in fresh media. Bacteria were subcultured on a weekly basis, i.e., the second subculture was inoculated from the first, and the third subculture from the second. (A) Wild type (circles), KOP55 (open squares), KO2333 (diamonds) and KOTap (inverted empty triangles). Growth curves are the means from the three subcultures and one representative of two experiments. Error bars represent standard errors of the mean (SEM). (B) Consecutive individual subcultivation growth curves are shown for KOTap (first subculture [squares], second subculture [triangles], and third subculture [open triangles]). The growth curve of the wild type is shown for comparison.
Fig 3.
Morphology of M. bovis BCG wild-type and KOTap cells. Samples correspond to the second subcultivation at day 14. Two fields with aggregates and individual cells are shown for comparison. Bars, 5 μm.
Inactivation of the Tap efflux pump in M. bovis BCG greatly alters global gene expression in stationary phase but not during exponential growth.
Efflux pump studies have traditionally focused on their roles in conferring intrinsic drug resistance in bacteria (26), a function having potential clinical implications (46). However, other documented roles in physiological homeostasis and virulence processes (47) suggest that antibiotic resistance genes, particularly those encoding multidrug efflux transporters, could have evolved from existing genes with other physiological functions (24).
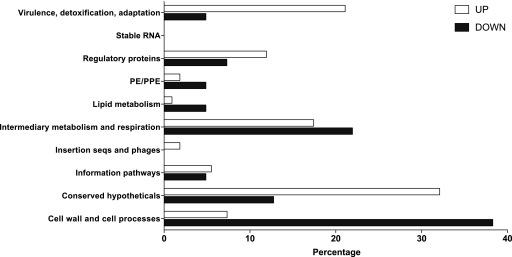
Because of the progressive growth defect observed in the KOTap strain, we used transcriptomic approaches to study the role of the Tap efflux pump during exponential and stationary growth and to define both in vitro growth dynamics and potential roles of Tap in TB latency. M. bovis BCG KOTap transformants were initially viable, indicating that tapTB is not essential for growth in vitro during exponential growth. When gene expression patterns of the tap mutant were compared to those of the wild-type strain during exponential growth (1 week; OD600 = 0.2 to 0.3), almost no differences were observed; only seven genes had slightly increased expression (Table 2; also see the supplemental material). In contrast, more than 100 genes had altered levels of expression in stationary-phase cultures; the induction or repression of a set of 18 relevant genes was validated by qPCR (Tables 2 and 3). A functional category analysis (http://tuberculist.epfl.ch/index.html) of the genes differentially expressed in stationary phase revealed a high proportion belonging to the virulence, detoxification, adaptation (VDA), intermediary metabolism and respiration (IMR), conserved hypotheticals (CH), and cell wall and cell processing (CWCP) groups, suggesting a major adaptation to a stress generated by the disruption of tapTB (Fig. 4). A comparative analysis of the genes up- or downregulated revealed a greater proportion of the upregulated genes in the VDA category, while downregulated genes were more abundant in the CWCP and lipid metabolism (LM) categories. Interestingly, Stewart et al. (58) reported a similar response of M. tuberculosis after heat shock, “with a bias toward adaptation/detoxification and regulatory genes, and away from cell wall-associated genes.” These results clearly indicated that Tap was needed to maintain balanced physiological functions during late stationary phase and pointed toward a potential role of the Tap efflux pump in latency. As bacterial growth is arrested during stationary phase, toxic waste products accumulate inside the cell and must be cleared from the cytoplasm. We suggest that normal physiological substrates of Tap are toxic by-products which accumulate during stationary phase when nutrients are limited. The inactivation of Tap would trigger a shift in cell metabolism to counteract the physiological stress produced by the accumulation of its endogenous physiological substrates. Detailed analyses of the genes and operons differentially expressed in the KOTap strain described in the following sections support this hypothesis (Table 3) (a complete list of differentially regulated genes is provided in the supplemental material).
Table 2.
Total number of genes differentially expressed after Tap inactivation in M. bovis BCG
| Growth phaseb | No. of genes with altered expressiona |
|||
|---|---|---|---|---|
| Increased by: |
Decreased by: |
|||
| ≥2 | ≥1.7 | ≤0.5 | ≤0.7 | |
| Exponential | 0 | 7 | 0 | 0 |
| Stationary | 103 | 109 | 30 | 40 |
Two cutoff values were used to define changes in gene expression (fold variation compared to wild-type levels of expression).
Gene expression was analyzed during exponential growth phase (1 week: OD600 = 0.2 to 0.3) and stationary phase (6 weeks: OD600 = 0.8 to 1.0).
Table 3.
Genes differentially regulated in M. bovis BCG during stationary phase due to inactivation of the Tap efflux pump
| Genea | Rv no.b | q valuec | Fold change in expressiond | qPCRe | Functional categoryf | Gene product |
|---|---|---|---|---|---|---|
| Upregulated | ||||||
| acr2 | Rv0251c | 0 | 12.03 | 18.14 ± 6.17 | VDA | Heat shock protein |
| dnaK | Rv0350 | 0 | 8.52 | 14.21 ± 0.98 | VDA | Chaperone |
| grpE | Rv0351 | 0 | 10.84 | VDA | Hsp-70 cofactor | |
| dnaJ | Rv0352 | NA | NA | 3.91 ± 0.82 | VDA | Chaperone |
| hspR | Rv0353 | 0 | 8.12 | VDA | Heat shock protein | |
| clpB | Rv0384c | 0 | 6.32 | VDA | Heat shock protein | |
| groEL2 | Rv0440 | 0 | 9.68 | 20.43 ± 13.50 | VDA | Chaperone |
| cysK2 | Rv0848 | 0 | 4.52 | 10.60 ± 9.23 | IMR | Cysteine synthase |
| trxB1 | Rv1471 | 0 | 2.56 | 4.92 ± 1.16 | IMR | Thioredoxin |
| nadA | Rv1594 | 0 | 1.97 | IMR | Quinolinate synthetase | |
| nadB | Rv1595 | 0 | 4.39 | IMR | l-Aspartate oxidase | |
| Rv1812c | 0 | 5.78 | IMR | NADH dehydrogenase | ||
| Rv1813c | 0 | 8.31 | CH | |||
| katG | Rv1908c | 0 | 2.54 | 3.0 ± 1.3 | VDA | Catalase-peroxidase |
| furA | Rv1909c | 0 | 4.94 | RP | Ferric uptake regulation | |
| Rv2466c | 0 | 5.78 | 5.16 ± 2.52 | CH | DsbA-like | |
| Rv2912c | 0 | 6.81 | RP | Transcriptional regulator | ||
| Rv2913c | 0 | 24.75 | IMR | d-Amino acid aminohydrolase | ||
| groEL1 | Rv3417c | 0 | 7.51 | 17.55 ± 11.64 | VDA | Chaperone |
| groES | Rv3418c | 0 | 6.35 | VDA | Chaperone | |
| bfrB | Rv3841 | 0 | 3.14 | IMR | Bacterioferritin | |
| whiB6 | Rv3862c | 0 | 6.00 | 4.39 ± 0.93 | RP | Transcriptional regulator |
| trxC | Rv3914 | 0 | 3.31 | IMR | Thioredoxin | |
| Downregulated | ||||||
| galE2 | Rv0501 | 0 | 0.44 | 0.50 ± 0.17 | IMR | UDP-glucose-4-epimerase |
| esxJ | Rv1038c | 0.95 | 0.45 | CWCP | ESAT-6-like | |
| sigE | Rv1221 | 0 | 0.37 | IP | Sigma factor | |
| eccB5 | Rv1782 | NA | NA | 0.43 ± 1.38 | CWCP | Esx conserved component |
| eccCb5 | Rv1784 | 0 | 0.44 | 0.36 ± 0.27 | CWCP | Esx conserved component |
| PPE26 | Rv1789 | NA | NA | 0.31 ± 0.11 | PE/PPE | PPE family |
| esxM | Rv1792 | 0.51 | 0.44 | 0.13 ± 0.01 | CWCP | ESAT-6-like |
| esxN | Rv1793 | NA | NA | 0.28 ± 0.02 | CWCP | ESAT-6-like |
| eccA5 | Rv1798 | NA | NA | 0.39 ± 0.12 | CWCP | Esx conserved component |
| mpt70 | Rv2875 | 0 | 0.36 | CWCP | Secreted immunogenic | |
| sugI | Rv3331 | 0 | 0.12 | 0.01 ± 0.00 | CWCP | Sugar transporter |
| nagA | Rv3332 | 0.38 | 0.38 | CWCP | GlcNAc6p deacetylase | |
| esxU | Rv3445c | 0 | 0.30 | CWCP | ESAT-6-like | |
| Asd | Rv3708c | 0.55 | 0.41 | IMR | Aspartate-semialdehyde dehydrogenase |
Genes were included in the table if their q value was ≤1 and the fold increase or decrease in gene expression was ≥2.0 or ≤0.5, respectively. Some genes with no expression data available (NA) were also included, provided they belonged to an induced or repressed gene cluster.
Genes are annotated as described by the Pasteur Institute on TubercuList (http://genolist.pasteur.fr/TubercuList/).
q value indicates the false discovery rate (FDR), i.e., the probability that the gene was falsely called (calculated by SAM). NA, not available.
Values of change in gene expression by microarray experiments.
Data represent mean values from two biologically independent experiments performed in duplicate. Standard deviation is shown.
Functional categories as per TubercuList (http://genolist.pasteur.fr/TubercuList/). CWCP, cell wall and cell processes; CH, conserved hypotheticals; IP, information pathways; ISP, Insertion seqs and phages; IMR, intermediary metabolism and respiration; LP, lipid metabolism; PE/PPE, PE/PPE; RP, regulatory proteins; SR, stable RNA; VDA, virulence, detoxification, adaptation.
Fig 4.
Functional categories of genes with increased (UP; cutoff value, 1.7) or decreased (DOWN; cutoff value, 0.7) expression in stationary phase in M. bovis BCG KOTap compared to wild-type strain. The total number of genes, either up- or downregulated (as shown in the supplemental material), is taken as 100%. The number of genes, as a percentage of the total, is depicted.
Genes upregulated during stationary phase upon tapTB disruption.
In general, the expression levels of stress response genes, including members of the HspR regulon (acr2, dnak, grpE, dnaJ, clpB, bfrB, and hspR), were highly upregulated. This set of genes responds to heat shock (58) and antibacterial agents produced by the immune system (57, 64). Increased expression of heat shock proteins is also promoted by the accumulation of unfolded polypeptides (39); in agreement with this, we observed that chaperone genes groEL1 and groEL2 were also highly upregulated (Table 3).
Genes involved in oxidative stress responses were also upregulated. Rv2466c is predicted to contain thioredoxin and disulfide oxidoreductase domains; it is a SigH-dependent gene induced under anaerobic cultures (38) that is associated with oxidative stress responses in M. tuberculosis (50). Other upregulated genes were involved in the biosynthesis of reducing equivalents, including nadA, nadB, and Rv1812c (encoding a putative NADH dehydrogenase); thioredoxins (trxB1 and trxC); or genes responding to stress conditions (katG [19] and furA [13]) (Table 3).
The gene Rv2913c, annotated as a d-amino acid aminohydrolase, displayed the highest increase in expression (25-fold) in the tapTB mutant. d-Amino acids are abundant components of peptidoglycan (PG) present in the peptide that cross-links the linear glycan chains in the cell wall (61). As discussed below, Tap could be involved in PG-recycling pathways that are growth rate dependent. In the tap mutant, toxic intermediates of cell wall assembly may accumulate during stationary phase. The increased expression of d-amino acid aminohydrolases might serve as an alternative detoxifying system.
The expression of whiB6 also increased; it is one of seven whiB-like genes in M. tuberculosis, which are thought to play roles in transcriptional regulation (56). Interestingly, whiB6 appears to be nonessential for the in vitro transition from exponential to stationary phase (63). However, in a study of the seven WhiB paralogs in M. tuberculosis, Geiman et al. (17) showed that whiB6 was the most highly induced by diverse stresses, including heat shock, oxidative stress, and treatments that affected cell wall integrity. Accordingly, WhiB6 might specifically respond to the stress cause by the inactivation of the Tap efflux pump. Interestingly, WhiB7 is thought to control the expression of tapTB (37), suggesting a regulatory link between these stress responsive WhiB proteins.
Genes downregulated during stationary phase upon tapTB disruption.
More than one-third of the genes downregulated in the KOTap strain belonged to the CWCP category (Fig. 4), suggesting that the stress conditions caused by tapTB disruption resulted in the slowdown of cell wall-related processes. Among these genes, sugI, encoding an efflux pump of the major facilitator superfamily (MFS) thought to be involved in sugar transport, was the most downregulated (Table 3). It forms an operon with the N-acetylglucosamine-6-phosphate (GlcNAc6p) deacetylase gene, nagA, also highly downregulated, which is involved in the pathway for recycling the amino sugar of the PG. It is estimated that 40% to 50% of the PG is degraded and reused each generation. In E. coli, NagA catalyzes the conversion of GlcNAc6p into glucosamine-6-phosphate (GlcN6P), which is then converted into UDP-GlcNAc, the main precursor of the PG biosynthetic pathway, by the GlmM and GlmU enzymes. Newton and coworkers (40) have suggested a role for SugI in the import of myo-inositol, a sugar needed for mycothiol biosynthesis and as a precursor to mycobacterial membrane and cell wall components. If so, why would inactivation of the Tap efflux pump have such a dramatic effect on the expression of the sugI-nagA operon during stationary phase but not during exponential growth? In addition to having a structural role, amino sugars are an important energy source for bacteria because they supply both carbon and nitrogen. This could be of particular importance in stationary phase, in which nutrients are limited. In this context, Tap could be involved in the maintenance of sugar phosphate homeostasis. The inactivation of Tap would lead to GlcN6P accumulation, causing toxicity (21, 48), which would explain the upregulation of genes involved in general stress responses. A model in which PG recycling is shut down to prevent the accumulation of sugar phosphates is supported by the observation that other genes involved in cell wall processes are also repressed (Table 3). These include the galE2 gene encoding a possible UDP-glucose-4-epimerase, involved in the biosynthetic arabinogalactan-peptidoglycan complex (MAPc) pathway of M. tuberculosis (7), and asd, encoding an l-aspartic-β-semialdehyde dehydrogenase. The asd gene forms an operon with ask, an aspartokinase; both genes are essential for the biosynthetic pathway of diaminopimelate (DAP), which forms essential interpeptide PG cross-linking (45). These observations suggest a general repression of genes involved in cell wall biosynthesis, in particular the formation of the PG.
Most sigma factors are involved in the regulation of stress responses, nutrient adaptation, and cell differentiation. In our analysis, sigma E was the only sigma factor whose expression was altered in the KOTap mutant strain (Table 3). The sigE gene is essential for growth in macrophages and virulence in mice (32, 34). In addition, a sigE mutant is more sensitive to membrane-disrupting agents and vancomycin as well as heat shock and oxidative compounds such as hydrogen peroxide or plumbagin (33). Interestingly, in addition to vancomycin the KOTap strain was more sensitive to the oxidative compounds diamide, hydrogen peroxide, and plumbagin (Tables 4 and 5), suggesting a role (direct or indirect) in cell wall stability and in the maintenance of the oxidative balance within the cell, as previously described for another efflux pump in mycobacteria (53).
Table 4.
Sensitivity of M. bovis BCG KOTap to redox compounds
| Redox compounda | Disc load (μmol) | Inhibition zoneb (mm) for M. bovis BCG strains |
|
|---|---|---|---|
| Wild type | KOTapc | ||
| Hydrogen peroxide | 80 | 15 | 18 |
| 40 | 13 | 14 | |
| Plumbagin | 0.01 | 26 | 30 |
| Diamide | 20 | 0 | 18 |
Dimethyl sulfoxide, which was used as the solvent for some of these compounds, had no effect on the growth of either strain (data not shown).
Zones of inhibition were recorded after 21 days at 37°C.
KOTap is M. bovis BCG with the tapTB gene inactivated.
Table 5.
Antimicrobial susceptibility of M. bovis BCG Tap-derivative strains
| Compound | MIC for M. bovis BCG strains (mg/liter)a |
|||
|---|---|---|---|---|
| Wild type | KOTapb | PAZ11c | AC48d | |
| 2′-N-Ethylnetilmicin | 1 | 0.5 | ND | ND |
| 6′-N-Ethylnetilmycin | 2 | 1 | ND | ND |
| Acriflavine | 1 | 1–0.5 | 4 | 2 |
| Chloramphenicol | 4 | 4 | 8 | 8 |
| Clarithromycin | 0.03 | 0.03–0.015 | 0.03 | 0.06–0.25 |
| Ethambutol | 4 | 4 | 8 | 4 |
| Gentamicin | 1 | 0.5 | 2–4 | 8 |
| Isoniazid | 0.2 | 0.2 | 0.4 | 0.4 |
| p-Aminosalicylate | 0.125 | 0.03 | 1 | 0.25 |
| Rifampin | 0.012 | 0.012 | 0.012 | 0.025 |
| Streptomycin | 0.2 | 0.2 | 0.4 | 1.6 |
| Spectinomycin | 1 | 0.25 | 4 | 4 |
| Tetracycline | 1 | 0.25 | 4 | 4 |
| Tetracycline + CCCPe | ND | ND | 1 | 1 |
| Triclosan | 8 | 4 | 32 | 8–16 |
| Vancomycin | 10 | 5 | 20–40 | 40 |
MICs were assayed over a range of 2-fold dilutions of antibiotics. No difference in sensitivity between wild-type and Tap-derivative strains was observed for the following compounds: amikacin (MICWT = 0.125 mg/liter), carbonyl cyanide m-chlorophenylhydrazone (MICWT = 5 mg/liter), ciprofloxacin (MICWT = 0.125 mg/liter), clofazimine (MICWT = 0.05 to 0.1 mg/liter), chlorpromazine (MICWT = 4 mg/liter), econazole (MICWT = 6.4 mg/liter), ethionamide (MICWT = 4 mg/liter), fluconazole (MICWT, >64 mg/liter), gatifloxacin (MICWT = 0.03 mg/liter), moxifloxacin (MICWT = 0.03 to 0.06 mg/liter), and ofloxacin (MICWT = 0.25 mg/liter). ND, not determined.
KOTap, M. bovis BCG with the tapTB gene inactivated.
PAZ11, M. bovis BCG strain with the pPAZ11 plasmid (tapTB cloned into pSUM36).
AC48, M. bovis BCG containing pAC48 plasmid (tapFR cloned in pSUM36).
CCCP was present at a concentration of 2 mg/liter.
Finally, several genes (eccB5, eccCb5, eccA5, esxJ, esxM, esxN, and esxU) encoding members of the immunologically active early secretory antigenic target-6 (ESAT-6) family of proteins, along with the major secreted immunogenic Mtp70 protein, were repressed in the KOTap strain (Table 3). The esat-6 gene is found in the RD1 (region of difference 1; absent from all M. bovis BCG strains), and it has been correlated with virulence (18). We confirmed the repression of the genes contained in the ESAT-6 gene cluster region 5 of M. tuberculosis (Table 3), involved in antigen secretion (18). These results suggest a link between the Tap efflux pump and virulence.
Mycobacterial Tap efflux pumps contribute to intrinsic antibiotic resistance in M. bovis BCG.
The mycobacterial Tap efflux pumps from M. fortuitum and M. tuberculosis were originally reported to confer tetracycline and aminoglycoside resistance in M. smegmatis, a fast-growing Mycobacterium (2). The levels of resistance conferred and the substrate specificities differed between the two efflux pumps; in general, TapFR provided higher levels of resistance to a larger number of drugs than TapTB (2). TapFR used the electrochemical gradient to extrude tetracycline from the cell, and this efflux activity could be inhibited by several compounds, such as the protonophore carbonyl cyanide m-chlorophenylhydrazone (CCCP) (51). In the present study, the contribution of these Tap efflux pumps to antimicrobial resistance was characterized in M. bovis BCG, a much closer relative to M. tuberculosis than M. smegmatis.
M. bovis BCG derivatives were constructed overexpressing tapTB or tapFR genes, or with the tapTB gene interrupted by the insertion of a hygromycin resistance cassette (Table 1; Fig. 1). Antimicrobial susceptibility to a set of compounds with diverse cellular targets was determined for these strains (Table 5). In general, variations in sensitivity due to the interruption of the tapTB gene were lower (2- to 4-fold) than those due to overexpressing either tapTB or tapFR (4- to 8-fold). The substrate specificity profile was also larger when either tap gene (tapTB or tapFR) was overexpressed; there are two possible explanations for these differences. First, the disruption of tapTB could lead to a compensatory increase in the levels of expression of other efflux pump genes with similar substrate recognition profiles. Second, it is well recognized that efflux pumps nonspecifically extrude a wide variety of chemically and structurally unrelated compounds and that constitutive overexpression may provide for increased resistance, a phenotype that would not be detected in studies of efflux pump mutants (42); here, tapTB and tapFR were overexpressed on a multicopy plasmid (5 to 10 copies per cell). A >4-fold change in sensitivity was observed for acriflavine, gentamicin, p-aminosalicylic acid (PAS), streptomycin, spectinomycin, tetracycline, triclosan, and vancomycin (Table 5). The degree of drug sensitivity probably correlates with the specificity of Tap for a particular substrate. In this regard, PAS (a second-line oral anti-TB agent), spectinomycin, and tetracycline were identified as specific Tap substrates in M. bovis BCG since a ≥4-fold change in sensitivity was observed in mutant and overexpressing strains compared to the wild-type strain.
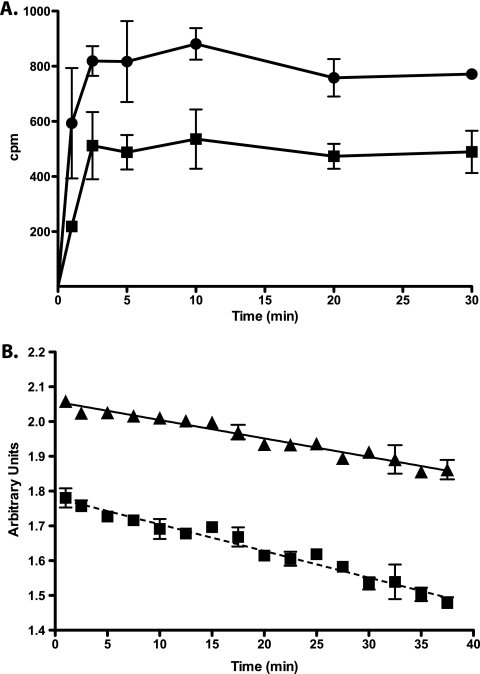
Two complementary approaches were used to confirm that this phenotype was due to an efflux mechanism. First, tetracycline susceptibility assays in the presence of CCCP were carried out using tap-overexpressing strains (Table 5); in M. bovis BCG, CCCP reduced Tap activity by inhibiting the electrochemical gradient across the membrane, similar to what was previously observed in M. smegmatis (51). Second, tetracycline and acriflavine accumulation experiments were performed (Fig. 5). [3H]tetracycline efflux experiments showed that KOTap cells accumulated approximately twice as much tetracycline as wild-type cells, achieving a steady-state level of accumulation within about 3 min (Fig. 5A). Similarly, acriflavine accumulation assays were performed with the M. bovis PAZ11 strain (tapTB overexpression). When acriflavine is accumulated inside the cells, its fluorescence emission is quenched upon binding to the DNA. The tapTB-overexpressing PAZ11 culture had higher levels of fluorescence (Fig. 5B) and a 4-fold increase in MIC to acriflavine (Table 5). Together, these data showed that TapTB provided tetracycline and acriflavine resistance in M. bovis BCG by active efflux.
Fig 5.
Tetracycline and acriflavine accumulation by intact cells of M. bovis Tap-derivative strains. (A) [3H]tetracycline uptake by wild-type (squares) and KOTap mutant (circles) cells. (B) Acriflavine uptake by SUM36 (squares) and the tapTB-overexpressing PAZ11 (triangles) cells. [3H]tetracycline and acriflavine were added to the cells at time zero. Acriflavine fluorescence emission is quenched upon binding to the DNA; thus, in contrast to [3H]tetracycline, a lower value reading indicates intracellular accumulation. The results are the average of three experiments, and error bars indicate standard deviations.
The contribution of Tap to intrinsic resistance to first-line anti-TB drugs ethambutol, rifampin, and isoniazid was first suggested by the fact that small increases in resistance were reproducibly observed in overexpressing strains (Table 5). To validate these results, 10-μl portions of 10-fold serial dilutions of the culture (original inoculum, 107 cell/ml) were inoculated on agar plates at a fixed subinhibitory concentration of isoniazid (0.05 mg/liter). Under these conditions, the wild-type strain grew at a 10−2 dilution; the overexpressing strain (tapTB or tapFR) grew at a 10−4 dilution (data not shown), documenting its ability to better adapt to or tolerate isoniazid.
The levels of drug resistance conferred by efflux pumps are generally low compared to what is seen for mutations in drug target genes and, unlike other bacteria (46), no clinically relevant efflux pump mutations have yet been identified in M. tuberculosis. While some authors have reported increases in tapTB expression after antibiotic treatment in clinical isolates (20, 55), a direct correlation between tapTB levels of expression and its contribution to antibiotic resistance has not been demonstrated in these clinical isolates. In fact, increased expression of an efflux pump gene upon antibiotic exposure does not necessarily mean that the antibiotic is a substrate of the efflux pump. Antibiotics might have deleterious cellular effects beyond their molecular targets (22) and thus generate other metabolic signals. It is possible that the induction of efflux pump genes might be a response to counteract antibiotic downstream effects, including oxidative stress (16). In addition, increased expression of efflux pumps may permit bacteria to survive otherwise lethal concentrations of substrate antibiotics and thus allow selection for target mutations that increase drug resistance levels. This phenomenon is known as “drug target resistance masking” (14). Together, this underlines the role of efflux pumps in the development of high levels of multidrug resistance.
Importantly, in an era in which tuberculosis (TB) therapeutic options are limited and multidrug-resistant (MDR) and extensively drug-resistant (XDR) TB strains are on the rise (23), a Tap inhibitor could provide new strategies for TB therapy allowing shortened treatment, perhaps by targeting the drug-tolerant subpopulation of cells (1). One interesting approach to accelerate this process would be to identify drugs clinically approved for other therapeutic applications as Tap inhibitors, such as verapamil (1). This approach, known as “repurposing” (3), would allow the timely introduction of new TB therapies. A recent report validated this concept in vivo. Louw et al. (28) infected mice with an MDR-TB strain and showed improved efficacy of rifampin when administered in combination with verapamil. However, the use of verapamil for TB therapy generates concerns about the pharmacological effects on cardiac conduction and potential extrapyramidal disorders associated with calcium channel blockers (59).
Perspectives.
In this study, we have characterized the important physiological role of Tap in stationary phase (probably through its contribution to the maintenance of the cell wall structure) and to intrinsic drug resistance. Our results build on the study by Adams et al. (1), suggesting the importance of Tap in drug tolerance. This subpopulation of M. tuberculosis cells tolerant to anti-TB drugs is one of the causes of prolonged TB therapy. Therefore, the development of Tap inhibitors could provide a valuable new weapon to combat TB. Such inhibitors would have important therapeutic applications. First, they would allow the introduction of already available although clinically non-TB-effective antibiotics such as spectinomycin or tetracycline and make second-line anti-TB drugs such as PAS and streptomycin more effective. Second, assuming that Tap has a primary role in the development of antibiotic tolerance, such an inhibitor would potentially reduce the duration of the current standard treatment. However, further research needs to be performed to validate these hypotheses, which could provide new avenues for TB therapy.
Supplementary Material
ACKNOWLEDGMENTS
We are grateful to Roberta Provvedi for helpful advice with the microarray experiment. We also thank Dmitry Apel, Jesús Gonzalo, and Ainhoa Arbués for microscopy technical support and Erin Gaynor for use of the fluorescence microscope.
This work was supported by Spanish Ministry of Science and Innovation grant BIO2009-09405 (J.A.A.), by FAR 2010 of the University of Pavia (E.D.R.), European Community's Seventh Framework Programme (FP7/2007–2013) under grant agreement N. 260872 (R.M.), and The Canadian Institute of Health Research to C.J.T. (MOP-82745 and MOP-82855). This work was also supported by fellowships from the Spanish Ministry of Science and Education (AP2001-1114), the European Molecular Biology Organization (ASTF 74.00-06), and the Programa Europa XXI de Estancias de Investigación from the Government of Aragon (Spain) and the Caja de Ahorros de la Immaculada (S.R.-G.).
Footnotes
Published ahead of print 9 January 2012
Supplemental material for this article may be found at http://aac.asm.org/.
REFERENCES
- 1. Adams KN, et al. 2011. Drug tolerance in replicating mycobacteria mediated by a macrophage-induced efflux mechanism. Cell 145:39–53 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Ainsa JA, et al. 1998. Molecular cloning and characterization of Tap, a putative multidrug efflux pump present in Mycobacterium fortuitum and Mycobacterium tuberculosis. J. Bacteriol. 180:5836–5843 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Boguski MS, Mandl KD, Sukhatme VP. 2009. Drug discovery. Repurposing with a difference. Science 324:1394–1395 [DOI] [PubMed] [Google Scholar]
- 4. Brosch R, et al. 2000. Comparative genomics uncovers large tandem chromosomal duplications in Mycobacterium bovis BCG Pasteur. Yeast 17:111–123 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Camacho LR, et al. 2001. Analysis of the phthiocerol dimycocerosate locus of Mycobacterium tuberculosis. Evidence that this lipid is involved in the cell wall permeability barrier. J. Biol. Chem. 276:19845–19854 [DOI] [PubMed] [Google Scholar]
- 6. Camacho LR, Ensergueix D, Perez E, Gicquel B, Guilhot C. 1999. Identification of a virulence gene cluster of Mycobacterium tuberculosis by signature-tagged transposon mutagenesis. Mol. Microbiol. 34:257–267 [DOI] [PubMed] [Google Scholar]
- 7. Crick DC, Mahapatra S, Brennan PJ. 2001. Biosynthesis of the arabinogalactan-peptidoglycan complex of Mycobacterium tuberculosis. Glycobiology 11:107R–118R [DOI] [PubMed] [Google Scholar]
- 8. De Rossi E, Ainsa JA, Riccardi G. 2006. Role of mycobacterial efflux transporters in drug resistance: an unresolved question. FEMS Microbiol. Rev. 30:36–52 [DOI] [PubMed] [Google Scholar]
- 9. De Rossi E, et al. 2002. The multidrug transporters belonging to major facilitator superfamily in Mycobacterium tuberculosis. Mol. Med. 8:714–724 [PMC free article] [PubMed] [Google Scholar]
- 10. De Rossi E, et al. 1998. Molecular cloning and functional analysis of a novel tetracycline resistance determinant, tet(V), from Mycobacterium smegmatis. Antimicrob. Agents Chemother. 42:1931–1937 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Dhamdhere G, Zgurskaya HI. 2010. Metabolic shutdown in Escherichia coli cells lacking the outer membrane channel TolC. Mol. Microbiol. 77:743–754 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Dziadek J, Madiraju MV, Rutherford SA, Atkinson MA, Rajagopalan M. 2002. Physiological consequences associated with overproduction of Mycobacterium tuberculosis FtsZ in mycobacterial hosts. Microbiology 148:961–971 [DOI] [PubMed] [Google Scholar]
- 13. Escolar L, Perez-Martin J, de Lorenzo V. 1999. Opening the iron box: transcriptional metalloregulation by the Fur protein. J. Bacteriol. 181:6223–6229 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Fange D, Nilsson K, Tenson T, Ehrenberg M. 2009. Drug efflux pump deficiency and drug target resistance masking in growing bacteria. Proc. Natl. Acad. Sci. U. S. A. 106:8215–8220 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Farrow MF, Rubin EJ. 2008. Function of a mycobacterial major facilitator superfamily pump requires a membrane-associated lipoprotein. J. Bacteriol. 190:1783–1791 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Fraud S, Poole K. 2011. Oxidative stress induction of the MexXY multidrug efflux genes and promotion of aminoglycoside resistance development in Pseudomonas aeruginosa. Antimicrob. Agents Chemother. 55:1068–1074 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Geiman DE, Raghunand TR, Agarwal N, Bishai WR. 2006. Differential gene expression in response to exposure to antimycobacterial agents and other stress conditions among seven Mycobacterium tuberculosis whiB-like genes. Antimicrob. Agents Chemother. 50:2836–2841 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Gey Van Pittius NC, et al. 2001. The ESAT-6 gene cluster of Mycobacterium tuberculosis and other high G+C Gram-positive bacteria. Genome Biol. 2:RESEARCH0044 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Heym B, Zhang Y, Poulet S, Young D, Cole ST. 1993. Characterization of the katG gene encoding a catalase-peroxidase required for the isoniazid susceptibility of Mycobacterium tuberculosis. J. Bacteriol. 175:4255–4259 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Jiang X, et al. 2008. Assessment of efflux pump gene expression in a clinical isolate Mycobacterium tuberculosis by real-time reverse transcription PCR. Microb. Drug Resist. 14:7–11 [DOI] [PubMed] [Google Scholar]
- 21. Kadner RJ, Murphy GP, Stephens CM. 1992. Two mechanisms for growth inhibition by elevated transport of sugar phosphates in Escherichia coli. J. Gen. Microbiol. 138:2007–2014 [DOI] [PubMed] [Google Scholar]
- 22. Kohanski MA, Dwyer DJ, Hayete B, Lawrence CA, Collins JJ. 2007. A common mechanism of cellular death induced by bactericidal antibiotics. Cell 130:797–810 [DOI] [PubMed] [Google Scholar]
- 23. Koul A, Arnoult E, Lounis N, Guillemont J, Andries K. 2011. The challenge of new drug discovery for tuberculosis. Nature 469:483–490 [DOI] [PubMed] [Google Scholar]
- 24. Krulwich TA, Lewinson O, Padan E, Bibi E. 2005. Do physiological roles foster persistence of drug/multidrug-efflux transporters? A case study. Nat. Rev. Microbiol. 3:566–572 [DOI] [PubMed] [Google Scholar]
- 25. Lau SY, Zgurskaya HI. 2005. Cell division defects in Escherichia coli deficient in the multidrug efflux transporter AcrEF-TolC. J. Bacteriol. 187:7815–7825 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Li XZ, Nikaido H. 2009. Efflux-mediated drug resistance in bacteria: an update. Drugs 69:1555–1623 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Lomovskaya O, et al. 2001. Identification and characterization of inhibitors of multidrug resistance efflux pumps in Pseudomonas aeruginosa: novel agents for combination therapy. Antimicrob. Agents Chemother. 45:105–116 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Louw GE, et al. 2011. Rifampicin reduces susceptibility to ofloxacin in rifampicin-resistant Mycobacterium tuberculosis through efflux. Am. J. Respir. Crit. Care Med. 184:269–276 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Louw GE, et al. 2009. A balancing act: efflux/influx in mycobacterial drug resistance. Antimicrob. Agents Chemother. 53:3181–3189 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Maciag A, et al. 2007. Global analysis of the Mycobacterium tuberculosis Zur (FurB) regulon. J. Bacteriol. 189:730–740 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Manganelli R, Dubnau E, Tyagi S, Kramer FR, Smith I. 1999. Differential expression of 10 sigma factor genes in Mycobacterium tuberculosis. Mol. Microbiol. 31:715–724 [DOI] [PubMed] [Google Scholar]
- 32. Manganelli R, et al. 2004. The extra cytoplasmic function sigma factor sigma(E) is essential for Mycobacterium tuberculosis virulence in mice. Infect. Immun. 72:3038–3041 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Manganelli R, Provvedi R. 2010. An integrated regulatory network including two positive feedback loops to modulate the activity of sigma(E) in mycobacteria. Mol. Microbiol. 75:538–542 [DOI] [PubMed] [Google Scholar]
- 34. Manganelli R, Voskuil MI, Schoolnik GK, Smith I. 2001. The Mycobacterium tuberculosis ECF sigma factor sigma(E): role in global gene expression and survival in macrophages. Mol. Microbiol. 41:423–437 [DOI] [PubMed] [Google Scholar]
- 35. McMurry L, Petrucci RE, Jr, Levy SB. 1980. Active efflux of tetracycline encoded by four genetically different tetracycline resistance determinants in Escherichia coli. Proc. Natl. Acad. Sci. U. S. A. 77:3974–3977 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Milano A, et al. 2009. Azole resistance in Mycobacterium tuberculosis is mediated by the MmpS5-MmpL5 efflux system. Tuberculosis (Edinb.) 89:84–90 [DOI] [PubMed] [Google Scholar]
- 37. Morris RP, et al. 2005. Ancestral antibiotic resistance in Mycobacterium tuberculosis. Proc. Natl. Acad. Sci. U. S. A. 102:12200–12205 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Muttucumaru DG, Roberts G, Hinds J, Stabler RA, Parish T. 2004. Gene expression profile of Mycobacterium tuberculosis in a non-replicating state. Tuberculosis (Edinb.) 84:239–246 [DOI] [PubMed] [Google Scholar]
- 39. Narberhaus F. 1999. Negative regulation of bacterial heat shock genes. Mol. Microbiol. 31:1–8 [DOI] [PubMed] [Google Scholar]
- 40. Newton GL, Ta P, Bzymek KP, Fahey RC. 2006. Biochemistry of the initial steps of mycothiol biosynthesis. J. Biol. Chem. 281:33910–33920 [DOI] [PubMed] [Google Scholar]
- 41. Nikaido H. 2009. Multidrug resistance in bacteria. Annu. Rev. Biochem. 78:119–146 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Nishino K, Nikaido E, Yamaguchi A. 2009. Regulation and physiological function of multidrug efflux pumps in Escherichia coli and Salmonella. Biochim. Biophys. Acta 1794:834–843 [DOI] [PubMed] [Google Scholar]
- 43. Parish T, Stoker NG. 1998. Mycobacterial protocols. In Parish T, Stoker NG. (ed), Methods in molecular biology, vol 101 Humana Press, Totowa, NJ [Google Scholar]
- 44. Parish T, Stoker NG. 2000. Use of a flexible cassette method to generate a double unmarked Mycobacterium tuberculosis tlyA plcABC mutant by gene replacement. Microbiology 146:1969–1975 [DOI] [PubMed] [Google Scholar]
- 45. Pavelka MS, Jr, Jacobs WR., Jr 1996. Biosynthesis of diaminopimelate, the precursor of lysine and a component of peptidoglycan, is an essential function of Mycobacterium smegmatis. J. Bacteriol. 178:6496–6507 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Piddock LJ. 2006. Clinically relevant chromosomally encoded multidrug resistance efflux pumps in bacteria. Clin. Microbiol. Rev. 19:382–402 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Piddock LJ. 2006. Multidrug-resistance efflux pumps—not just for resistance. Nat. Rev. Microbiol. 4:629–636 [DOI] [PubMed] [Google Scholar]
- 48. Plumbridge J. 2009. An alternative route for recycling of N-acetylglucosamine from peptidoglycan involves the N-acetylglucosamine phosphotransferase system in Escherichia coli. J. Bacteriol. 191:5641–5647 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Prentki P, Krisch HM. 1984. In vitro insertional mutagenesis with a selectable DNA fragment. Gene 29:303–313 [DOI] [PubMed] [Google Scholar]
- 50. Raman S, et al. 2001. The alternative sigma factor SigH regulates major components of oxidative and heat stress responses in Mycobacterium tuberculosis. J. Bacteriol. 183:6119–6125 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Ramon-Garcia S, Martin C, Ainsa JA, De Rossi E. 2006. Characterization of tetracycline resistance mediated by the efflux pump Tap from Mycobacterium fortuitum. J. Antimicrob. Chemother. 57:252–259 [DOI] [PubMed] [Google Scholar]
- 52. Ramon-Garcia S, Martin C, De Rossi E, Ainsa JA. 2007. Contribution of the Rv2333c efflux pump (the Stp protein) from Mycobacterium tuberculosis to intrinsic antibiotic resistance in Mycobacterium bovis BCG. J. Antimicrob. Chemother. 59:544–547 [DOI] [PubMed] [Google Scholar]
- 53. Ramon-Garcia S, Martin C, Thompson CJ, Ainsa JA. 2009. Role of the Mycobacterium tuberculosis P55 efflux pump in intrinsic drug resistance, oxidative stress responses, and growth. Antimicrob. Agents Chemother. 53:3675–3682 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Sambrook J, Russell DW. 2001. Molecular cloning: a laboratory manual, 3rd ed Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY [Google Scholar]
- 55. Siddiqi N, et al. 2004. Mycobacterium tuberculosis isolate with a distinct genomic identity overexpresses a tap-like efflux pump. Infection 32:109–111 [DOI] [PubMed] [Google Scholar]
- 56. Soliveri JA, Gomez J, Bishai WR, Chater KF. 2000. Multiple paralogous genes related to the Streptomyces coelicolor developmental regulatory gene whiB are present in Streptomyces and other actinomycetes. Microbiology 146:333–343 [DOI] [PubMed] [Google Scholar]
- 57. Stewart GR, et al. 2005. The stress-responsive chaperone alpha-crystallin 2 is required for pathogenesis of Mycobacterium tuberculosis. Mol. Microbiol. 55:1127–1137 [DOI] [PubMed] [Google Scholar]
- 58. Stewart GR, et al. 2002. Dissection of the heat-shock response in Mycobacterium tuberculosis using mutants and microarrays. Microbiology 148:3129–3138 [DOI] [PubMed] [Google Scholar]
- 59. Sweetman SC, ed. 2011. Martindale: the complete drug reference. Pharmaceutical Press, London, United Kingdom [Google Scholar]
- 60. Tusher VG, Tibshirani R, Chu G. 2001. Significance analysis of microarrays applied to the ionizing radiation response. Proc. Natl. Acad. Sci. U. S. A. 98:5116–5121 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61. van Heijenoort J. 2001. Formation of the glycan chains in the synthesis of bacterial peptidoglycan. Glycobiology 11:25R–36R [DOI] [PubMed] [Google Scholar]
- 62. Vaquerizas JM, et al. 2005. GEPAS, an experiment-oriented pipeline for the analysis of microarray gene expression data. Nucleic Acids Res. 33:W616–W620 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63. Voskuil MI, Visconti KC, Schoolnik GK. 2004. Mycobacterium tuberculosis gene expression during adaptation to stationary phase and low-oxygen dormancy. Tuberculosis (Edinb.) 84:218–227 [DOI] [PubMed] [Google Scholar]
- 64. Wilkinson KA, et al. 2005. Infection biology of a novel alpha-crystallin of Mycobacterium tuberculosis: Acr2. J. Immunol. 174:4237–4243 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.