Abstract
We investigated the outcome of the interaction of Cryptococcus neoformans with murine macrophages using laser scanning cytometry (LSC). Previous results in our lab had shown that phagocytosis of C. neoformans promoted cell cycle progression. LSC allowed us to simultaneously measure the phagocytic index, macrophage DNA content, and 5-ethynyl-2′-deoxyuridine (EdU) incorporation such that it was possible to study host cell division as a function of phagocytosis. LSC proved to be a robust, reliable, and high-throughput method for quantifying phagocytosis. Phagocytosis of C. neoformans promoted cell cycle progression, but infected macrophages were significantly less likely to complete mitosis. Hence, we report a new cytotoxic effect associated with intracellular C. neoformans residence that manifested itself in impaired cell cycle completion as a consequence of a block in the G2/M stage of the mitotic cell cycle. Cell cycle arrest was not due to increased cell membrane permeability or DNA damage. We investigated alveolar macrophage replication in vivo and demonstrated that these cells are capable of low levels of cell division in the presence or absence of C. neoformans infection. In summary, we simultaneously studied phagocytosis, the cell cycle state of the host cell and pathogen-mediated cytotoxicity, and our results demonstrate a new cytotoxic effect of C. neoformans infection on murine macrophages: fungus-induced cell cycle arrest. Finally, we provide evidence for alveolar macrophage proliferation in vivo.
INTRODUCTION
The interaction of the human pathogenic fungus Cryptococcus neoformans with macrophages is thought to be a key event in the outcome of cryptococcal infection (10, 13, 17, 28, 29). C. neoformans is a facultative intracellular pathogen and, once within a macrophage, C. neoformans can replicate intracellularly with outcomes that range from host cell lysis to nonlytic exocytosis (2, 3, 23). Previous work in our laboratory has established that phagocytosis of C. neoformans by murine macrophages could lead macrophages into cell cycle progression, namely, into the S phase of the cell cycle (21). Later work established that Fcγ Receptor (FcγR) cross-linking triggered cell cycle progression resulting in increased proliferation of murine macrophages (20). However, while FcγR cross-linking (20) or ingestion of antibody-coated beads led to cyclin D1 activation, phagocytosis of live yeasts suppressed cyclin D1 activation (18), possibly reflecting fungal-mediated host cell damage. Cyclin D1 is a major checkpoint in the passage from G1 to S phase. Hence fungus-macrophage interaction could influence host cell cycle machinery. Consequently, there is considerable interest in the relationship between macrophage cell cycle and phagocytic function.
Macrophages are derived from monocytes that migrate into tissues, where they acquire tissue-specific characteristics and can live as resident tissue cells for years (25, 26). Evidence for resident macrophage proliferation has been available for some time (30), but this phenomenon plays an unknown role in the maintenance of tissue specific macrophages. In the lung, specifically, the site of the initial infection in human cryptococcosis, there is evidence for in vivo alveolar macrophages (AM) proliferation (4, 31, 32). AM recovered from mice exposed to cigarette smoke manifested increased proliferation in vitro, suggesting that cell division could be increased in response to a damaging stimuli (12). Nevertheless, many questions still remain as to the relative contribution of local macrophage proliferation versus influx of blood monocytes in response to infection. In recent months, several studies have appeared in the literature investigating this phenomenon and how macrophage life, replication and death are balanced at the onset and resolution of tissue insult and damage (1, 9, 14, 15). These studies suggest that macrophage proliferation contributes to normal tissue homeostasis and that macrophages can replicate at the site of inflammation. In the present study we show that laser scanning cytometry (LSC) can be adapted to study phagocytosis and have used this technique to explore how phagocytosis of yeast cells influenced macrophage cell cycle progression and mitosis. LSC was developed to scan, analyze, and compare images from microscopic preparations in a fully automated form. It enables users to perform analysis similar to flow cytometry using various quantitative parameters extracted from the scanned images (11). The ability of LSC to perform microscopic correlations of cellular and subcellular events in a large population of cells makes it a highly attractive technique for studying intracellular processes. LSC is routinely used in several applications, such as microarray analysis, tissue section analysis, and immunophenotyping, and in cell cycle or mitosis analysis (16, 42). It is generally accepted that the results of LSC are equivalent to flow cytometry and that for certain assays it can be more sensitive than flow cytometry, such as detecting cells in transition between mitotic phases or in very early apoptosis (33, 42). Hence, LSC can be a powerful tool for studying pathogen interactions with macrophages.
(The data in the present study are from a thesis to be submitted by Carolina Coelho in partial fulfillment of the requirements for a Ph.D. degree from the Faculty of Medicine, University of Coimbra, Coimbra, Portugal.)
MATERIALS AND METHODS
Yeast strains, cell lines, and reagents.
C. neoformans var. grubii strain H99 (serotype A) was obtained from John Perfect (Durham, NC), and C. neoformans var. neoformans strain 24067 (serotype D) was obtained from the American Type Tissue Collection (Rockville, MD). Strain H99 was used for all in vitro studies. Both strains were cultured in Sabouraud dextrose broth (Difco, Carlsbad, CA) for 2 days at 37°C with agitation (150 to 180 rpm). Yeast cells were washed three times with sterile phosphate-buffered saline (PBS), counted on a hemocytometer, and suspended at the appropriate cell density in cell culture media.
The IgG1 monoclonal antibody (MAb) 18B7 was described previously (5) and was used as an opsonin at 10 μg/ml, unless otherwise noted. The macrophage-like murine cell line J774.16, which was originally derived from a reticulum sarcoma, was used for most experiments. Macrophages were grown in media consisting of Dulbecco minimal essential medium (DMEM; CellGro; Mediatech, Manassas, VA), 10% NCTC-109 Gibco medium (Invitrogen, Carlsbad, CA), 10% heat-inactivated fetal calf serum (FCS; Atlanta Biologicals, Lawrenceville, GA), and 1% nonessential amino acids (CellGro).
Bone marrow-derived macrophages (BMDM) were obtained from wild-type 6- to 8-week-old BALB/c female mice (National Cancer Institute, Bethesda, MD). Briefly, mice were killed by CO2 asphyxiation, and bone marrow cells were harvested from the hind leg bones by flushing them with DMEM. The harvested cells were cultured at 37°C with 5% CO2 in DMEM with 20% L-929 cell conditioned medium, 10% fetal bovine serum (FBS), 2 mM l-glutamine (CellGro), 1% nonessential amino acids (CellGro), 1% HEPES buffer (CellGro), and β-mercaptoethanol (Gibco, Carlsbad, CA). Macrophages were allowed to grow for 6 to 8 days before plating at the desired density for the experiments.
All animal experiments were conducted according to ethical guidelines, with the approval of the Institutional Animal Care and Use Committee of Albert Einstein College of Medicine.
In vitro phagocytosis assays.
For LSC experiments, J774.16 and BMDM cells were seeded in a 96-well glass bottom plate with dark edges (MGB096-1-2-HG-L; Matrical Biosciences, Spokane, WA). The macrophages were plated at a density of 2.4 × 104 cells/well and used for phagocytosis studies after adhering to the microtiter plate for either 2 h or overnight. Yeasts were added at a multiplicity of infection (MOI) of 1:2 and the opsonic MAb was added at 10 μg/ml in a final volume of 200 μl per well. When necessary, C. neoformans was heat killed (HK C. neoformans) by incubation at 56°C for 60 min. Phagocytosis was allowed to proceed for 2 h at 37°C under a 5% CO2 atmosphere.
After phagocytosis, the wells were washed twice with 200 μl of PBS, fixed in ice-cold methanol for 30 min at −20°C, and washed again. Wheat germ agglutinin (WGA) conjugated to Alexa 633 (Invitrogen) was used at 10 μg/ml and incubated overnight at 4°C. We found no instances of C. neoformans staining with WGA in our experimental conditions. Uvitex 2B (Polysciences, Inc., Warrington, PA) was added at a 0.1 μg/ml and allowed to stain for 1 min. Propidium iodide (PI; Sigma-Aldrich, St. Louis, MO) was added at a concentration of 5 μg/ml in a total volume of 400 μl per well. The cells were analyzed in PI solution.
Alternative protocols were designed according to experimental conveniences. When using antibodies for detection, preparations were blocked for 30 min at room temperature with 2% FCS in PBS. In one protocol, detection of yeasts was done by detecting the opsonizing antibody bound to the capsule with an Alexa 488-conjugated goat antibody to murine IgG (Invitrogen) at a 1:50 dilution. The macrophage contour could also be successfully detected using Alexa 633-conjugated antibody to mouse F4/80 (Invitrogen) at a dilution of 1:25.
Confirmation of successful internalization can be achieved by immunostaining extracellular C. neoformans previous to permeabilization, followed by Uvitex staining of the total C. neoformans. Staining of C. neoformans nuclei with PI occurred but, given the smaller size of C. neoformans nuclei and the dimmer fluorescent signal, it was not enough to interfere with DNA quantification. Irrespective of this, we established an independent nucleus contour, such that the staining of the C. neoformans nuclei is automatically excluded from the macrophage nuclei contour, to assure that it did not interfere with our determination of macrophage nuclei and DNA quantification. In addition, cell cycle plots were prepared with the nucleus contour that corresponded only to the macrophage nuclei.
Images were acquired in LSC by imaging 35 to 60 fields in each well with the ×40 objective lens at a 0.5-μm resolution, allowing a field size of 500 μm by 192 μm. For phagocytosis assays, at least 1,000 cells were imaged for each experimental replicate.
After the acquisition of fluorescent images in LSC, the preparations were stained by incubation with Giemsa dye solution for 2 h at room temperature and then washed with PBS to remove the excess stain. The phagocytic index was quantified by direct observation using an inverted microscope for a total of three fields per well, with at least 100 cells/field. Macrophages with internalized C. neoformans were readily distinguishable from cells that had not ingested C. neoformans or from cells where C. neoformans was simply attached to the outside, due to the visible vacuole containing engulfed C. neoformans.
In vitro replication studies.
A protocol to study replication in vitro was adapted from the work of Darzynkiewicz et al. (8). Briefly, J774.16 macrophages were plated at 2.4 × 104 cells/well and allowed to adhere for 2 h. In all conditions, a 10 μM solution of 5-ethynyl-2′-deoxyuridine (EdU) in cell medium was added for 2 h, unless otherwise noted, and then removed by washing with warm media. For prelabeling studies, the cells were labeled with EdU and then allowed to ingest C. neoformans for 15 min at an MOI of 1:5; afterward, the extracellular C. neoformans was removed by two washes with warm media. For postlabeling studies, C. neoformans phagocytosis was performed in the same manner, but EdU was added after the C. neoformans was removed. After 2 h, the cell medium was replaced with fresh medium, and the cells were allowed to cycle for an additional 2 h. Coincubation studies consisted of phagocytosis of C. neoformans at an MOI of 1:2 in the presence of EdU for 2 h. The cells were fixed in methanol, and nonspecific signal was blocked with 2% FBS in PBS. EdU labeling was performed according to the manufacturer's instructions. Briefly, this labeling strategy coupled the thymidine analogue EdU with an azide-conjugated Alexa 488 for detection, via the Click-it Chemistry reaction (Invitrogen). Macrophages were counterstained with WGA-Alexa 633 and PI, and yeast were detected with Uvitex. LSC images were obtained with a ×40 objective lens at a 0.5-μm resolution, allowing a field size of 500 μm by 192 μm. For cell cycle analysis, 49 to 100 fields were imaged to information from at least 2,000 cells in each experimental replicate.
Experiments with BMDM were performed alike, except that the time for EdU incubation was extended to 6 h due to the slower replication rate of these cells relative to J774.16 cells. When indicated, BMDM were incubated with gamma interferon (IFN-γ) at 400 U/ml or lipopolysaccharide (LPS) at 1 ng/ml for 18 h before phagocytosis, and the stimuli were renovated after each medium change. Taxol (Tocris Bioscience, Ellisville, MO) was added at 250 nM for 18 h.
DNA fragmentation and permeability assays.
Phagocytic experiments were carried out as described above for 2, 6, and 18 h. Valinomycin (Tocris Bioscience) was added at a concentration of 400 nM for 18 h to induce apoptosis through DNA damage, serving as a positive control for both experiments. Cell membrane permeability was measured by adding 100 nM Image-iT DEAD Green (Invitrogen) to cells 30 min before fixation with 4% paraformaldehyde. Cells were counterstained with Alexa 633-conjugated WGA and DAPI (4′,6′-diamidino-2-phenylindole). The experiment was performed twice for J774.16 cells and once for BMDM in triplicate wells.
TUNEL (terminal deoxynucleotidyltransferase-mediated dUTP-biotin nick end labeling) staining (Invitrogen) was performed according to the manufacturer's instructions. Briefly, the wells were fixed with 4% paraformaldehyde, and 100 μl of TdT reaction cocktail was added for 1 h at 37°C; the samples were then washed and developed by incubation with a Click-iT reaction cocktail that couples TdT to Alexa 647, allowing fluorescence detection. The cells were counterstained with PI, and yeast cells were labeled with Uvitex. This experiment was performed twice for J774.16 cells and once for BMDM with a single experimental replicate. LSC images were obtained with a ×40 objective lens at a 0.5-μm resolution, allowing a field size of 500 μm by 192 μm. At least 1,000 cells were imaged for each experimental replicate.
In vivo replication studies.
AM labeling was performed as described previously (17, 19). Briefly, 100 μl of 40 mM PKH26 (PKH) was dissolved into 300 μl of diluent B (Sigma-Aldrich) in order to label phagocytic cells and injected into the tail vein of BALB/c mice. After 2 days, mice were infected with the virulent strain 24067. In vivo studies were performed with strain 24067 because this strain is better characterized in our lab in the context of the mouse model of intratracheal infection (41). Mice were anesthetized with ketamine (10 mg/kg) and xylazine (125 mg/kg) intraperitoneally (i.p.) in PBS, a midline incision over the trachea was performed, and an inoculum of 106 C. neoformans in 50 to 100 μl was injected into the trachea. The incision was closed with VetBOND (3M, St. Paul, MN). At 4 days postinfection, mice were injected i.p. with 100 μg of EdU in PBS–20% dimethyl sulfoxide or vehicle alone, as described previously (24). Mice were sacrificed 6 h after EdU administration by CO2 asphyxiation, and bronchoalveolar lavage (BAL) was performed to extract the AM. Briefly, the trachea was exposed, and the lungs were lavaged 10 times with 0.8 ml of sterile calcium and magnesium-free HBSS (Life Technologies, Grand Island, NY) supplemented with 1 mM EGTA (Sigma-Aldrich). The BAL was spun, resuspended in cell culture medium, and allowed to adhere for 2 h at 37°C in 5% CO2. The cells were treated with a hypotonic solution of NH4Cl for 10 min on ice to remove red blood cells and subsequently fixed for 30 min at −20°C. The preparations were then blocked with 2% FCS in PBS, and EdU staining was developed according to the manufacturer's instructions (Invitrogen). Nuclei were counterstained with DAPI. Images were acquired in LSC studies by imaging 156 fields/well with a ×60 objective lens that allowed 0.15-μm resolution. Additional confirmation was performed by inspecting the entire area of the negative control wells in an inverted epifluorescence microscope at ×20 magnification. No positive events were ever found in the negative control samples.
Data collection and analysis.
An iCys research imaging cytometer (CompuCyte Corp., Westwood, MA) was used for these experiments. Instrument control, data acquisition, and analysis were performed using the iCys cytometric analysis software provided with the instrument. The instrument was calibrated using an empty 96-well plate to allow use of the autofocus feature.
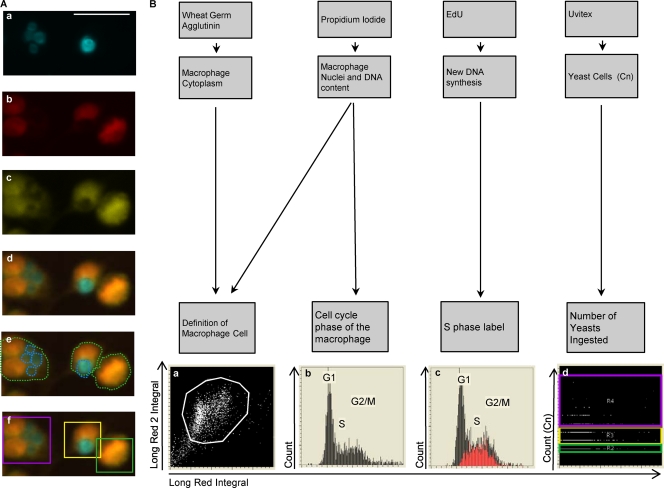
For image analysis, three different type of events were defined: the “Cell” event was the combined image of the nuclear PI signal plus the cell membrane WGA, the nuclear signal generated the “Nucleus” event, and the “Uvitex” event was used for the Uvitex-stained yeasts (Fig. 1).
Fig 1.
Quantification of phagocytosis and cell cycle phase in macrophages by LSC. (A) Phagocytic quantification. Fluorescence images of Uvitex-stained C. neoformans (Cn) (a), PI-stained nuclei (b), WGA-stained cytoplasm (c), and merged Uvitex, PI, and WGA (d) are shown. (e and f) Contours encircling fluorescent areas (light green line) define the macrophage area, and contours encircling Uvitex signal define the C. neoformans area (cyan line) (e); the C. neoformans yeast (C. neoformans) subcontours inside each macrophage are also quantified (f). Categories were created and translated into color-coded boxes—no C. neoformans, green; 1 to 2 C. neoformans, yellow; and >3 C. neoformans, magenta—allowing verification of the software identification. Scale bar, 20 μm. (B) Cell cycle status and correlation with phagocytosis. The association of different fluorescent markers allowed the identification of macrophages and study of intracellular events within each macrophage. Macrophages were classified according to the number of yeasts that they had ingested and then subclassified according to their cell cycle phases. (a) Cytoplasmic signal and nuclear signals were merged to delineate the macrophage and define the macrophage population (white box). (b) PI staining quantifies the amount of DNA, producing a histogram with three regions that reflected the cell cycle stage, i.e., the G1, S, and G2/M phase. (c) EdU labeling (Red) identifies cells actively synthesizing DNA (S phase). (d) The number of C. neoformans events inside each macrophage was plotted and divided into three categories: no C. neoformans, green; 1 to 2 C. neoformans, yellow; and >3 C. neoformans, magenta. The experiment was performed with primary macrophages infected with C. neoformans strain H99 at an MOI of 1:2.
The creation of these channels was the first step in analyzing cell fluorescence so cells could be recognized and contoured by the software and translated into events. The “Cell” channel contoured macrophages using a merged PI and WGA image. Clusters of positive pixels were delimited, creating the threshold contour, which allowed the software to identify a single macrophage cell as one “event.” Association of separate events such as “Cell,” “Nucleus,” or “Yeast” was the last step, allowing quantification of the number of “Yeast” events localized within the contour of the “Cell” event, i.e., the number of phagocytosed particles inside each macrophage.
Events are represented in a scatter plot or histograms, where different regions could be defined. The statistics of the number of events occurring in each of the regions of interest were obtained and used to calculate the phagocytic index and fungal burden.
Statistical analysis and plotting.
Graphs and statistical analysis were performed in Prism, version 5.00, for Mac OS X (GraphPad Software, San Diego, CA).
RESULTS
Development of the LSC detection protocol.
The first objective of the present study was to establish whether LSC could be adapted to study the phagocytosis of C. neoformans by macrophages. The irregular shape of a macrophage cell was successfully detected by merging PI nuclear staining with WGA (Fig. 1). Yeast cells were successfully detected with either the fungal cell wall stain Uvitex or specific capsule-binding antibody staining. The method was first established in the macrophage-like immortalized cell line J774.16 and then adapted to primary cells, including BMDM and AM, and subsequently to in vivo phagocytosis after intratracheal instillation of C. neoformans (data not shown).
We ascertained LSC accuracy by inspecting the images created by the instrument (see Fig. S1 in the supplemental material). LSC correctly identified more than 90% of the macrophages. In addition to detecting number of phagocytic macrophages, LSC quantified the number of yeasts that each macrophage ingested with 85% accuracy and distinguished up to 10 yeasts inside each cell. LSC could analyze more than 1,000 cells within the time frame in which a human operator would only be able to analyze 100 to 300 cells, while greatly reducing operator time investment.
Cell cycle analysis was performed by two measurements: DNA content quantification after PI staining and new DNA synthesis through nucleoside analogue EdU incorporation, as recently published (4) and as illustrated in Fig. 1B. Hence, we concluded that LSC quantification was a fast, versatile, and reliable method for studying the cell cycle in phagocytic macrophages.
LSC measurements using different C. neoformans staining protocols.
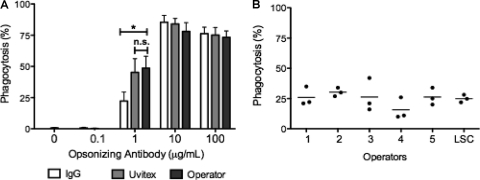
To ensure that LSC was compatible with a wide range of fluorescent tools, we established and compared two separate C. neoformans staining protocols. The cell wall-specific stain Uvitex is available in only one fluorescent color. Since a capsular antibody was used as an opsonin to mediate phagocytosis, we investigated whether the same antibody could also be used for C. neoformans detection. The advantage of antibody detection is that it would allow us to choose a wide range of commercially available fluorophores. Consequently, we compared immunostaining to Uvitex staining and compared both to human operator visual quantification after Giemsa staining (Fig. 2A) over a range of opsonizing MAb concentrations. Uvitex staining was effective independently of the concentration of opsonizing antibody used. In contrast, immunostaining was effective only when opsonizing MAb concentrations of ≥10 μg/ml were used. Concentrations of opsonizing antibody of <10 μg/ml promoted effective phagocytosis, but the phagocytic index determined by antibody staining was half the value detected by other stains, since this method is dependent on a certain amount of antibody bound to fungal cells. Concerns that adhered yeasts could be mistakenly measured as internalized yeasts were addressed by combining both C. neoformans detection techniques, where one of the stains was applied before methanol permeabilization and the other was applied after permeabilization. We determined that at 2 h the measurement of total yeasts (single staining) or differentiation of bound and internalized yeast (double staining) produced similar results (for example, both methods quantified 19% of nonphagocytic macrophages) (see Fig. S2 in the supplemental material), thus eliminating the need to use differential staining when performing phagocytic measurements for 2 h or longer. However, when shorter time intervals were used or when internalization of C. neoformans was blocked (incubation of cells at 4°C promotes binding without internalization), measurement of total yeasts had a higher phagocytic rate than when double staining was performed. Hence, only in some conditions was it necessary to perform double staining to differentiate adherent C. neoformans and avoid biasing phagocytic index.
Fig 2.
Comparison of LSC staining strategies with reference methods. LSC quantification correlated with human operator quantification of a Giemsa-stained preparation. (A) The BMDM phagocytic rate was detected by LSC by staining with a fluorescence-labeled antibody against the opsonizing antibody and simultaneously by Uvitex cell wall staining. The same preparations were subsequently stained with Giemsa stain and quantified by a human operator. The experiment was performed twice counting triplicate wells. Means ± the standard deviations (SD) of triplicate wells of a representative experiment are shown. Two-tailed t test with a 95% confidence interval (*, P < 0.01; n.s., not significant). (B) Human operator quantification was compared to LSC in J774.16 macrophages. Triplicate wells were analyzed by LSC and subsequently stained by Giemsa and quantified by five independent human operators. The operators were blind to the conditions of the assay. The pperators were instructed to choose 50% confluent fields but were left free to define their fields of analysis. Means ± the SD of triplicate wells are shown. Groups were compared using one-way analysis of variance. P = 0.04 with a 95% confidence interval.
We inquired whether LSC quantification correlated with microscopic counting performed by a human operator, which is currently the standard in the field. Furthermore, we investigated human-to-human variation in phagocytic index determinations given the same preparation to quantify. A comparison of the LSC count to that obtained by five human operators was done using in the same triplicate wells of one phagocytic assay (Fig. 2B). The phagocytic rate for triplicate wells obtained by LSC protocol (31%) was comparable to that measured by different human operators (from 15 to 30%), as was the standard deviation obtained. Curiously, there was no statistical difference between the counts between operators, as assessed by one-way analysis of variance (P = 0.4). These results show that LSC was adaptable to a wide variety of phagocytic indexes and experimental conditions and that operator bias should not be a concern in fungal phagocytic determinations.
Cell cycle correlation with ingestion of yeast cells.
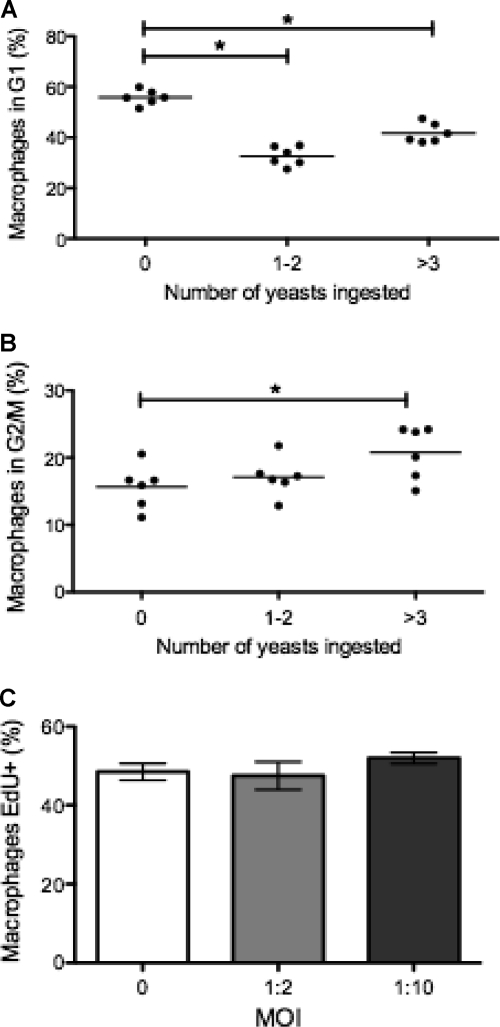
Previous work in our lab had established that phagocytosis of C. neoformans or inert latex beads drives macrophage cells into S phase (15). We investigated this result further by studying cell cycle progression as a function of intracellular fungal burden. In concordance with our prior study, ingestion of heat-killed (HK) or live C. neoformans cells was associated with a decrease in the proportion of G1-phase cells (56% in cells with no C. neoformans versus 33% in cells with 1 to 2 yeasts and 42% when cells contained ≥3 yeasts) and a concomitant increase in the proportion of cells in the S and G2/M phases for live C. neoformans (15% in cells with no C. neoformans versus 17% in 1 to 2 yeasts and 20% when ≥3 yeasts were ingested) (Fig. 3A and B). Note that cell cycle progression increased as fungal burden increased, suggesting that a higher fungal burden had a stronger effect in driving the macrophage into S phase.
Fig 3.
Association of cell cycle state in J774.16 macrophages with fungal burden. Macrophage progression to the G2/M phase of cell cycle correlated with fungal burden within the macrophage. Macrophages were grouped according to the number of yeasts ingested and their cell cycle phases. (A and B) Percentage of macrophages in the G1 phase (A) and G2/M phase (B). Experiments were performed three times, with six replicate wells. The data points and averages are shown. (C) Percentage of EdU+ macrophages during phagocytosis. Macrophages were allowed to incorporate EdU for 2 h at an MOI (macrophage/yeast ratio) of 0 (no yeasts), 1:2, or 1:10. The experiment was repeated twice in duplicate wells with similar results. Shown are means ± the SD of duplicate wells of a representative experiment.
To expand and confirm our findings, we used EdU incorporation as an alternative to DNA quantification. In contrast to PI staining, which reflects the total DNA content, EdU incorporation labels cells with active DNA synthesis, i.e., cells in the S phase of the cell cycle (Fig. 3C). When J774.16 macrophages ingested C. neoformans, the rate of EdU incorporation was equal to that of macrophages that did not phagocytose C. neoformans. In other experiments, macrophages were allowed to ingest more C. neoformans at an MOI of 1:10 (macrophage/yeast ratio), and still there was no significant increase in Edu+ cells relative to cells with no C. neoformans. Furthermore, when phagocytosis was allowed to proceed for 4 h, there was still no increase in the rate of EdU+ cells (results not shown).
In vitro J774.16 macrophage proliferation after ingestion of yeast cells.
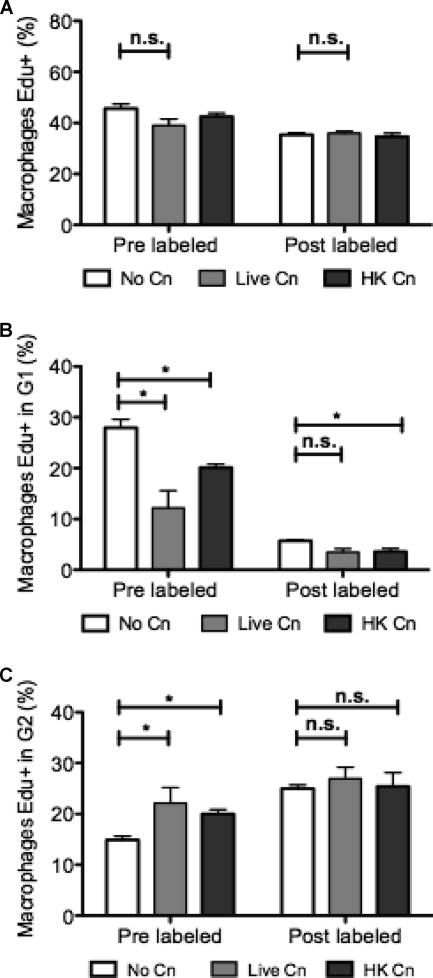
To investigate the apparent discordance between DNA quantification and EdU incorporation, we carried out pulse-labeling experiments to track cycling macrophages. Macrophages were labeled with EdU for 2 h, EdU was removed, and the cells were left to complete the cell cycle, such that EdU-labeled macrophages would now be in the G1 phase of the cell cycle. Some macrophages were prelabeled with EdU for 2 h, allowed to phagocytose C. neoformans for 15 min, and left for an additional 4 h to complete cell cycle. Postlabeled cells were allowed to phagocytose first for 2 h and then incubated with EdU for 2 h and given an additional 2 h to progress through the cycle. Both pre- and postlabeled cells that phagocytosed C. neoformans had a percentage of Edu+ cells that was similar to that of uninfected macrophages (44% in cells with no C. neoformans versus 43% in live C. neoformans and 44% for HK C. neoformans) (Fig. 4A), findings similar to the results in Fig. 3C. In uninfected macrophages, 27% of the prelabeled macrophages were now in the G1 gate of the DNA histogram, but when live C. neoformans was present, only 12% of the Edu+ macrophages had reached the G1 phase (Fig. 4B). These results were consistent with an increase in Edu+ cells located in the G2/M phase (Fig. 4C), when macrophages ingest C. neoformans, such that 22% of the macrophages that ingested live C. neoformans were still in the G2/M phase, while uninfected macrophages had only 11%. Both results combined reflect an accumulation of macrophages at the G2/M phase and a decreased rate of successful mitosis. To validate our experimental approach, we treated J774.16 cells with taxol, which is known to arrest cells at the G2/M phase. We observed that taxol treatment does not affect overall EdU incorporation but does result in a marked increase in the number of Edu+ cells in G2/M phase (data not shown). Hence, the apparent discordance between PI and EdU was caused by cell cycle block at G2/M phase, such that cells are able to proceed through the S phase and stop in the G2/M phase. Our experimental data implied that antibody-opsonized C. neoformans ingestion caused macrophage cell cycle arrest at the G2/M phase.
Fig 4.
EdU incorporation and J774.16 macrophage completion of cycle. Ingestion of C. neoformans decreased the number of macrophages that successfully divided and returned to the G1 phase. These macrophages were retained at the G2/M phase. Macrophages that had not ingested C. neoformans (No Cn) after 2 h of C. neoformans phagocytosis were allowed to incorporate EdU for 6 h. (A) Percentage of total macrophages that incorporated EdU. (B) Percentage of macrophages that incorporated EdU, underwent mitotic division and subsequently reached the G1 phase. (C) Percentage of macrophages that incorporated EdU and reached the G2/M phase. Experiments were performed three times. Shown are means ± the SD of triplicate wells of a representative experiment. Groups were compared using a two-tailed t test with a 95% confidence interval (*, P < 0.05; n.s., not significant).
BMDM proliferation after ingestion of yeast cells.
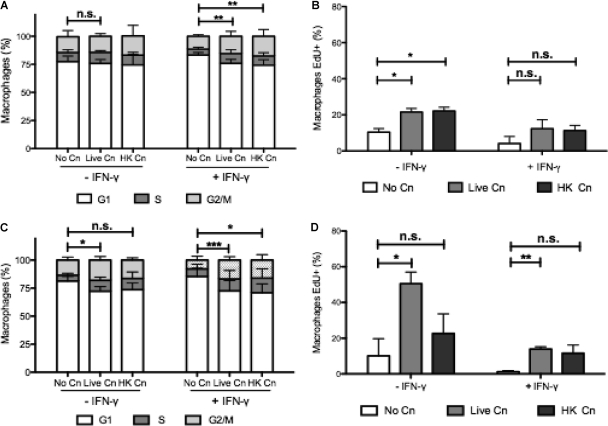
To investigate the relevance of our results in primary cells, our experiments were adapted to BMDM, which have a much slower rate of proliferation than the J774.16 macrophage-like cell line. Some of these BMDM were stimulated with IFN-γ, which is known to arrest macrophages in the G1 phase (38). After 6 h of phagocytosis, there was a decrease in the G1 phase accompanied by an increase in the G2/M phase (Fig. 5A). This difference, however, was not statistically significant (P = 0.3), possibly because the slower rate of replication of primary macrophages resulted in far fewer events. We studied macrophages treated with IFN-γ, which is known to suppress cell cycle, and in these cells phagocytosis of C. neoformans overcame IFN-γ-induced cell cycle arrest such that macrophages to progress into the S phase, resulting in a statistically significant difference (P = 0.0078). EdU was added to BMDM under the same conditions, and for these cells we measured increased EdU incorporation when macrophages ingested C. neoformans (10% versus 21% for cells with no C. neoformans versus live C. neoformans conditions). In IFN-γ-stimulated macrophages there was a higher fold increase in the percentage of Edu+ cells (4% versus 12% in cells with no C. neoformans versus live C. neoformans) (Fig. 5B). Next, we allowed C. neoformans infection to proceed for 24 h (Fig. 5C) and labeled new DNA synthesis for the last 6 h of infection (Fig. 5D). The percentage of macrophages in the G1 phase was reduced when macrophages ingested C. neoformans compared to control conditions (P = 0.02). In addition, in the presence of IFN-γ, there was a decrease in the percentage of macrophages in the G1 phase and a significant increase in the percentage of macrophages in the G2/M phase (P < 0.0001), suggesting that cell cycle arrest occurred in BMDM 24 h after phagocytosis. Similarly, EdU incorporation was increased at 24 h when macrophages had ingested live C. neoformans or HK C. neoformans (10.1% in noninfected macrophages versus 50.5% in macrophages with live C. neoformans). In IFN-γ-treated wells the fold increase in EdU incorporation was more drastic (1.19% versus 13.9%). We repeated these experiments for LPS-activated macrophages. C. neoformans phagocytosis was able to override the cell cycle arrest provoked by LPS (data not shown). In the absence of growth factors, there was always a complete cell cycle arrest and no EdU incorporation (data not shown). We conclude that C. neoformans infection provokes a sustained proliferative response in BMDM, as measured by the incorporation of labeled DNA nucleotides. However, at 24 h postphagocytosis, IFN-γ-treated BMDM are arrested at the G2/M phase, as measured in cell cycle plots, implying that the proliferative response was arrested and replication was not successful. Unlike the J774.16 cells, in BMDM there is an increase in the rate of EdU incorporation, mainly because BMDM have a much slower replication rate that might be subject to modulation. In contrast, J774.16 cells replicate more rapidly. However, for both J774.16 and BMDM, our results indicate cell cycle arrest in the G2/M phase after C. neoformans phagocytosis.
Fig 5.
Progression in cell cycle and new DNA synthesis in BMDM. BMDM progressed into S phase, as demonstrated by cell cycle analysis and EdU incorporation, when C. neoformans was ingested. (A) Cell cycle plot after 6 h of phagocytosis. (B) Incorporation of EdU after 6 h of phagocytosis. (C) Cell cycle plot after 24 h of phagocytosis. (D) Incorporation of EdU 24 h after phagocytosis. Macrophages were infected with live and HK C. neoformans (indicated as Live Cn and HK Cn, respectively) for 6 or 24 h, and EdU was added in the last 6 h of the experiment. Phagocytosis occurred in the presence of growth factors, with or without IFN-γ (to induce cell cycle arrest in G1 phase). The experiment was repeated four times using triplicate wells. Shown are means ± the SD of triplicate wells of two experiments. Groups were compared using a two-tailed t test with a 95% confidence interval (***, P < 0.001; **, P < 0.01; *, P < 0.05; n.s., not significant).
Alteration of nuclear morphology.
Cyclomodulins are bacterial products that interfere with host cell cycle by mechanisms that usually involve direct effects to cell cycle machinery or indirectly by disturbing the cytoskeletal organization. Cyclomodulin usually results in host cell endonucleation (multiple nuclei inside the same cell), enlargement of the nuclei or of the cell, or inhibition of cytokinesis that ultimately leads to cell death (27). Hence, we inspected LSC images for altered nuclear morphology. We observed several events of nuclear enlargement, binucleate cells, and cells that seemed to result from fusion of several macrophages (see Fig. S3 in the supplemental material) in both uninfected and infected wells. Cell cycle plots did not show an increase in ploidy over >4n (data not shown) for any of the conditions studied. Hence, the cell cycle arrest observed for C. neoformans does not appear to be comparable to that observed with cyclomodulins.
Causes of cell cycle arrest.
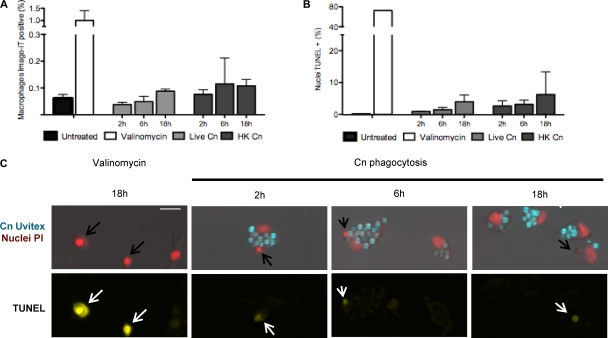
Cell cycle arrest could be caused by interference directly in the cell cycle or by C. neoformans indirect toxic effects. To investigate the cause of the cell cycle arrest, we stained macrophages with the nuclear stain Image-iT green, which is impermeable for intact membranes. Bright Image-iT fluorescence indicates a permeable cellular membrane, characteristic of either late apoptosis or necrosis (Fig. 6A). In both J774.16 and BMDM there was an increase in the number of membrane-permeable macrophages when C. neoformans was ingested. However, very few macrophages were membrane permeable, suggesting that cell damage involving cell membrane integrity does not explain cell cycle arrest.
Fig 6.
DNA fragmentation and membrane permeability in J774.16 macrophages. J774.16 macrophages did not manifest increased membrane permeability or DNA fragmentation after phagocytosis of C. neoformans. (A and B) Quantification of the number of cells positive for membrane permeability (A) or DNA fragmentation (B). Treatment with valinomycin at 400 nM for 18 h was used as a positive control, while uninfected macrophages were used as the negative control. (C) Representative images of macrophages positive for TUNEL staining after treatment with 400 nM valinomycin (positive control) or after infection with live C. neoformans (data for HK C. neoformans are not shown). Nuclei positive for DNA fragmentation have apoptotic cell morphology and are rapidly engulfed by a neighboring cell (arrows). Scale bar, 20 μm. Experiments were performed in duplicate wells for DNA fragmentation or in triplicate wells for membrane permeability measurements. Experiments were performed twice in J774.16 macrophages, and the results of one experiment are shown. Experiments were performed once for BMDM, and same results as observed for J774.16 were obtained.
Next, we measured DNA fragmentation by the TUNEL assay. DNA damage will activate cell cycle checkpoints and prevent cell cycle progression and, if the DNA damage is extensive enough, it can trigger apoptosis. We observed an increase in TUNEL+ cells for macrophages with ingested C. neoformans relative to control macrophages: 0% in uninfected macrophages versus 1.4% in live C. neoformans versus 3.2% in HK C. neoformans (Fig. 6B). Most frequently, we found TUNEL+ nuclei already ingested by a neighboring macrophage (Fig. 6C, note arrows), suggesting that, in our system, the apoptotic cells are promptly removed by neighboring cells. The images show that every TUNEL+ nucleus has a morphology consistent with very late apoptosis or even nuclear debris. From this, we conclude cell cycle arrest is unlikely to be caused by direct DNA damage, as measured by DNA fragmentation.
In vivo macrophage proliferation in the presence or absence of C. neoformans infection.
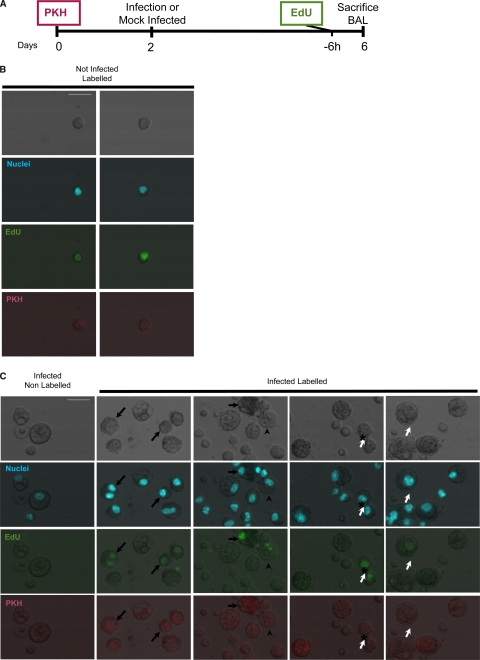
AM are considered the first line of defense for C. neoformans infection. The lipophilic dye PKH, when suspended in a buffer that causes its aggregation, can be used to label phagocytic cells. This strategy was used by Maus et al. (24) to label AM in vivo. At the same time, i.p. injection of EdU will label proliferating cells in vivo (28). Hence, we decided to combine these strategies to investigate AM replication in the lung.
A suspension of PKH was administered to mice. Blood, bone marrow, and alveolar macrophages from BAL were collected 2 and 6 days after PKH injection (Fig. 7A). At both time intervals, only BAL cells showed PKH staining. PKH+ cells comprised 30 to 50% of the recovered population, with 15% being very brightly stained (Table 1). For mice that were intratracheally infected with C. neoformans, only BAL cells had PKH+ staining. Due to infection, there is a massive immune infiltration into the bronchoalveolar space leading to a decrease in the percentage of PKH+ cells to 4 to 20%. These cells could be assumed to be AM. However, the absolute number of PKH+ cells was higher in infected mice. The reason for this discrepancy is unknown. In our view, this could be due to PKH labeling lung macrophages other than AM. These cells could infiltrate the bronchoalveolar space in response to infection or, alternatively, infection could change the adhesion characteristics of the AM, allowing a greater recovery of AM. Other explanations are also possible. Consequently, we suggest caution in the interpretation of PKH as an AM label in a setting of infection and inflammation.
Fig 7.
AM proliferation in vivo. Macrophages in the bronchoalveolar space replicated both in steady state and in the context of intratracheal C. neoformans infection. (A) Timeline of the experiment. PKH was administered to mice to label AM macrophages, 2 days before mice were infected with C. neoformans strain 24067 intratracheally. Mock-infected mice (sterile PBS) and infected mice were administered EdU to label cells in S phase at day 4, and 6 h later the mice were sacrificed, and BAL was performed. (B) Representative image of AM replication recovered in a mock-infected mouse. (C) Representative images of BAL recovered cells from C. neoformans-infected mice. The first column displays cells from nonlabeled mouse. The following columns show cells from labeled mice. AM (PKH+) can incorporate EdU (arrows). Cells with macrophage morphology but PKH− can also incorporate EdU (open arrows). Note that one of the macrophages initiating replication has ingested C. neoformans (arrowheads) and adjacent positive cells, an observation suggestive of recently completed mitosis (*). See Table 1 for the frequency of events. Scale bars, 20 μm. The experiment was repeated three times with two to four mice per group, and representative images are shown.
Table 1.
AM incorporation of EdU in vivo
| Expt no. | Mouse no. | No. of cellsa |
|||
|---|---|---|---|---|---|
| Total | PKH+ (%) | EdU+ | PKH+ EdU+ | ||
| Uninfected mice | |||||
| 1 | 1 | 63 | 35 (55) | 1 | 1 |
| 2 | 134 | 46 (34) | 2 | 2 | |
| 2 | 1 | 178 | 55 (30) | 1 | 1 |
| 2 | 209 | 41 (30) | 4 | 2 | |
| C. neoformans-infected mice | |||||
| 1 | 1 | 4,730 | 205 (4) | 16 | 1 |
| 2 | 2,787 | 663 (23) | 8 | 2 | |
| 2 | 1 | 875 | 191 (22) | 13 | 2 |
| 2 | 1,896 | 181 (10) | 13 | 1 | |
| 3 | 3,539 | 751 (21) | 15 | 2 | |
| 4 | 2,817 | 574 (20) | 18 | 2 | |
Analyzed cells correspond to 1/60 of the BAL-recovered cells. The Edu+ PKH− cells are of unknown origin.
To study proliferation, mice were administered EdU at 4 days postinfection (or mock infection with sterile PBS). Mice were sacrificed 6 h later, and BAL was performed. We estimate that we imaged 1/20 and 1/60 of the recovered BAL cells for mock-infected and infected mice, respectively. In mock-infected mice (steady-state conditions) we found one to four EdU+ cells per mouse (Table 1 and Fig. 7B). In C. neoformans-infected mice, we found 13 to 18 events per mouse and, of these, only one to two were double positive (PKH+ Edu+) (Table 1 and Fig. 7C). These results show that the AM in noninfected mice proliferate, suggesting that resident macrophage proliferation plays a role in tissue homeostasis. Furthermore, AM proliferation was found in the cells recovered from the bronchoalveolar space after C. neoformans infection. The rates of division seem to not be altered in the infected mice, although the low number of events does not allow us to make definitive conclusions.
DISCUSSION
This study established the usefulness and adaptability of LSC for the study of macrophage-fungal interactions. Microscopy protocols usually require a significant amount of operator input, both in the acquisition of images and in the analysis of these images. LSC automates data acquisition but, more importantly, it allows for automated and quantitative data analysis (11, 33, 37). LSC was shown to be comparable to human counting for measuring phagocytosis and provides significant advantages that were exploited for the study of the relationship of phagocytosis and cell cycle progression. LSC allowed the use of images to quantify DNA content, in other words, and thus to study the cell cycle in adherent cells and correlate it with intracellular processes (6, 22).
Previous results from our laboratory showed that FcγR-mediated ingestion of yeast cells was associated with increased cell cycle progression (18, 20, 21). Both complement-mediated C. neoformans phagocytosis and ingestion of inert latex beads were effective at stimulating cell cycle progression (21). These effects could be reproduced using a model of frustrated phagocytosis mediated by FcγR (20), whereby continuous stimulation of receptors without particle ingestion triggered cell cycle progression and allowed dissection of signaling pathways involved in the proliferative response. It is known that macrophages can divide after C. neoformans ingestion and even divide after nonlytic exocytosis (3, 23), but there has been no study of how macrophage replication is affected by C. neoformans ingestion. Later work showed that ingestion of live yeast cells, but not latex beads, inhibited cyclin D1 expression (18), producing an apparent paradox not resolved in their work. Cyclin D1 is a major checkpoint in the transition from G1 to S phase, and cyclin D1 expression is inhibited in response to LPS, triggering mitotic arrest (35). In addition, there is evidence that the cell cycle phase influences macrophage behavior: cell cycle arrest in the G1 phase influences major histocompatibility complex class II expression (39) or prevents LPS-induced apoptosis (40).
In the present study, we revisited this problem using LSC. Our experiments confirmed cell cycle progression in both J774.16 cells and primary macrophages using LSC. However, LSC provided additional information on this phenomenon by establishing that the likelihood of the effect increased with increasing intracellular fungal burden. Furthermore, cell cycle progression was observed after the ingestion of live and HK C. neoformans, demonstrating that cell cycle progression does not require fungal viability but is a consequence of FcγR activation and subsequent phagocytosis (20). DNA content quantification and a pulse-labeling strategy revealed a reduction in the number of cells that cycled back into the G1 phase, suggesting that macrophage-like cells containing C. neoformans were less likely to complete mitosis or, in other words, were arrested at the G2/M phase of the cell cycle. Pulse-labeling experiments could not be performed in primary cells due to longer replication times, but when primary cells were treated with IFN-γ, which is known to produce cell cycle arrest in dividing cells, we observed that phagocytosis was associated with an initial cell cycle progression that proceeded to arrest in the G2/M phase. We conjectured that cell division impairment could be a consequence of fungal mediated host cell damage or a by-product of the macrophage's attempt to degrade a high load of large foreign particles. It is known that the phagosome of macrophages becomes permeabilized after C. neoformans infection (34), and it is conceivable that the spill of phagosomal contents into the cytoplasm produces damage that translates into cell cycle arrest. Consequently, we explored several potential mechanisms of macrophage damage. Analysis of TUNEL staining and of membrane permeability in both J774.16 and BMDM cells revealed that neither of these mechanisms was likely responsible for cell cycle arrest. We considered the possibility that the mitotic defect was a consequence of a fungal product analogous to bacterial cyclomodulins (27), which are known to provoke cell cycle arrest and alter DNA content or nuclear morphology. However, we found no evidence of such an effect in our experiments. In this regard, we note that taxol prevents mitosis by stabilizing cytoskeletal microtubules and that C. neoformans has been reported to interfere with host cell cytoskeleton (7, 19). We conclude that despite C. neoformans phagocytosis causing macrophages to progress in the cell cycle, the yeast cells produced toxic effects that had widespread effects in the host cell cycle, resulting in G2/M-phase arrest (the present study) and cyclin D1 inhibition (18). Defining the mediator of the cell cycle interference remains an outstanding question for future studies.
In the present study, we made the unexpected but relevant observation was that IFN-γ- and LPS-mediated cell cycle arrest could be overcome by phagocytosis-derived cell cycle progression. Both IFN-γ and LPS are known to arrest cell division in the presence of macrophage–colony-stimulating factor (36, 40). This observation raised the possibility that microbial ingestion and macrophage effector functions in vivo in conditions where IFN-γ is made do not necessarily lead to cell cycle arrest. Furthermore, macrophages might respond to infection in a way similar to other types of immune cells that undergo clonal expansion to generate additional effector cells to fight infection. We note that an increase in the proliferative ability of AM was reported in humans following exposure to asbestos or in chronic inflammatory disorders (4, 31) and in mice exposed to cigarette smoke (12).
To date, replication of AM in vivo has not been definitively established given the difficulties of distinguishing between resident AM and the infiltrating inflammatory cells by standard methods. A recently published study reported macrophage proliferation in response to infection in the peritoneal and pleural cavities after an inflammatory stimulus (15). These studies were quickly followed by reports of macrophage proliferation in other experimental models: zymosan-induced peritonitis (9) and autoimmune encephalitis (1). Given that C. neoformans establishes chronic pulmonary infection, we attempted to search for AM proliferation in a mouse model of cryptococcal pneumonia. Consequently, we developed a double-labeling strategy to investigate the problem of AM replication in the presence or absence of C. neoformans infection. Our labeling strategy took advantage of the relative specificity of PKH for AM and EdU for replicating cells (24, 28). Our results demonstrated synthesis of new DNA in AM in both infected and uninfected mice. Both the resident population and the newly arrived blood-derived macrophages incorporated EdU and showed features of mitotic division. This observation provides an important confirmation of the findings of Jenkins et al. (15). Our results, albeit in a different model, suggest that macrophage proliferation occurs in the bronchoalveolar space after C. neoformans infection, as reported in macrophages from other tissues (1, 9, 15). Furthermore, we show that the resident AM population is capable of proliferation as part of normal tissue homeostasis, meaning in the absence of any infection. We were unable to demonstrate an increased rate of AM replication in vivo as a consequence of C. neoformans infection. One explanation for the low rate of replication in vivo could be the impairment of mitosis completion, meaning that what we observed in vitro for C. neoformans-containing macrophages also occurs in vivo. Hence, it is conceivable that the rate of lung macrophage proliferation would be higher in other inflammatory conditions where there is no mitotic arrest, such as the presence of inert particles in the lungs (4, 31) or the inhalation of cigarette smoke (12). The proliferation of AM occurred at a low rate in the presence or absence of infection but is likely to make only a small contribution to inflammatory cell numbers during pulmonary cryptococcosis. Our observations can also be integrated with the recent report that upon resolution of lung infection newly recruited cells die by apoptosis while leaving PKH-labeled resident AM untouched (14). Interpretation of our data in conjunction with the aforementioned reports (1, 9, 15) suggests that resident macrophage numbers are supported by both local proliferation and the infiltration of new cells. In the event of an injury or inflammation, the equilibrium between these processes is changed until a new equilibrium is established.
In summary, LSC confirmed that macrophage-like cells progressed in cell cycle after phagocytosis, as reported earlier (21), and allowed us to explore this process in significantly greater detail. We now report that despite increased cell cycle progression there completion of mitosis is less efficient, possibly as a result of phagocytosis-dependent toxicity after the ingestion of large microbial particles. Furthermore, we observed that phagocytosis of C. neoformans is sufficient to suppress IFN-γ and LPS cell cycle arrest. Lastly, we established a protocol to study proliferation of resident phagocytic cells in vivo that allowed us to demonstrate that resident AM are capable of proliferation, both as part of tissue homeostasis and as part of the inflammatory process. These findings enhance our understanding of macrophage biology in the setting of phagocytosis and C. neoformans infection.
Supplementary Material
ACKNOWLEDGMENTS
This study was supported by Ph.D. grant SFRH/BD/33471/2008 from Fundação Ciência e Tecnologia (C.C.), NIH awards 5R01HL059842, 5R01AI033774, 5R37AI033142, and 5R01AI052733 (A.C.), and the Center for AIDS Research at Einstein.
We acknowledge the Analytical Imaging Facility and, in particular, Vera DesMarais and Peng Guo for their technical assistance.
Footnotes
Published ahead of print 17 January 2012
Supplemental material for this article may be found at http://iai.asm.org/.
REFERENCES
- 1. Ajami B, Bennett JL, Krieger C, McNagny KM, Rossi FM. 2011. Infiltrating monocytes trigger EAE progression, but do not contribute to the resident microglia pool. Nat. Neurosci. 14:1142–1149 [DOI] [PubMed] [Google Scholar]
- 2. Alvarez M, Burn T, Luo Y, Pirofski LA, Casadevall A. 2009. The outcome of Cryptococcus neoformans intracellular pathogenesis in human monocytes. BMC Microbiol. 9:51. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Alvarez M, Casadevall A. 2006. Phagosome extrusion and host-cell survival after Cryptococcus neoformans phagocytosis by macrophages. Curr. Biol. 16:2161–2165 [DOI] [PubMed] [Google Scholar]
- 4. Bitterman PB, Saltzman LE, Adelberg S, Ferrans VJ, Crystal RG. 1984. Alveolar macrophage replication: one mechanism for the expansion of the mononuclear phagocyte population in the chronically inflamed lung. J. Clin. Invest. 74:460–469 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Casadevall A, et al. 1998. Characterization of a murine monoclonal antibody to Cryptococcus neoformans polysaccharide that is a candidate for human therapeutic studies. Antimicrob. Agents Chemother. 42:1437–1446 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Chakraborty AA, Tansey WP. 2009. Inference of cell cycle-dependent proteolysis by laser scanning cytometry. Exp. Cell Res. 315:1772–1778 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Chen SH, et al. 2003. Cryptococcus neoformans induces alterations in the cytoskeleton of human brain microvascular endothelial cells. J. Med. Microbiol. 52:961–970 [DOI] [PubMed] [Google Scholar]
- 8. Darzynkiewicz Z, Traganos F, Zhao H, Halicka HD, Li J. 2011. Cytometry of DNA replication and RNA synthesis: historical perspective and recent advances based on “click chemistry.” Cytometry A 79:328–337 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Davies LC, et al. 2011. A quantifiable proliferative burst of tissue macrophages restores homeostatic macrophage populations after acute inflammation. Eur. J. Immunol. 41:2155–2164 [DOI] [PubMed] [Google Scholar]
- 10. Feldmesser M, Kress Y, Novikoff P, Casadevall A. 2000. Cryptococcus neoformans is a facultative intracellular pathogen in murine pulmonary infection. Infect. Immun. 68:4225–4237 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Henriksen M, Miller B, Newmark J, Al-Kofahi Y, Holden E. 2011. Laser scanning cytometry and its applications: a pioneering technology in the field of quantitative imaging cytometry. Methods Cell Biol. 102:161–205 [DOI] [PubMed] [Google Scholar]
- 12. Hornby SB, Kellington JP. 1990. DNA synthesis in alveolar macrophages and other changes in lavaged cells following exposure of CBA/H mice to cigarette smoke. Environ. Health Perspect. 85:107–112 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Ieong MH, Reardon CC, Levitz SM, Kornfeld H. 2000. Human immunodeficiency virus type 1 infection of alveolar macrophages impairs their innate fungicidal activity. Am. J. Respir. Crit. Care Med. 162:966–970 [DOI] [PubMed] [Google Scholar]
- 14. Janssen WJ, et al. 2011. Fas determines differential fates of resident and recruited macrophages during resolution of acute lung injury. Am. J. Respir. Crit. Care Med. 184:547–560 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Jenkins SJ, et al. 2011. Local macrophage proliferation, rather than recruitment from the blood, is a signature of TH2 inflammation. Science 332:1284–1288 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Kamentsky L. 2001. Laser scanning cytometry. Methods Cell Biol. 63:51–87 [DOI] [PubMed] [Google Scholar]
- 17. Kechichian TB, Shea J, Del Poeta M. 2007. Depletion of alveolar macrophages decreases the dissemination of a glucosylceramide-deficient mutant of Cryptococcus neoformans in immunodeficient mice. Infect. Immun. 75:4792–4798 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Luo Y, Casadevall A. 2010. Intracellular cryptococci suppress Fc-mediated cyclin D1 elevation. Commun. Integr. Biol. 3:390–391 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Luo Y, Isaac BM, Casadevall A, Cox D. 2009. Phagocytosis inhibits F-actin-enriched membrane protrusions stimulated by fractalkine (CX3CL1) and colony-stimulating factor 1. Infect. Immun. 77:4487–4495 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Luo Y, Pollard JW, Casadevall A. 2010. Fcγ receptor cross-linking stimulates cell proliferation of macrophages via the ERK pathway. J. Biol. Chem. 285:4232–4242 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Luo Y, Tucker SC, Casadevall A. 2005. Fc- and complement-receptor activation stimulates cell cycle progression of macrophage cells from G1 to S. J. Immunol. 174:7226–7233 [DOI] [PubMed] [Google Scholar]
- 22. Luther E, Kamentsky LA. 1996. Resolution of mitotic cells using laser scanning cytometry. Cytometry 23:272–278 [DOI] [PubMed] [Google Scholar]
- 23. Ma H, Croudace JE, Lammas DA, May RC. 2006. Expulsion of live pathogenic yeast by macrophages. Curr. Biol. 16:2156–2160 [DOI] [PubMed] [Google Scholar]
- 24. Maus U, et al. 2001. Monocytes recruited into the alveolar air space of mice show a monocytic phenotype but upregulate CD14. Am. J. Physiol. Lung Cell. Mol. Physiol. 280:L58–L68 [DOI] [PubMed] [Google Scholar]
- 25. Mosser DM, Edwards JP. 2008. Exploring the full spectrum of macrophage activation. Nat. Rev. Immunol. 8:958–969 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Murphy J, Summer R, Wilson AA, Kotton DN, Fine A. 2008. The prolonged life-span of alveolar macrophages. Am. J. Respir. Cell Mol. Biol. 38:380–385 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Nougayrede JP, Taieb F, De Rycke J, Oswald E. 2005. Cyclomodulins: bacterial effectors that modulate the eukaryotic cell cycle. Trends Microbiol. 13:103–110 [DOI] [PubMed] [Google Scholar]
- 28. Osterholzer JJ, et al. 2011. Chemokine receptor 2-mediated accumulation of fungicidal exudate macrophages in mice that clear cryptococcal lung infection. Am. J. Pathol. 178:198–211 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Osterholzer JJ, et al. 2009. Role of dendritic cells and alveolar macrophages in regulating early host defense against pulmonary infection with Cryptococcus neoformans. Infect. Immun. 77:3749–3758 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Spector WG, Wynne KM. 1976. Proliferation of macrophages in inflammation. Agents Actions 6:123–126 [DOI] [PubMed] [Google Scholar]
- 31. Spurzem JR, Saltini C, Rom W, Winchester RJ, Crystal RG. 1987. Mechanisms of macrophage accumulation in the lungs of asbestos-exposed subjects. Am. Rev. Respir. Dis. 136:276–280 [DOI] [PubMed] [Google Scholar]
- 32. Tarling JD, Lin HS, Hsu S. 1987. Self-renewal of pulmonary alveolar macrophages: evidence from radiation chimera studies. J. Leukoc. Biol. 42:443–446 [PubMed] [Google Scholar]
- 33. Telford WG, Komoriya A, Packard BZ. 2002. Detection of localized caspase activity in early apoptotic cells by laser scanning cytometry. Cytometry 47:81–88 [DOI] [PubMed] [Google Scholar]
- 34. Tucker SC, Casadevall A. 2002. Replication of Cryptococcus neoformans in macrophages is accompanied by phagosomal permeabilization and accumulation of vesicles containing polysaccharide in the cytoplasm. Proc. Natl. Acad. Sci. U. S. A. 99:3165–3170 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Vadiveloo PK. 1999. Macrophages: proliferation, activation, and cell cycle proteins. J. Leukoc. Biol. 66:579–582 [DOI] [PubMed] [Google Scholar]
- 36. Vadiveloo PK, Keramidaris E, Morrison WA, Stewart AG. 2001. Lipopolysaccharide-induced cell cycle arrest in macrophages occurs independently of nitric oxide synthase II induction. Biochim. Biophys. Acta 1539:140–146 [DOI] [PubMed] [Google Scholar]
- 37. Wojcik E, Saraga S, Jin J, Hendricks J. 2001. Application of laser scanning cytometry for evaluation of DNA ploidy in routine cytologic specimens. Diagn. Cytopathol. 24:200–205 [DOI] [PubMed] [Google Scholar]
- 38. Xaus J, et al. 1999. Interferon gamma induces the expression of p21waf-1 and arrests macrophage cell cycle, preventing induction of apoptosis. Immunity 11:103–113 [DOI] [PubMed] [Google Scholar]
- 39. Xaus J, et al. 2000. The expression of MHC class II genes in macrophages is cell cycle dependent. J. Immunol. 165:6364–6371 [DOI] [PubMed] [Google Scholar]
- 40. Xaus J, et al. 2000. LPS induces apoptosis in macrophages mostly through the autocrine production of TNF-α. Blood 95:3823–3831 [PubMed] [Google Scholar]
- 41. Zaragoza O, Alvarez M, Telzak A, Rivera J, Casadevall A. 2007. The relative susceptibility of mouse strains to pulmonary Cryptococcus neoformans infection is associated with pleiotropic differences in the immune response. Infect. Immun. 75:2729–2739 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Zbigniew D, Holden E, Orfao A, Telford W, Wlodkowic D. (ed). 2011. Recent advances in cytometry. A. Instrumentation and methods, 5th ed, vol 102 Academic Press, London, United Kingdom [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.