Abstract
Bacteria growing in biofilms are physiologically heterogeneous, due in part to their adaptation to local environmental conditions. Here, we characterized the local transcriptome responses of Pseudomonas aeruginosa growing in biofilms by using a microarray analysis of isolated biofilm subpopulations. The results demonstrated that cells at the top of the biofilms had high mRNA abundances for genes involved in general metabolic functions, while mRNA levels for these housekeeping genes were low in cells at the bottom of the biofilms. Selective green fluorescent protein (GFP) labeling showed that cells at the top of the biofilm were actively dividing. However, the dividing cells had high mRNA levels for genes regulated by the hypoxia-induced regulator Anr. Slow-growing cells deep in the biofilms had little expression of Anr-regulated genes and may have experienced long-term anoxia. Transcripts for ribosomal proteins were associated primarily with the metabolically active cell fraction, while ribosomal RNAs were abundant throughout the biofilms, indicating that ribosomes are stably maintained even in slowly growing cells. Consistent with these results was the identification of mRNAs for ribosome hibernation factors (the rmf and PA4463 genes) at the bottom of the biofilms. The dormant biofilm cells of a P. aeruginosa Δrmf strain had decreased membrane integrity, as shown by propidium iodide staining. Using selective GFP labeling and cell sorting, we show that the dividing cells are more susceptible to killing by tobramycin and ciprofloxacin. The results demonstrate that in thick P. aeruginosa biofilms, cells are physiologically distinct spatially, with cells deep in the biofilm in a viable but antibiotic-tolerant slow-growth state.
INTRODUCTION
Bacterial biofilms are communities of microorganisms that grow in association with surfaces. The cells are often encased in protective matrices composed of extracellular polysaccharides with embedded proteins and DNA (48, 57, 72). Infections with bacterial biofilms on artificial implant devices or on host tissues are a leading cause of health-related problems, since the cells within the biofilms are often tolerant to antimicrobial treatments and to host defenses (15, 16). For example, Pseudomonas aeruginosa is a bacterium that causes biofilm-associated infections on the pulmonary tissues of cystic fibrosis patients (39). P. aeruginosa biofilms have been shown to impair the activity of neutrophils (1, 31) and to tolerate the antibiotics ceftazidime, ciprofloxacin, and tobramycin at concentrations far greater than those necessary to kill free-swimming, planktonic bacteria (5, 44, 47, 69). This antibiotic tolerance may be due in part to variant subpopulations within the biofilms that are protected from treatment (6, 22, 23, 33, 35, 37, 45). A particular antimicrobial agent may effectively target certain populations of cells but leave the remaining cells viable, allowing them to repopulate the biofilms when the treatment is stopped. These antibiotic-tolerant subpopulations have been termed persister cells (37) and are likely important for chronic infections. P. aeruginosa isolates from chronic pulmonary infections are often clonal over time (11, 46, 70), even though patients undergo extensive antibiotic treatments. Since most antibiotics target actively growing cells, the persister subpopulations may include metabolically inactive bacteria (37).
Bacteria within biofilms differ in cell physiology, depending on the spatial location of the cells within the community (64). As nutrients and oxygen diffuse into the biofilms and are utilized by the bacteria, chemical concentration gradients of the nutrients are established. Cell metabolism results in generation of waste products, secondary metabolites, and signaling compounds that also form chemical gradients within biofilm strata. These gradients may intersect and overlap, creating many unique microenvironments within biofilms. Adaptive variability allows the cells to respond to their local environmental conditions (29, 30, 75). In addition, genetic alterations, including mutations, genetic recombination, and stochastic gene expression, have been identified as mechanisms that generate phenotypically and genetically distinct variant subpopulations within biofilms (2, 7, 9, 12, 20, 25, 27, 34, 41, 42, 67, 76). Microscopic evidence has verified the presence of spatially segregated and differentially antibiotic susceptible subpopulations of cells (24, 32, 49). For example, the antibiotics tobramycin and ciprofloxacin preferentially kill cells at the periphery of the biofilms, while other chemical agents, including gallium, sodium dodecyl sulfate, and colistin, are effective against cells in the cluster centers.
In order to characterize physiological features of biofilm bacteria and to gain a better understanding of mechanisms for enhanced antibiotic resistances, transcriptomic and proteomic profiles of microbial biofilms have been determined and compared to the profiles of planktonic cells (21, 28, 43, 50, 58, 68, 73). For these global approaches, it is often necessary to harvest the entire biofilm community to obtain sufficient RNA or protein for analysis. Therefore, these approaches provide transcriptome or proteome information as an average of that for the entire biofilm and may not be adequate for discovering differences in gene expression that occur at localized sites and due to various environmental conditions or stochastic gene expression events. Previously, we used laser capture microdissection (LCM) to isolate clusters of cells from within P. aeruginosa biofilms and quantified the rRNA and mRNA abundances of individual genes from the isolated cell groups (36, 51). The results indicated that the abundances of individual mRNA transcripts may vary by several orders of magnitude, even for cells in close proximity to each other. The previous results also suggested that the top of P. aeruginosa biofilms contains cells in transition to stationary phase, while the cells at the bottom of the biofilm may be in a slow-growth or dormant state (51).
In this study, we used LCM to isolate cells from different regions within P. aeruginosa biofilms and then performed Affymetrix microarray analysis to characterize the global transcriptome response of the subpopulations. We focused on transcription of cells isolated from the aerobic periphery of thick biofilms compared to that of cells obtained from the bottom of these biofilms. To test further the hypothesis that cells at the bottom of the biofilm are in a slow-growth and antibiotic-tolerant state, we experimentally discriminated the metabolic status and antibiotic susceptibility of these bacteria. The results showed low metabolic activity of cells in the deeper regions of the biofilms, as indicated by low mRNA abundances. The cells at the base of the biofilms also had reduced sensitivity to the antibiotics ciprofloxacin and tobramycin. Cells in the bottom portion of the biofilms maintained a high abundance of ribosomal RNAs as well as mRNA for genes associated with ribosome hibernation factors, suggesting that the slow-growing and antibiotic-tolerant subpopulation may protect ribosomes from degradation for later resuscitation when nutrients or space become available.
MATERIALS AND METHODS
Bacterial strains and biofilm growth conditions.
Pseudomonas aeruginosa PAO1 and a PAO1 derivative, MH475 (provided by Matthew Parsek) were used for these studies. MH475 contains a gene (the gfp gene) encoding the arabinose-inducible green fluorescent protein (GFP) on a chromosomally encoded mini-Tn7 element (14). Markerless deletion mutations of the rmf and PA4463 genes were constructed with P. aeruginosa PAO1 using the allelic exchange approach (13) and the PCR primers shown in Table 1. For epifluorescence microscopy, P. aeruginosa PAO1 and its deletion mutant strains contain the GFP expressed from plasmid pMF230 (48). Most biofilm experiments were performed using colony biofilms on tryptic soy agar (TSA; Becton, Dickinson and Company [BD], Franklin Lakes, NJ) (4). For these experiments, an overnight P. aeruginosa culture was diluted into fresh tryptic soy broth (TSB) to an optical density at 600 nm (OD600) of 0.05 and a 10- or 25-μl aliquot was used to inoculate sterile, black, polycarbonate membranes (0.22-μm pore size, 13- or 25-mm diameter; GE Water & Process Technologies, Fairfield, CT) positioned on TSA plates. Plates were incubated at 37°C until the membranes were dry and then inverted and incubated for up to 72 h. Membranes containing colony biofilms were aseptically transferred to fresh TSA plates every 12 h for 52 or 72 h, with the final transfer occurring 4 h prior to cryoembedding. For propidium iodide (PI) staining, biofilms were transferred every 24 h. Drip flow biofilm experiments were performed as described previously (75) with a flow of one-tenth-strength TSB medium at 1 ml/min for 72 h.
Table 1.
Primer sequences
| Primer | Sequence | Concn (nm) | PCR product size (bp) |
|---|---|---|---|
| Deletion mutation primers | |||
| RMF ECOR15′ UP | GAATTCTCCACCGGGAATCGCGCC | ||
| RMF-T7UPR-BAM | CTATAGTGAGTCGTATTAGGATCCAACGGATCACGCTTAAGTC | ||
| RMF-T7DOWNF-BAM | GGATCCTAATACGACTCACTATAGGCTTACAACGTCTCAATCAAC | ||
| RMF ECORRI 3′ | GAATTCGATACCCCGGTGGAGCGCG | ||
| PA4463ECORI5′UP | GAATTCGCAGATCGAATCGAGCG | ||
| PA4463T7UR-BAM | CTATAGTGAGTCGTATTAGGATCCGCCACTGATGTTGACTTGCAT | ||
| PA4463T7DOWNF | GGATCCTAATACGACTCACTATAGCAAGGCGTAGGCGCCCGCTGA | ||
| PA4463ECORRI3′ | GAATTCGGGGAAGCGCTGCAACTG | ||
| qRT-PCR primers | |||
| PA4823-For | ACCTGCTGATGGTGGTCAG | 300 | 108 |
| PA4823-Rev | TGAGCGATGCACGGTTGAG | 300 | |
| PA1089-For | GAAGACGAGATCGAGGTGCT | 300 | 74 |
| PA1089-Rev | CGCAGCTTGAAGGATTTGTC | 300 | |
| PA4865-For | CGAGCTGATGCACTACGG | 200 | 128 |
| PA4865-Rev | GTGGACGGTCACCAGCTT | 300 | |
| PA0059-For | GACGGTTTCGCCATCACT | 300 | 128 |
| PA0059-Rev | GCGTTGAGCACCTTGGATAC | 200 | |
| PA2146-For | GCACAGCATCAAGGTGGTAAA | 200 | 109 |
| PA2146-Rev | GCGGATCGTTCTTGAAGTTG | 300 | |
| PA0997-For | CCCTCTACGTGGTCGATACC | 300 | 98 |
| PA0997-Rev | CTCGCTGTCCACTTCCAATC | 300 | |
| PA3531-For | TCATCCAGCACCTCAACAAG | 300 | 150 |
| PA3531-Rev | TGTCGGCATGCTTCATCTC | 300 | |
| PA4220-For | CCAGCGGCTATCTGTTGACT | 200 | 183 |
| PA4220-Rev | ACAACGCCAAGCCCAGTA | 200 | |
| PA16s-For | CAAAACTACTGAGCTAGAGTACG | 200 | 215 |
| PA16s-Rev | TAAGATCTCAAGGATCCCAACGGCT | 200 | |
| acp-For | ACTCGGCGTGAAGGAAGAAG | 250 | 80 |
| acp-Rev | CGACGGTGTCAAGGGAGT | 250 | |
| PA3049-For (rmf) | AAGCGTGATCCGTTGGAAAG | 250 | 89 |
| PA3049 -Rev | GGATGGGTGAACGGACAAAG | 250 | |
| PA4463-For | GCCATTTCGACAAGATCACC | 250 | 204 |
| PA4463-Rev | GCTGCCGTTCGAGATACTTC | 250 | |
| LucI-For | GTGTTGGGCGCGTTATTTATC | 400 | 78 |
| LucI-Rev | ACTGTTGAGCAATTCACGTTCA | 400 | |
| PA3126-For (IbpA) | CTGTTCCGTCATTCCGTAGG | 250 | 135 |
| PA3126-Rev | GGCGATAACGATGCGATACT | 250 |
GFP labeling of active and dormant cell subpopulations.
Prior to biofilm studies, P. aeruginosa MH475 planktonic cultures were analyzed by flow cytometry for induction of the GFP in the presence or absence of the inducing agent, l-arabinose. Any cells exhibiting GFP intensity over 100 on an arbitrary scale were scored as GFP-expressing (GFP+) cells, while any events registering an intensity below 100 were scored as non-GFP expressing (GFP−) cells. The arbitrary threshold of 100 was determined by first exploring the intensities of noninduced, negative-control P. aeruginosa MH475 cells, which were almost exclusively less than 100. To label the active-cell subpopulation of biofilms with the GFP, colony biofilms of P. aeruginosa MH475 were grown for 48 h in the absence of l-arabinose, transferred to medium supplemented with 2% l-arabinose, and incubated for an additional time as indicated in Results. To label metabolically dormant cells with the GFP, P. aeruginosa MH475 was first cultured in biofilms in the presence of 2% l-arabinose for 48 h. Biofilms were then transferred to medium without arabinose and incubated for an additional 12 to 48 h. Cells that expressed GFP were characterized microscopically after cryoembedding in Tissue-Tek optimum cutting temperature (OCT) compound (Miles, Inc., Elkhart, IN) (75) and thin sectioning of biofilms as described previously (36). GFP-expressing cells were also characterized by flow cytometry and cell sorting, for which colony biofilms were disaggregated by placing the biofilm in 9 ml of phosphate-buffered water (PBW) and vortexing the mixture on high for 1 min. The resulting suspensions were serially diluted and analyzed using a BD FACSAria flow cytometer. Cells were collected and scored as fluorescent counts versus the overall population. Viable cell numbers were determined as CFU on TSA plates.
The following antibiotics were used as needed: gentamicin sulfate (Sigma, St. Louis, MO), 15 μg ml−1; tobramycin sulfate (Sigma), 10 μg ml−1; ciprofloxacin hydrochloride (gift of the Bayer Corporation, Leverkusen, Germany), 1 μg ml−1.
Analysis of antibiotic tolerances of biofilm subpopulations.
P. aeruginosa MH475 cells were treated with ciprofloxacin or tobramycin during induction of GFP with l-arabinose or during GFP repression in the absence of l-arabinose. For GFP induction experiments, 48-h P. aeruginosa MH475 colony biofilms were transferred to medium containing both 2% arabinose and antibiotic (10 μg ml−1 tobramycin sulfate or 1 μg ml−1 ciprofloxacin hydrochloride) and incubated for additional time as indicated in Results. Biofilms were disaggregated, and viable cells were enumerated as CFU on TSA plates. In addition, parallel samples were sorted by flow cytometry based on GFP expression. GFP+ and GFP− cells were plated separately to determine CFU of each subpopulation. In GFP repression experiments, colony biofilms were incubated for 48 h in the presence of 2% l-arabinose to generate a population containing all fluorescently labeled cells. Biofilms were transferred to TSA plates without arabinose and incubated for 12 h. Biofilms were then transferred to plates without arabinose but containing an antibiotic and incubated for additional time as indicated in Results. Flow cytometry and cell sorting were used to determine viable cell counts from the fluorescent and nonfluorescent biofilm subpopulations.
For cells processed using flow cytometry, log reductions were calculated using the formula −log10 [(%Pt × %pt)/(%Pu × %pu)], where %Pt is the percentage of the total antibiotic-treated population (either GFP+ or GFP−), %pt is the percentage of the antibiotic-treated population (either GFP+ or GFP−) that was viable as plated after sorting by flow cytometry, %Pu is the percentage of the total untreated population (either GFP+ or GFP−), and %pu is the percentage of the untreated population (either GFP+ or GFP−) that was viable as plated after sorting by flow cytometry.
Staining of biofilm cells with propidium iodide.
Propidium iodide staining was performed by adding 110 μl of a 0.06 mM propidium iodide solution to a 13-mm membrane support (Millipore). Biofilms (at 52 h) were aseptically transferred to the support and incubated at room temperature in the dark for 25 to 35 min. Biofilms were then cryoembedded and thin sectioned. For epifluorescence microscopy, sections were mounted on Superfrost Plus glass slides (Fisher Scientific, Waltham, MA) and examined using a Nikon E-800 upright epifluorescence microscope with a tetramethyl rhodamine isocyanate (TRITC) filter set (546/10 excitation [ex], 575 dichromatic mirror (DM), 590LP emission [em]) for visualizing propidium iodide staining, and with a fluorescein isothiocyanate (FITC) filter set (480/30 ex, 515 DM, 535/40 em) for visualizing GFP fluorescence. Images were captured using a Photometrics CoolSnap fx digital camera with a 600-ms exposure time. Images in the green and red channels were overlaid using MetaMorph (Molecular Devices, Downingtown, PA).
Laser capture microdissection.
Biofilms were cryoembedded and thin sectioned as described previously (36). To examine vertical biofilm strata, frozen samples were thin-sectioned (5.0- to 15-μm sections) using a Leica CM-1850 cryostat. For LCM experiments, cryoembedded biofilm sections were placed on membrane-coated microscope slides and maintained on dry ice. Microscope slides containing biofilm sections were thawed on the microscope stage for 5 s and then examined using a 10× or 20× objective lens. LCM (Zeiss/P.A.L.M. Laser-MicroBeam system) was used to dissect and capture 24,000- to 100,000-μm2 sections from different regions of the biofilms (36). Dissected areas were captured in lysis buffer contained in the caps of 0.5-ml microcentrifuge tubes using the laser catapult parameter (LCP).
RNA extraction and qRT-PCR for microarray validation.
Quantitative reverse transcription-PCR (qRT-PCR) was performed with LCM samples to assess microarray results for the eight genes shown in Fig. S2 in the supplemental material. For qRT-PCR of these mRNAs, after the addition of 1.7 × 104 transcript copies of spike-in control luciferase (lucI gene) RNA (Promega), total RNA was extracted using the hot phenol approach described previously (36). Resuspended RNA was treated with DNase using a Turbo DNA-free kit (Ambion, Inc.) in 10-μl reaction mixtures for 20 min. All samples were stored at −80°C until analysis by qRT-PCR, at which time they were diluted to a final volume of 120 μl. qRT-PCR primer sequences are shown in Table 1. Primers, designed using the Primer3 program (56), were evaluated using Mfold (40, 80) and Oligo Analyzer version 3.0 (Integrated DNA Technologies, Inc.). Primer concentrations were determined by performing the optimization protocol recommended by the Rotor Gene system (real-time summary, version 1.7). One-step qRT-PCR, using 5 μl of template RNA per reaction, was performed as described previously (51) using a QuantiTect SYBR green RT-PCR kit and a Rotor-Gene 6000 instrument (Corbett Research). For each gene, qRT-PCR was performed for five biological replicates (five independently cultivated biofilms) from the top 30 μm of biofilms and five biological replicates from the bottom 30 μm of biofilms.
RNA extraction and qRT-PCR of acpP, rmf, PA4463, and ibpA genes.
qRT-PCR was also performed to assay local mRNA abundances of the acpP, rmf, PA4463, and ibpA genes. For these experiments, samples were laser catapulted into 35 μl of RLT lysis buffer (Qiagen), vortexed for 5 s, and then centrifuged before the addition of 310 μl RLT. Five microliters of carrier RNA and 1.7 × 104 transcript copies of luciferase (lucI) RNA (Promega) were added as a spike-in control. RNA extraction then proceeded with an RNeasy Microkit (Qiagen) as described by the microdissected cyrosection protocol. Following the addition of 20 units of RNasin Plus (Promega), RNA samples were treated with 0.8 μl DNase using a Turbo DNA-free kit (Ambion, Inc.) in 30-μl reaction mixtures for 20 min. All samples were stored at −80°C until analysis by qRT-PCR. Primers (shown in Table 1) were evaluated in silico using the IDT Oligoanalyzer tool and NCBI's primer blast tool. Primer sets were validated by determining efficiencies obtained from 10-fold serial dilution curves of RNA extracted from Pseudomonas aeruginosa cultured in biofilm. Efficiencies were calculated with the equation E = 10−1/slope − 1, with an E of 1 indicating exponential amplification of the product. For each primer set, efficiencies ranged from 1.00 to 1.03, and dilution curves gave correlation coefficients (R2) of ≥0.98. One-step qRT-PCRs were assembled according to the Rotor-Gene SYBR green RT-PCR handbook (Qiagen). Cycling parameters used on the Rotor-Gene 6000 (Corbett Research) were as follows: one cycle of 55°C for 12 min, denaturation at 95°C for 6 min, and then 40 cycles of 95°C for 5 s and 60°C for 12 s. Data were acquired during the 60°C annealing step, and a melt curve was generated at the conclusion of each run. For each gene and for each biofilm subsection, five biological replicates from different biofilms were assayed in duplicate or triplicate. Intraassay variation, as calculated from threshold cycle (CT) values, was less than 3%. Every assay contained negative controls lacking reverse transcriptase or a template to control for potential DNA contamination.
qRT-PCR data analysis.
Biological sample size was standardized by laser microdissection of equal volumes of biofilm, and a common threshold was set for analysis of all genes. An exogenous reference RNA (spiked-in lucI RNA) was selected as a calibrator to control for sample variability (18, 36, 79). Heat maps were created using Genesis (65) and display log2 background-corrected signal intensity of microarray data and 2−ΔCT RT-PCR data, where ΔCT = (CT gene of interest) − (CT lucI control) (59). For bar graphs displaying average expression levels, the 2−ΔCT qRT-PCR data were scaled by a factor of 108 (36). Samples producing erroneous melt curves or no amplification were considered to be below the limit of detection (BD). Normalized values were plotted in Excel for visualization of the difference in expression levels between the tops and bottoms of the biofilms (n = 5 samples each). Fold change analysis, calculated by REST 2009 software (52), indicated no significant difference in mRNA levels between the top and bottom biofilm subsections for the ibpA (P = 0.832), rmf (P = 0.206), and PA4463 (P = 0.165) genes but did indicate a significant difference in acpP gene abundance between the top and bottom of the biofilm (P = 0.001).
RNA extraction and poly(A) tailing for microarray analysis.
For each biological replicate, nine LCM samples, obtained by catapulting into 36 μl RLT buffer (Qiagen), were combined into one 1.5-ml microcentrifuge tube, resulting in a total captured area of 1.5 × 107 μm3. RNA was extracted with an RNeasy Microkit (Qiagen) following the microdissected cyrosection protocol. Turbo DNase (Ambion) treatment was performed to remove any traces of residual DNA. For this, 0.8 μl of Turbo DNase, 2 μl of 10× Turbo buffer, and 1 μl of 0.5× RNaseIn (Promega) were added to the 16-μl RNA sample. After a 20-min 37°C incubation, 3 μl of inactivation reagent was added and the RNA was recovered according to the manufacturer's instructions. A 1.5-μl aliquot was removed for RNA quantitation with a RiboGreen (Invitrogen) assay. The RNA was poly(A) tailed using a poly(A) polymerase tailing kit (Epicentre) according to the manufacturer's instructions with a 20-μl final reaction volume and a 25-min incubation time. The reaction was stopped by freezing at −20°C. Poly(A)-tailed RNA was then cleaned according to the RNA cleanup protocol in the RNeasy Microkit. The extraction and processing method was validated by evaluation of RNA integrity of representative samples using the Bioanalyzer 2100 Pico chip. Five biological samples were obtained from the top 30 μm and from the bottom 30 μm of five different biofilms, which were cultured independently.
RNA amplification for microarray analysis.
A QuantiTect whole-transcriptome kit (Qiagen), which uses a method based on multiple displacement amplification, was used to uniformly amplify the poly(A)-tailed RNA samples. One microliter of a 1:2,000,000 dilution of poly(A) controls (Affymetrix) was added, and amplification proceeded per the manufacturer's instructions. Briefly, cDNA was reverse transcribed from the poly(A)-tailed RNA using random and oligo(dT) primers and then ligated and amplified using REPLI-g DNA polymerase with an 8-h (high-yield reaction) incubation time. The resulting double-stranded DNA was purified using a QIAamp DNA Minikit (Qiagen). For this, the DNA was brought to a 200-μl volume with nuclease-free water before the addition of 200 μl of buffer AL and 200 μl of 100% ethyl alcohol (EtOH). The mixture was applied to a QIAamp column, washed with 500 μl buffer AW1, washed again with 500 μl buffer AW2, and eluted with 45 μl buffer EB following a 5-min incubation period. The purified DNA was then fragmented in One-Phor-All buffer (GE Healthcare) with DNase 1 (Amersham) for 1 min at 37°C according to an experimentally determined formula, y = 0.287x + 1.253, where y is the volume of 1:10-diluted DNase enzyme (in microliters) and x is the amount of DNA to be fragmented (in micrograms). The reaction was stopped by incubation at 98°C for 10 min. DNA fragment size was then assessed with an Agilent bioanalyzer using a DNA chip. Fragmented double-stranded cDNA was terminally labeled by incubation with 10 μl of 5× reaction buffer, 2 μl GeneChip DNA labeling reagent (Affymetrix), and 2 μl terminal deoxynucleotidyl transferase (Promega) for 60 min at 37°C. Two microliters of 0.5 M EDTA was added to stop the reaction.
Hybridization, scanning, and analysis.
Labeled cDNA was hybridized for 16 h at 50°C with constant rotation to Affymetrix Pseudomonas aeruginosa microarrays (part 900640). Microarrays were washed and stained using a GCOS Fluidics Station 450 and scanned with an Affymetrix 7G scanner. Affymetrix GCOS v1.4 was used to generate CEL and CHP files. CEL files were imported into FlexArray v1.5.1 for quality control (QC) and data analysis (8).
Microarray data accession number.
Microarray data have been deposited in the GEO database with accession number GSE34762.
RESULTS AND DISCUSSION
Transcriptomic analysis of biofilm subpopulations.
LCM was used to isolate and capture biofilm subpopulations from the top 30 μm and bottom 30 μm of the P. aeruginosa PAO1 biofilms (average thickness of ∼350 μm). High-quality RNA, confirmed by the integrity of the rRNAs (see Fig S1 in the supplemental material), was purified from the samples for Affymetrix microarray analysis. In order to obtain sufficient nucleic acid for microarray hybridization from LCM samples, RNA amplification was necessary. Several RNA amplification strategies were tested, and the QuantiTect whole-transcriptome amplification protocol was ultimately chosen. In this approach, following reverse transcription, cDNA fragments are ligated together, forming long cDNA chimeras. The cDNA chimeras are then amplified by DNA multiple-strand displacement (17). This strategy helps reduce amplification bias that may be caused by variable transcript size. However, as amplification bias is inherent in any RNA amplification approach, several methods were used to assess and reduce this bias. Diluted Affymetrix spike-in controls were added to the samples and amplified along with the RNA. Five biological replicates were performed for each biofilm subsection (top and bottom of biofilms), and the mean and variance for each gene expression level were evaluated (see Table S1 in the supplemental material). In addition, eight genes with differing expression levels, as assayed by microarrays, were further analyzed by using LCM and qRT-PCR (36), a technique that does not require RNA amplification prior to analysis. Fig. S2A in the supplemental material is a heat map of individual samples for the eight genes as well as for the acpP gene and 16S rRNA, showing the relative signal intensities of the mRNA abundances and comparing the microarray and qRT-PCR approaches. Fig S2B in the supplemental material shows the average relative signal intensity assayed by both techniques. The results show that PA0059 (osmC), PA2146, and PA3531 (bfrB) genes had high mRNA abundances at the top of the biofilms and very low levels of expression at the bottom of the biofilm, as assayed by both techniques. PA4220, PA4823, and PA4865 (ureA) genes had very low expression levels at both the top and the bottom of the biofilms, as assayed by microarray and confirmed by qRT-PCR. PA0997 and PA1089 had intermediate levels of expression at the top of the biofilms and very low expression levels at the bottom of the biofilms. The results show consistency between the microarray approach, which requires RNA amplification, and the qRT-PCR studies of selected genes, for which no RNA amplification is necessary prior to analysis but only a limited number of genes may be analyzed.
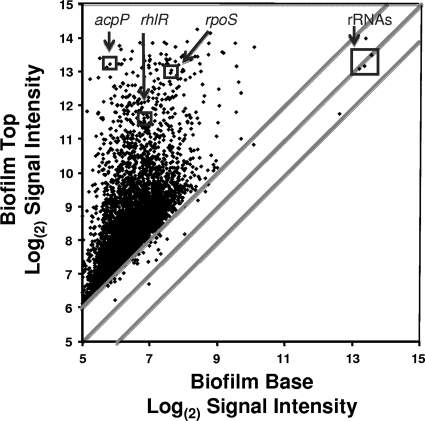
The results of microarray analysis with the mean (n = 5) expression level for each gene plotted for the top 30 μm of the biofilm versus the bottom 30 μm of the biofilms are shown in Fig. 1 and in Table S1 in the supplemental material. Highlighted in Fig. 1 are genes that we analyzed previously for local mRNA abundances in biofilms by using LCM and qRT-PCR (36, 51). Previously, we demonstrated that mRNA for the housekeeping gene involved in fatty acid biosynthesis, the acpP gene, was orders of magnitude greater at the top than in the middle or at the bottom of the biofilms. The microarray results here are consistent with those results and show that acpP mRNA had an approximately 120-fold-greater signal intensity at the top of the biofilm than at the biofilm base. Similarly, mRNA abundances for rpoS and rhlR genes, previously shown to be higher at the top than the bottom of the biofilm (51) also showed much higher mRNA abundances at the top of the biofilms by using microarray analysis. The abundance of 16S rRNA, shown previously to be at relatively similar levels at both the top and bottom of the biofilms (36, 51), also showed similar abundances by the microarray studies observed here.
Fig 1.
Microarray analysis of the P. aeruginosa biofilm transcriptome, with the log(2) signal intensity for each gene transcript plotted for the top of the biofilm versus that for the bottom of the biofilm (n = 5 for each gene). For averages and variances of the signal intensity for each gene, see Table S1 in the supplemental material.
Metabolic activity of biofilm cell subpopulations.
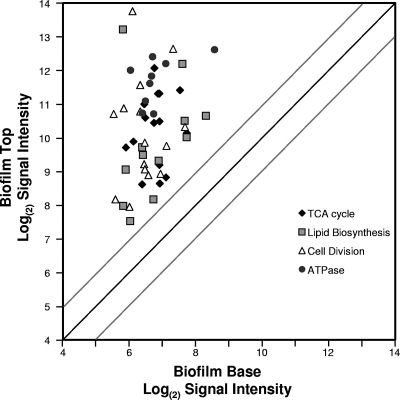
The microarray results shown in Fig. 1 demonstrate a skew in the data, with most genes showing much higher mRNA abundances at the top of the biofilms than at the bottom. This skew suggests that cells at the top of the biofilm have higher overall levels of gene expression and metabolic activity than those cells at the bottom of the biofilms. In support of this, genes showing very high mRNA abundances at the top of the biofilms but little mRNA deep in the biofilms are genes involved in cell growth and general metabolism. Figure 2 and Table S2 in the supplemental material show mRNA abundance data for four groups of genes associated with housekeeping functions, including ATP metabolism (atpI-atpC operon), tricarboxylic acid (TCA) cycle enzymes, genes for proteins involved in cell division (fts and min operons), and genes coding for enzymes involved in fatty acid biosynthesis. Since the half-life of most mRNAs is short, the high abundance of transcripts for housekeeping functions suggests that most cells near the top of the biofilms are metabolically active for growth and cell division. Cells at the bottom of these biofilms are not expressing high levels of these genes and therefore may be in a slow-growth state or even metabolically dormant.
Fig 2.
Microarray results of mRNA signal intensities of metabolic housekeeping genes plotted for the top of the biofilm versus the those for the bottom of the biofilms. Specific genes are listed in Table S2 in the supplemental material, along with the averages and variances of the microarray results (n = 5). The dark diagonal line is for genes with equal mRNA abundances at the top and bottom of the biofilms. The gray lines show the results of a 2-fold difference in mRNA abundance for the top versus the bottom of the biofilms.
Selective GFP labeling of active and dormant subpopulations of P. aeruginosa biofilms.
To characterize further the metabolic activity of cells at different regions within the biofilms, we used selective green fluorescent protein (GFP) labeling of active and inactive cells. P. aeruginosa MH475, which contains an l-arabinose-inducible gfp gene, was first cultured planktonically in medium without arabinose. Subculturing in medium with arabinose caused initially nonfluorescent cells to become fluorescent over the next several hours (see Fig. S3A in the supplemental material). After 12 h of growth under inducing conditions, 96% of the cells were scored as GFP+ by using the flow cytometry assay described above. When a stationary-phase planktonic culture was induced with arabinose, there was little induction of GFP (see Fig. S3A); only 7% of the bacteria were scored as GFP+, demonstrating that most cells had entered an inactive state. The observation of a few bright cells is interesting and suggests that even under conditions in which most of the bacteria appeared inactive, there was a small subpopulation that was actively synthesizing new protein. In the converse experiment, cells were originally cultured in the presence of arabinose to obtain fluorescent cells. These cells were then transferred to medium lacking the inducing agent. As the number of cells increased, the GFP was diluted and/or degraded in the daughter cells, resulting in a rapid loss of fluorescence. This population, which was initially scored as containing 93% GFP+ cells, was completely devoid of bright cells after 8 h of growth (see Fig. S3B). The time constant for the decay of fluorescence in this population was 1.3 ± 0.4 h−1 (mean ± SE), which is similar to the specific growth rate of the organism in this medium, 1.2 ± 0.03 h−1 (mean ± SE) (55).
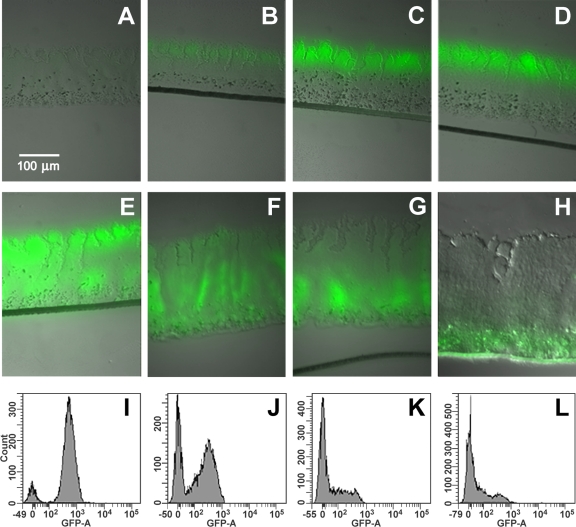
Similar experiments were performed with P. aeruginosa MH475 colony biofilms by transfer to medium with or lacking arabinose. When biofilms were first cultured in the absence of l-arabinose for 48 h and then induced with arabinose, approximately 25% of the cells were scored as GFP+ (Fig. 3A to D). Although the intensity of the fluorescence increased over time, the percentage of cells did not increase significantly upon prolonged incubation with arabinose. The GFP+ cells formed a 65-μm-thick band along the biofilm periphery at the air-biofilm interface. In a previous study, we analyzed gfp mRNA abundances of similar biofilms by LCM and qRT-PCR (36). Very little gfp mRNA was observed in the middle or bottom portions of the biofilm, indicating that the lack of fluorescence in the deeper regions of the biofilms was due to little gfp expression in these regions of the biofilms and not due to lack of activation of the GFP by depletion of oxygen. While the cells at the base of the biofilms are nonfluorescent, they are still viable. Microscopic direct counts (DC) and CFU counts were similar and indicated that 95% ± 6% of all cells remained viable. CFU counts with and without gentamicin were also similar, indicating that the cells in the deeper portion of the biofilm maintained the gfp genetic construct.
Fig 3.
Selective GFP labeling of active and inactive bacteria. (A) Colony biofilm of P. aeruginosa MH475 without induction by arabinose. Colony biofilm after 48 h without arabinose and then induced with 2% arabinose for (B) 4 h, (C) 12 h, or (D) 24 h. (E) Colony biofilm of P. aeruginosa MH475 cultured in the presence of 2% arabinose for 48 h. (E to H) Colony biofilms cultured in the presence of 2% arabinose for 48 h and then transferred to medium without arabinose for an additional (F) 24 h, (G) 36 h, or (H) 48 h. FACS analysis of l-arabinose-cultured biofilms, followed by the removal of arabinose and incubational for an additional (I) 0 h, (J) 24 h, (K) 36 h, (L) 48 h.
To label the inactive cells with GFP, colony biofilms were first cultured in the continuous presence of arabinose for 48 h, resulting in most cells being scored as GFP+ by fluorescence-activated cell sorter (FACS) analysis (Fig. 3E and I). These biofilms were then subcultured in medium lacking l-arabinose. As the biofilms continued to grow without an inducing agent, the GFP was diluted due to distribution of the GFP to each daughter cell or by degradation. In contrast to the results with the planktonic cultures, the loss of fluorescence across the biofilm population was not uniform. Instead, fluorescence first diminished in the upper regions of the biofilm (Fig. 3E to H). After 48 h of growth in the absence of the inducer, a GFP+ subpopulation was located predominantly along the biofilm-substratum interface at the base of the biofilm (Fig. 3H). FACS showed two peaks, representing fluorescent and nonfluorescent cells (Fig. 3I to L). In biofilms, the percentage of cells scoring as GFP+ by flow cytometry in arabinose depletion experiments decreased from 85%, when the cells were first transferred to medium lacking the inducer, to less than 1% after 72 h in the absence of arabinose. The time constant for this loss of fluorescence was 0.05 ± 0.01 h−1, which was 26 times slower than the decay observed for planktonic cells for the overall population. However, this growth rate is the average for the entire population, which contains both actively dividing and inactive cells. Taken together with the microarray analysis, which showed very little mRNA for housekeeping genes, these results demonstrate that these biofilms contained at least two distinct subpopulations, an actively growing population of cells near the air-biofilm interface and a population of cells with a very low growth rate in the deeper regions of the biofilms.
Response of cells to oxygen depletion in thick P. aeruginosa biofilms.
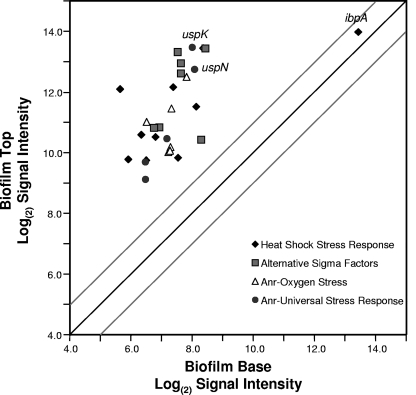
One explanation for the low gene expression activity and low growth rate of cells in the deeper regions of these biofilms is that these cells are oxygen limited. Previous oxygen microelectrode measurements of similar P. aeruginosa colony biofilms have shown that the oxygen concentration progressively decreases from the air interface to the bottom of the biofilm (10, 71). Other studies have suggested that P. aeruginosa may be exposed to anoxic or hypoxic conditions in the cystic fibrosis (CF) lung environment (26, 60, 74, 77). Several transcriptome studies have characterized gene induction during microaerophilic and anaerobic respiratory conditions for P. aeruginosa (3, 53, 61, 66). Microarray results here show that some of the most abundant mRNA transcripts at the top of these biofilms are for genes that are induced by oxygen limitation stress (Fig. 4) (see Table S3 in the supplemental material). Among these are the low oxygen sensor/regulator Anr and several genes with Anr-regulatory boxes in their promoter regions, such as the arcDABC arginine metabolism genes and the oprG gene for the outer membrane protein. These genes were shown to be highly upregulated during the transition from aerobic to hypoxic conditions (66). The upregulation of genes responsive to low oxygen stress at the top of these biofilms is not initially intuitive, since cells at the biofilm periphery are in close proximity to the air-biofilm interface. It would be reasonable to expect that cells in the bottom fraction would be more likely to respond to oxygen limitation than cells in the top fraction. However, these results suggest that cells at the bottom of the biofilms have experienced prolonged anoxia, whereas the cells at the top of the biofilm may be in a transition state from aerobic to hypoxic conditions. This is consistent with the microarray results of Trunk et al. (66), which characterized gene induction during the transition from aerobic to hypoxic conditions. While genes regulated by Anr are among the most abundant mRNA transcripts at the top of the biofilm, not all Anr-regulated genes are upregulated in the present study. Among the genes not showing increased expression at the top of these biofilm are genes involved in nitrate/nitrite reduction (see Table S1 in the supplemental material).
Fig 4.
Microarray results for mRNA amounts of stress-responsive genes. The signal intensities for individual genes with means and variances are shown in Table S3 in the supplemental material. The dark diagonal line would indicate genes with equal mRNA abundances at the top and bottom of the biofilms. The gray lines show the results of a 2-fold difference in mRNA abundance for the top versus the bottom of the biofilms.
Expression of stress-responsive genes.
The universal stress-responsive (usp) genes are regulated by Anr (66). Transcripts for these genes, including PA1789, PA3309, PA4328, PA4352, and PA5027, are highly abundant at the top of the biofilms (Fig. 4) (see Table S3 in the supplemental material). The two most abundant usp mRNA transcripts are the uspK (PA3309) and uspN (PA4352) genes. Proteomic analysis of whole biofilms showed that UspK and UspN were abundant in P. aeruginosa biofilms (50). The previous proteomics study comparing two different P. aeruginosa strains in a flow-through biofilm system demonstrated that uspK and uspN induction and stress response may be a general feature of P. aeruginosa biofilms. PA3309 (uspK) encodes a Usp-type universal stress protein required for pyruvate fermentation and long-term anaerobic survival in P. aeruginosa (61). Schreiber et al. (61) used a PA3309 promoter-GFP reporter gene fusion integrated into the P. aeruginosa chromosome to study the spatial distribution of PA3309 expression in flow-cell biofilms. In contrast to the results here, where expression was highest in the top fraction, they detected expression in the deeper layers of the biofilm. However, the geometry of these two biofilm systems differs, which may account for the differences in layers where this gene is expressed. The three studies (references 50 and 61 and the present study) indicate that biofilms cultivated using different systems contain oxygen-limited zones and result in the expression of Anr-regulated stress response genes. The location of these zones may differ depending on the biofilm cultivation system.
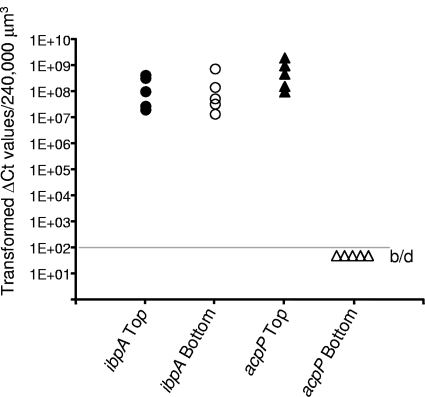
In addition to Anr-controlled stress response, other stress-responsive genes are expressed at high levels at the top of these P. aeruginosa biofilms. Alternative sigma factors, including mRNA for the rpoH heat shock sigma factor, are abundant at the top of the biofilms (Fig. 4) (see Table S3 in the supplemental material). Our previous proteomics study using whole biofilms showed high protein amounts of chaperones and proteases generally associated with the heat shock response (50). The transcriptomics results here are consistent with those of the proteomics study and also indicate that proteases and chaperones regulated by RpoH and misfolded protein stress are highly induced at the top of these biofilms. The mRNA transcript for the ibpA gene was the most abundant transcript at both the top and bottom of the biofilms (Fig. 4). IbpA has been shown to be upregulated in P. aeruginosa whole biofilms in a variety of different studies (21). IbpA plays a role in the heat shock response by binding misfolded proteins for delivery to refolding or degradation proteins (54). Lindner et al. (38) demonstrated that in Escherichia coli, IbpA localizes to the cell pole and proposed that IbpA may bind misfolded proteins for segregation into the aging members of the population. To verify local mRNA abundances of the ibpA gene, we performed LCM and qRT-PCR on the ibpA transcripts from the top and bottom of the P. aeruginosa biofilms. The results show high ibpA mRNA abundance at both the top and bottom of the biofilms (Fig. 5). As a control, we performed an analysis for but did not detect mRNA for the acpP housekeeping gene in the bottom of the biofilms from the same samples used to test ibpA mRNA levels. Since both microarrays and qRT-PCR measure RNA abundances, it is difficult to discern whether the high ibpA mRNA abundance at the bottom of these biofilms is due to local gene expression or to unusual stability of the ibpA transcript.
Fig 5.
qRT-PCR analysis of ibpA mRNA levels at the top (filled circles) and bottom (open circles) of P. aeruginosa biofilms. Samples used to measure ibpA mRNA were also used to measure acpP mRNA, which showed a high abundance at the top of the biofilms (filled triangles) but was below the limit of detection (b/d) at the bottom of the biofilms (open triangles).
Expression of genes involved in stationary phase and quorum sensing.
Previously, we characterized the local expression of the rpoS stationary-phase sigma factor and the rhlR quorum-sensing (QS) regulator in P. aeruginosa biofilms (51). Based on those results and on the local abundances of the 16S rRNA, we proposed that cells at the top of the biofilms were in a transition state from exponential to stationary phase. Here, using microarrays, we expanded on those results by analyzing local mRNA abundances of genes proposed to be in the RpoS or QS regulons (62, 63). The QS regulators, the rhlR and lasR genes, and genes proposed to be members of these regulons had high mRNA abundances at the top of the biofilms and very low abundances at the bottom of the biofilm (see Fig S4 and Table S4 in the supplemental material). While it may be expected that QS and stationary phase would be properties of cells deeper in the biofilms, where cells are aging and cell density may be the greatest, these processes appear to be occurring at the top of thick P. aeruginosa biofilms. The expression of the rpoS and QS regulons at the top of the biofilm likely reflects the transition of these cells to stationary phase as the cells become oxygen stressed. Little QS and rpoS regulon expression in the deep region of these biofilms reflects the overall low metabolic activity of those cells.
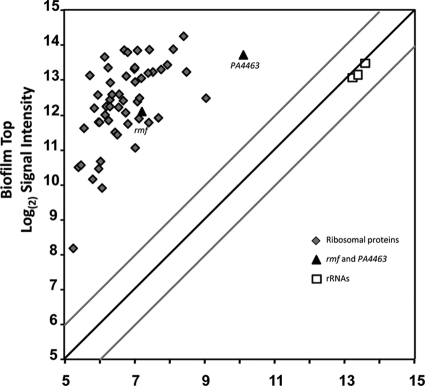
The ribosome and ribosome hibernation factors.
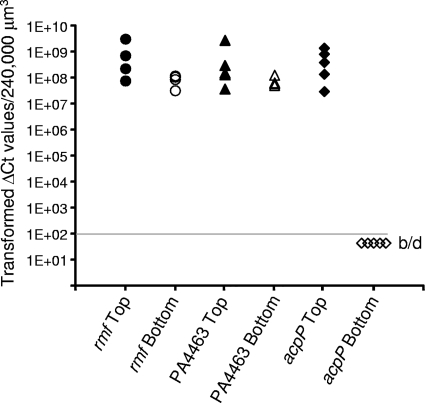
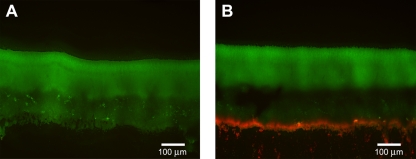
The ribosomal RNAs remain at relatively constant levels throughout these biofilms, suggesting that even the dormant cells maintain a minimum number of ribosomes per cell (51). The results here are consistent with those of our previous studies and show approximately equivalent amounts of the 23S, 16S, and 5S rRNAs at the top and the bottom of the biofilms (Fig. 6) (see Table S5 in the supplemental material). In contrast, mRNAs for the ribosomal proteins are among the most abundant transcripts at the top of the biofilms and are generally two orders of magnitude greater in abundance there than at the bottom of the biofilms (Fig. 6). Therefore, ribosomes are synthesized by the metabolically active cells at the top of the biofilms. However, since rRNA is rapidly degraded if it is not associated with a ribosome (19), the ribosomes and rRNAs are stably maintained in the metabolically dormant cells at the bottom of the biofilms. These cells may be capable of resuscitation once the appropriate nutrients or space becomes available. Interestingly, an mRNA transcript that is abundant at the bottom of the biofilms (in addition to large amounts at the top of the biofilms) is that for PA4463. PA4463 is homologous to the ribosome-associated inhibitor RaiA of E. coli. Along with ribosome modulation factor, Rmf, RaiA is involved in dimerizing of ribosomes into a 100S unit, maintaining the ribosomes in a stable but hibernating state (78). LCM and qRT-PCR experiments confirmed the local abundance of PA4463 mRNA transcripts at the bottom of the biofilms and also show that mRNA for the rmf gene is above that of the acpP housekeeping gene (Fig. 7). These protein products may be important in maintaining viability of the dormant subpopulation. To test this possibility, we constructed deletion mutations of the rmf and PA4463 genes and assayed the phenotype of cells lacking these hibernation factors. The wild-type and mutant strains were cultured in colony biofilms and stained with propidium iodide, which is used to detect cells with compromised membrane integrity. The wild-type cells as well as the PA4463 mutant cells showed very little propidium iodide uptake, indicating that most cells with a PA4463 deletion remain intact (Fig. 8A). However, the rmf mutants consistently showed a band of propidium iodide staining for cells at the bottom of the biofilms (Fig. 8B), suggesting that the rmf gene may be required for maintenance of cell integrity in the older dormant cells in the biofilm population.
Fig 6.
Microarray results for mRNA amounts of ribosomal proteins (diamonds), ribosomal RNAs (squares), and ribosome hibernation factors (triangles). The dark diagonal line would indicate genes with equal mRNA abundances at the top and bottom of the biofilms. The gray lines show the results of a 2-fold difference in mRNA abundance for the top versus the bottom of the biofilms. Means and variances of the signal intensities are shown in Table S5 in the supplemental material.
Fig 7.
qRT-PCR analysis of rmf and PA4463 mRNA levels at the top and bottom of P. aeruginosa biofilms. Samples used to measure rmf and PA4463 mRNA were also used to measure acpP mRNA, which showed a high abundance at the top of the biofilms but was below the limit of detection (b/d) at the bottom of the biofilms.
Fig 8.
Propidium iodide (PI) staining of P. aeruginosa colony biofilms, Each strain constitutively expressed the GFP from plasmid pMF230 for visualization of the colonies. Cells were stained by placing colony biofilms on filters containing PI for 30 min, as described in Materials and Methods. Biofilms were imaged following cryoembedding and thin sectioning. (A) Very little PI staining was observed for P. aeruginosa PAO1. (B) The P. aeruginosa Δrmf strain consistently showed a band of red fluorescence, indicative of PI uptake, along the bottom of the biofilm. The P. aeruginosa ΔPA4463 strain had variable levels of staining but generally did not stain with PI. PI uptake is often used to indicate cells with compromised cell membranes.
Dormant biofilm cells are more tolerant to antibiotics than active cells.
Dormant cells in P. aeruginosa biofilms should be less susceptible to killing by antibiotics than the actively growing population. Here, we used the selective GFP labeling approaches described above and FACS to determine the tolerance of the active and dormant cell subpopulations to tobramycin and ciprofloxacin. Coincident with the addition or removal of arabinose, cells were exposed to an antibiotic (10 mg ml−1 tobramycin or 1 mg ml−1 ciprofloxacin). Biofilms were then incubated in the presence of the antibiotic for 24 h. The biofilms were dispersed and sorted by FACS into fluorescent and nonfluorescent subpopulations. Each subpopulation was tested for viability as CFU of the dispersed subpopulations (Table 2). When the active population was selectively labeled with GFP by transferring biofilms from medium lacking arabinose to medium with arabinose, a 1.2- to 3.2-log reduction was observed for the GFP fluorescent cells following exposure to the antibiotic (Table 2). In contrast, no reduction in viability was observed for the nonfluorescent cells. In the converse experiment, in which the dormant cells were selectively labeled with GFP as described above, the addition of antibiotics resulted in a 1.7- and 2.8-log reduction for the nonfluorescent cells. No reduction in viability was observed for the GFP-labeled cells. We also performed similar experiments using a continuous-drip flow biofilm system and obtained similar results (Table 2). The results support the hypothesis that the subpopulation of inactive bacteria harbored in P. aeruginosa biofilms is less susceptible to killing by tobramycin and ciprofloxacin. However, the actively growing population remains sensitive to antibiotic killing.
Table 2.
Differential antibiotic susceptibility of P. aeruginosa biofilm subpopulations
| Biofilm type | Antibiotic | Log reduction |
P value | |
|---|---|---|---|---|
| GFP+ cells | GFP− cells | |||
| Arabinose-induced exptsa | ||||
| Colony | Ciprofloxacin | 1.21 ± 0.12 | −0.48 ± 0.21 | 0.002 |
| Colony | Tobramycin | 3.27 ± 0.61 | −1.02 ± 0.22 | 0.003 |
| Arabinose removal exptsb | ||||
| Colony | Ciprofloxacin | 0.39 ± 0.03 | 2.78 ± 0.06 | 0.001 |
| Colony | Tobramycin | 0.08 ± 0.12 | 2.78 ± 0.02 | 0.001 |
| Drip flow | Ciprofloxacin | 1.32 ± 0.47 | 2.48 ± 0.58 | 0.184 |
| Drip flow | Tobramycin | 0.63 ± 0.38 | 1.72 ± 0.28 | 0.019 |
The population was sorted by flow cytometry into GFP+ (active cell) and GFP− (dormant cell) fractions.
The population was sorted by flow cytometry into GFP+ (dormant cell) and GFP− (active cell) fractions.
The results here demonstrate that in thick P. aeruginosa biofilms, cells are in at least two distinct physiological states with respect to gene expression, growth status, and antibiotic tolerance. Cells at the top of these biofilms are active with respect to gene expression and protein biosynthetic activity (GFP production). The cells at the top of the biofilm, although active, are likely stressed, perhaps due to bacterial utilization of oxygen and transition to an hypoxic environment. Based on results showing induction of the QS and rpoS regulons, the cells at the top of the biofilm appear to be in transition to a growth status that resembles the stationary phase of planktonic bacteria. The cells at the bottom of the biofilm are generally inactive with respect to expression of most genes or protein production. However, the cells at the bottom of these biofilms are viable and contain relatively high levels of mRNA for the ibpA gene. Since IbpA is a chaperone and part of the protein refolding/degradation complex, it likely helps protect cells from protein misfolding in the aging cell subpopulation. The bottom of the biofilm also has high abundances of mRNA for the Rmf and PA4463 ribosome hibernation factors. One mechanism for maintaining viability in these dormant cells is to inactivate ribosomes in the dimerized state, with Rmf and PA4463, keeping protein synthesis in an inactive state until proper nutrients or space becomes available. Presently, it is not clear if these genes that show high mRNA abundance at the bottom of the biofilms are actively expressed at the biofilm base or if they are expressed by the cells entering stationary phase, and the mRNA maintained by unusually long half-lives of these specific transcripts. The biofilm subpopulation with a low growth rate is tolerant to the antibiotics ciprofloxacin and tobramycin, while the faster-growing population is more susceptible to killing by these antibiotics, suggesting that while bacteria growing in biofilms are more resistant to antibiotics than those grown planktonically, it is only subpopulations of the biofilms that have increased antibiotic resistances.
Supplementary Material
ACKNOWLEDGMENTS
This work was supported by Public Health Service grant AI-094268 from the National Institute of Allergy and Infectious Diseases (M.J.F.). Microscopy equipment was purchased with funds provided by the M. J. Murdock Charitable Trust and the NSF Major Research Instrumentation program. We acknowledge the Montana State University INBRE Functional Genomics Core Facility, which is supported by NIH grant P20-RR16455.
Footnotes
Published ahead of print 17 February 2012
Supplemental material for this article may be found at http://jb.asm.org/.
REFERENCES
- 1. Alhede M, et al. 2009. Pseudomonas aeruginosa recognizes and responds aggressively to the presence of polymorphonuclear leukocytes. Microbiology 155: 3500– 3508 [DOI] [PubMed] [Google Scholar]
- 2. Allegrucci M, Sauer K. 2007. Characterization of colony morphology variants isolated from Streptococcus pneumoniae biofilms. J. Bacteriol. 189: 2030– 2038 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Alvarez-Ortega C, Harwood CS. 2007. Responses of Pseudomonas aeruginosa to low oxygen indicate that growth in the cystic fibrosis lung is by aerobic respiration. Mol. Microbiol. 65: 153– 165 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Anderl JN, Franklin MJ, Stewart PS. 2000. Role of antibiotic penetration limitation in Klebsiella pneumoniae biofilm resistance to ampicillin and ciprofloxacin. Antimicrob. Agents Chemother. 44: 1818– 1824 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Anwar H, Costerton JW. 1990. Enhanced activity of combination of tobramycin and piperacillin for eradication of sessile biofilm cells of Pseudomonas aeruginosa. Antimicrob. Agents Chemother. 34: 1666– 1671 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Balaban NQ, Merrin J, Chait R, Kowalik L, Leibler S. 2004. Bacterial persistence as a phenotypic switch. Science 305: 1622– 1625 [DOI] [PubMed] [Google Scholar]
- 7. Baty AM, III, et al. 2000. Differentiation of chitinase-active and non-chitinase-active subpopulations of a marine bacterium during chitin degradation. Appl. Environ. Microbiol. 66: 3566– 3573 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Blazejczyk M, Miron M, Nadon R. 2007. FlexArray: a statistical data analysis software for gene expression microarrays. Génome Québec, Montreal, Canada: http://www.gqinnovationcenter.com/services/bioinformatics/flexarray/index.aspx [Google Scholar]
- 9. Boles BR, Thoendel M, Singh PK. 2004. Self-generated diversity produces “insurance effects” in biofilm communities. Proc. Natl. Acad. Sci. U. S. A. 101: 16630– 16635 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Borriello G, et al. 2004. Oxygen limitation contributes to antibiotic tolerance of Pseudomonas aeruginosa in biofilms. Antimicrob. Agents Chemother. 48: 2659– 2664 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Burns JL, et al. 2001. Longitudinal assessment of Pseudomonas aeruginosa in young children with cystic fibrosis. J. Infect. Dis. 183: 444– 452 [DOI] [PubMed] [Google Scholar]
- 12. Chai Y, Chu F, Kolter R, Losick R. 2008. Bistability and biofilm formation in Bacillus subtilis. Mol. Microbiol. 67: 254– 263 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Choi K-H, Schweizer H. 2005. An improved method for rapid generation of unmarked Pseudomonas aeruginosa deletion mutants. BMC Microbiol. 5: 30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Choi KH, et al. 2005. A Tn7-based broad-range bacterial cloning and expression system. Nat. Methods 2: 443– 448 [DOI] [PubMed] [Google Scholar]
- 15. Costerton JW, Lewandovski Z, Caldwell DE, Korber DR, Lappin-Scott HM. 1995. Microbial biofilms. Annu. Rev. Microbiol. 49: 711– 745 [DOI] [PubMed] [Google Scholar]
- 16. Costerton JW, Stewart PS, Greenberg EP. 1999. Bacterial biofilms: a common cause of persistent infections. Science 284: 1318– 1322 [DOI] [PubMed] [Google Scholar]
- 17. Dean FB, et al. 2002. Comprehensive human genome amplification using multiple displacement amplification. Proc. Natl. Acad. Sci. U. S. A. 99: 5261– 5266 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. DeCoste NJ, Gadkar VJ, Filion M. 2011. Relative and absolute quantitative real-time PCR-based quantifications of hcnC and phlD gene transcripts in natural soil spiked with Pseudomonas sp. strain LBUM300. Appl. Environ. Microbiol. 77: 41– 47 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Deutscher MP. 2003. Degradation of stable RNA in bacteria. J. Biol. Chem. 278: 45041– 45044 [DOI] [PubMed] [Google Scholar]
- 20. Elowitz MB, Levine AJ, Siggia ED, Swain PS. 2002. Stochastic gene expression in a single cell. Science 297: 1183– 1186 [DOI] [PubMed] [Google Scholar]
- 21. Folsom JP, et al. 2010. Physiology of Pseudomonas aeruginosa in biofilms as revealed by transcriptome analysis. BMC Microbiol. 10: 294. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Gefen O, Balaban NQ. 2009. The importance of being persistent: heterogeneity of bacterial populations under antibiotic stress. FEMS Microbiol. Rev. 33: 704– 717 [DOI] [PubMed] [Google Scholar]
- 23. Gefen O, Gabay C, Mumcuoglu M, Engel G, Balaban NQ. 2008. Single-cell protein induction dynamics reveals a period of vulnerability to antibiotics in persister bacteria. Proc. Natl. Acad. Sci. U. S. A. 105: 6145– 6149 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Haagensen JA, et al. 2007. Differentiation and distribution of colistin- and sodium dodecyl sulfate-tolerant cells in Pseudomonas aeruginosa biofilms. J. Bacteriol. 189: 28– 37 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Hansen SK, et al. 2007. Characterization of a Pseudomonas putida rough variant evolved in a mixed-species biofilm with Acinetobacter sp. strain C6. J. Bacteriol. 189: 4932– 4943 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Hassett DJ, et al. 2009. Pseudomonas aeruginosa hypoxic or anaerobic biofilm infections within cystic fibrosis airways. Trends Microbiol. 17: 130– 138 [DOI] [PubMed] [Google Scholar]
- 27. Hasty J, Pradines J, Dolnik M, Collins JJ. 2000. Noise-based switches and amplifiers for gene expression. Proc. Natl. Acad. Sci. U. S. A. 97: 2075– 2080 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Hentzer M, Eberl L, Givskov M. 2005. Transcriptome analysis of Pseudomonas aeruginosa biofilm development: anaerobic respiration and iron limitation. Biofilms 2: 37– 62 [Google Scholar]
- 29. Huang C, Yu FP, McFeters G, Stewart P. 1995. Nonuniform spatial patterns of respiratory activity within biofilms during disinfection. Appl. Environ. Microbiol. 61: 2252– 2256 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Huang CT, Xu KD, McFeters GA, Stewart PS. 1998. Spatial patterns of alkaline phosphatase expression within bacterial colonies and biofilms in response to phosphate starvation. Appl. Environ. Microbiol. 64: 26– 1531 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Jesaitis AJ, et al. 2003. Compromised host defense on Pseudomonas aeruginosa biofilms: characterization of neutrophil and biofilm interactions. J. Immunol. 171: 4329– 4339 [DOI] [PubMed] [Google Scholar]
- 32. Kaneko Y, Thoendel M, Olakanmi O, Britigan BE, Singh PK. 2007. The transition metal gallium disrupts Pseudomonas aeruginosa iron metabolism and has antimicrobial and antibiofilm activity. J. Clin. Invest. 117: 877– 888 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Keren I, Shah D, Spoering A, Kaldalu N, Lewis K. 2004. Specialized persister cells and the mechanism of multidrug tolerance in Escherichia coli. J. Bacteriol. 186: 8172– 8180 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Koh KS, et al. 2007. Phenotypic diversification and adaptation of Serratia marcescens MG1 biofilm-derived morphotypes. J. Bacteriol. 189: 119– 130 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Kussell E, Kishony R, Balaban NQ, Leibler S. 2005. Bacterial persistence: a model of survival in changing environments. Genetics 169: 1807– 1814 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Lenz AP, Williamson KS, Pitts B, Stewart PS, Franklin MJ. 2008. Localized gene expression in Pseudomonas aeruginosa biofilms. Appl. Environ. Microbiol. 74: 4463– 4471 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Lewis K. 2007. Persister cells, dormancy and infectious disease. Nat. Rev. Microbiol. 5: 48– 56 [DOI] [PubMed] [Google Scholar]
- 38. Lindner AB, Madden R, Demarez A, Stewart EJ, Taddei F. 2008. Asymmetric segregation of protein aggregates is associated with cellular aging and rejuvenation. Proc. Natl. Acad. Sci. U. S. A. 105: 3076– 3081 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Lyczak JB, Cannon CL, Pier GB. 2002. Lung infections associated with cystic fibrosis. Clin. Microbiol. Rev. 15: 194– 222 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Mathews DH, Sabina J, Zuker M, Turner DH. 1999. Expanded sequence dependence of thermodynamic parameters improves prediction of RNA secondary structure. J. Mol. Biol. 288: 911– 940 [DOI] [PubMed] [Google Scholar]
- 41. McAdams HH, Arkin A. 1997. Stochastic mechanisms in gene expression. Proc. Natl. Acad. Sci. U. S. A. 94: 814– 819 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. McEllistrem MC, Ransford JV, Khan SA. 2007. Characterization of in vitro biofilm-associated pneumococcal phase variants of a clinically relevant serotype 3 clone. J. Clin. Microbiol. 45: 97– 101 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Mikkelsen H, Duck Z, Lilley KS, Welch M. 2007. Interrelationships between colonies, biofilms, and planktonic cells of Pseudomonas aeruginosa. J. Bacteriol. 189: 2411– 2416 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Moriarty TF, Elborn JS, Tunney MM. 2007. Effect of pH on the antimicrobial susceptibility of planktonic and biofilm-grown clinical Pseudomonas aeruginosa isolates. Br. J. Biomed. Sci. 64: 101– 104 [DOI] [PubMed] [Google Scholar]
- 45. Mulcahy LR, Burns JL, Lory S, Lewis K. 2010. Emergence of Pseudomonas aeruginosa strains producing high levels of persister cells in patients with cystic fibrosis. J. Bacteriol. 192: 6191– 6199 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Nguyen D, Singh PK. 2006. Evolving stealth: genetic adaptation of Pseudomonas aeruginosa during cystic fibrosis infections. Proc. Natl. Acad. Sci. U. S. A. 103: 8305– 8306 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Nickel JC, Ruseska I, Wright JB, Costerton JW. 1985. Tobramycin resistance of Pseudomonas aeruginosa cells growing as a biofilm on urinary catheter material. Antimicrob. Agents Chemother. 27: 619– 624 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Nivens DE, Ohman DE, Williams J, Franklin MJ. 2001. Role of alginate and its O acetylation in formation of Pseudomonas aeruginosa microcolonies and biofilms. J. Bacteriol. 183: 1047– 1057 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Pamp SJ, Gjermansen M, Johansen HK, Tolker-Nielsen T. 2008. Tolerance to the antimicrobial peptide colistin in Pseudomonas aeruginosa biofilms is linked to metabolically active cells, and depends on the pmr and mexAB-oprM genes. Mol. Microbiol. 68: 3– 240 [DOI] [PubMed] [Google Scholar]
- 50. Patrauchan MA, Sarkisova SA, Franklin MJ. 2007. Strain-specific proteome responses of Pseudomonas aeruginosa to biofilm-associated growth and to calcium. Microbiology 153: 3838– 3851 [DOI] [PubMed] [Google Scholar]
- 51. Pérez-Osorio AC, Williamson KS, Franklin MJ. 2010. Heterogeneous rpoS and rhlR mRNA levels and 16S rRNA/rDNA (rRNA gene) ratios within Pseudomonas aeruginosa biofilms, sampled by laser capture microdissection. J. Bacteriol. 192: 2991– 3000 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Pfaffl MW, Horgan GW, Dempfle L. 2002. Relative expression software tool (REST) for group-wise comparison and statistical analysis of relative expression results in real-time PCR. Nucleic Acids Res. 30: e36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Platt MD, et al. 2008. Proteomic, microarray, and signature-tagged mutagenesis analyses of anaerobic Pseudomonas aeruginosa at pH 6.5, likely representing chronic, late-stage cystic fibrosis airway conditions. J. Bacteriol. 190: 2739– 2758 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Ratajczak E, Zietkiewicz S, Liberek K. 2009. Distinct activities of Escherichia coli small heat shock proteins IbpA and IbpB promote efficient protein disaggregation. J. Mol. Biol. 386: 178– 189 [DOI] [PubMed] [Google Scholar]
- 55. Roostalu J, Joers A, Luidalepp H, Kaldalu N, Tenson T. 2008. Cell division in Escherichia coli cultures monitored at single cell resolution. BMC Microbiol. 8: 68. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Rozen S, Skaletsky H. 2000. Primer3 on the WWW for general users and for biologist programmers. Methods Mol. Biol. 132: 365– 386 [DOI] [PubMed] [Google Scholar]
- 57. Sarkisova S, Patrauchan MA, Berglund D, Nivens DE, Franklin MJ. 2005. Calcium-induced virulence factors associated with the extracellular matrix of mucoid Pseudomonas aeruginosa biofilms. J. Bacteriol. 187: 4327– 4337 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58. Sauer K, Camper AK, Ehrlich GD, Costerton JW, Davies DG. 2002. Pseudomonas aeruginosa displays multiple phenotypes during development as a biofilm. J. Bacteriol. 184: 1140– 1154 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59. Schmittgen TD, Livak KJ. 2008. Analyzing real-time PCR data by the comparative C(T) method. Nat. Protoc. 3: 1101– 1108 [DOI] [PubMed] [Google Scholar]
- 60. Schobert M, Jahn D. 2010. Anaerobic physiology of Pseudomonas aeruginosa in the cystic fibrosis lung. Int. J. Med. Microbiol. 300: 549– 556 [DOI] [PubMed] [Google Scholar]
- 61. Schreiber K, et al. 2006. Anaerobic survival of Pseudomonas aeruginosa by pyruvate fermentation requires an Usp-type stress protein. J. Bacteriol. 188: 659– 668 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62. Schuster M, Hawkins AC, Harwood CS, Greenberg EP. 2004. The Pseudomonas aeruginosa RpoS regulon and its relationship to quorum sensing. Mol. Microbiol. 51: 973– 985 [DOI] [PubMed] [Google Scholar]
- 63. Schuster M, Lostroh CP, Ogi T, Greenberg EP. 2003. Identification, timing, and signal specificity of Pseudomonas aeruginosa quorum-controlled genes: a transcriptome analysis. J. Bacteriol. 185: 2066– 2079 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64. Stewart PS, Franklin MJ. 2008. Physiological heterogeneity in biofilms. Nat. Rev. Microbiol. 6: 199– 210 [DOI] [PubMed] [Google Scholar]
- 65. Sturn A, Quackenbush J, Trajanoski Z. 2002. Genesis: cluster analysis of microarray data. Bioinformatics 18: 207– 208 [DOI] [PubMed] [Google Scholar]
- 66. Trunk K, et al. 2010. Anaerobic adaptation in Pseudomonas aeruginosa: definition of the Anr and Dnr regulons. Environ. Microbiol. 12: 1719– 1733 [DOI] [PubMed] [Google Scholar]
- 67. Valle J, Vergara-Irigaray M, Merino N, Penades JR, Lasa I. 2007. σB regulates IS256-mediated Staphylococcus aureus biofilm phenotypic variation. J. Bacteriol. 189: 2886– 2896 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68. Waite RD, et al. 2006. Clustering of Pseudomonas aeruginosa transcriptomes from planktonic cultures, developing and mature biofilms reveals distinct expression profiles. BMC Genomics 7: 162. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69. Walters MC, III, Roe F, Bugnicourt A, Franklin MJ, Stewart PS. 2003. Contributions of antibiotic penetration, oxygen limitation, and low metabolic activity to tolerance of Pseudomonas aeruginosa biofilms to ciprofloxacin and tobramycin. Antimicrob. Agents Chemother. 47: 317– 323 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70. Warren AE, et al. 2011. Genotypic and phenotypic variation in Pseudomonas aeruginosa reveals signatures of secondary infection and mutator activity in certain cystic fibrosis patients with chronic lung infections. Infect. Immun. 79: 4802– 4818 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71. Werner E, et al. 2004. Stratified growth in Pseudomonas aeruginosa biofilms. Appl. Environ. Microbiol. 70: 6188– 6196 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72. Whitchurch CB, Tolker-Nielsen T, Ragas PC, Mattick JS. 2002. Extracellular DNA required for bacterial biofilm formation. Science 295: 1487. [DOI] [PubMed] [Google Scholar]
- 73. Whiteley M, et al. 2001. Gene expression in Pseudomonas aeruginosa biofilms. Nature 413: 860– 864 [DOI] [PubMed] [Google Scholar]
- 74. Worlitzsch D, et al. 2002. Effects of reduced mucus oxygen concentration in airway Pseudomonas infections of cystic fibrosis patients. J. Clin. Invest. 109: 317– 325 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75. Xu KD, Stewart PS, Xia F, Huang C-T, McFeters GA. 1998. Spatial physiological heterogeneity in Pseudomonas aeruginosa biofilm is determined by oxygen availability. Appl. Environ. Microbiol. 64: 4035– 4039 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76. Yildiz FH, Schoolnik GK. 1999. Vibrio cholerae O1 El Tor: identification of a gene cluster required for the rugose colony type, exopolysaccharide production, chlorine resistance, and biofilm formation. Proc. Natl. Acad. Sci. U. S. A. 96: 4028– 4033 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77. Yoon SS, et al. 2002. Pseudomonas aeruginosa anaerobic respiration in biofilms: relationships to cystic fibrosis pathogenesis. Dev. Cell 3: 593– 603 [DOI] [PubMed] [Google Scholar]
- 78. Yoshida H, Ueta M, Maki Y, Sakai A, Wada A. 2009. Activities of Escherichia coli ribosomes in IF3 and RMF change to prepare 100S ribosome formation on entering the stationary growth phase. Genes Cells 14: 1– 280 [DOI] [PubMed] [Google Scholar]
- 79. Zhang Y, et al. 2009. Transcription level of messenger RNA per gene copy determined with dual-spike-in strategy. Anal. Biochem. 394: 202– 208 [DOI] [PubMed] [Google Scholar]
- 80. Zuker M. 2003. Mfold web server for nucleic acid folding and hybridization prediction. Nucleic Acids Res. 31: 3406– 3415 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.