Abstract
The proteins encoded by the spoVA operon, including SpoVAD, are essential for the uptake of the 1:1 chelate of pyridine-2,6-dicarboxylic acid (DPA2,6) and Ca2+ into developing spores of the bacterium Bacillus subtilis. The crystal structure of B. subtilis SpoVAD has been determined recently, and a structural homology search revealed that SpoVAD shares significant structural similarity but not sequence homology to a group of enzymes that bind to and/or act on small aromatic molecules. We find that molecular docking placed DPA2,6 exclusively in a highly conserved potential substrate-binding pocket in SpoVAD that is similar to that in the structurally homologous enzymes. We further demonstrate that SpoVAD binds both DPA2,6 and Ca2+-DPA2,6 with a similar affinity, while exhibiting markedly weaker binding to other DPA isomers. Importantly, mutations of conserved amino acid residues in the putative DPA2,6-binding pocket in SpoVAD essentially abolish its DPA2,6-binding capacity. Moreover, replacement of the wild-type spoVAD gene in B. subtilis with any of these spoVAD gene variants effectively eliminated DPA2,6 uptake into developing spores in sporulation, although the variant proteins were still located in the spore inner membrane. Our results provide direct evidence that SpoVA proteins, in particular SpoVAD, are directly involved in DPA2,6 movement into developing B. subtilis spores.
INTRODUCTION
Spores of various Bacillus species are metabolically dormant and extremely resistant to a variety of stress factors, including heat, radiation, and a host of toxic chemicals (31, 32). A characteristic feature of these spores is the presence of high levels (∼12% of spore dry weight) of pyridine-2,6-decarboxylic acid (dipicolinic acid) (DPA2,6) in their central core, and this DPA is important for spore stability and spore resistance to heat, desiccation, and UV radiation (20, 29, 31). Most of the DPA2,6 exists in the spore core as a 1:1 chelate with divalent cations, predominantly Ca2+ (Ca-DPA2,6). Ca-DPA2,6 is accumulated by the developing spore late in sporulation from the mother cell (4, 5). In Bacillus subtilis, Ca-DPA2,6 accumulation requires the SpoVA proteins encoded by the heptacistronic spoVA operon, which is expressed just prior to Ca-DPA2,6 uptake by the developing spore; mutations in any of the first six cistrons of the spoVA operon, but not spoVAF, eliminate Ca-DPA2,6 transport (5, 7, 8, 32, 33). The SpoVA proteins are also needed for Ca-DPA2,6 uptake by spores of Clostridium perfringens (24). However, SpoVA proteins are not involved in DPA2,6 synthesis (6). The amino acid sequences of the SpoVA proteins are not similar to those of proteins with known function, except for that of SpoVAF, which is similar to that of the A subunits of spores' germinant receptors (5, 8). However, the sequences of many of the SpoVA proteins suggest that they are membrane proteins, with some predicted to be integral membrane proteins (4, 8). Indeed, even the two SpoVA proteins that appear likely to be soluble based on their amino acid sequences, SpoVAD and SpoVAEa, have been localized to the B. subtilis spore's inner membrane (5, 8, 11, 37).
In addition to being involved in Ca-DPA2,6 uptake in sporulation, the SpoVA proteins have also been implicated in the Ca-DPA2,6 release that takes place rapidly in the first minutes of spore germination (30, 33, 35, 36, 38). Indeed, overexpression of the spoVA operon results in an increased rate of Ca-DPA2,6 release during spore germination, while spores with a temperature-sensitive mutation in the spoVA operon are defective in Ca-DPA2,6 release at the nonpermissive temperature. In addition, there is evidence that at least some SpoVA proteins may associate with the germinant receptors to which nutrient germinants bind to trigger spore germination (35).
Despite the evidence linking SpoVA proteins to Ca-DPA2,6 uptake during sporulation and its release during spore germination, this evidence is largely circumstantial, and there is no direct evidence that these proteins may (i) associate to form a Ca-DPA2,6 channel in the spore's inner membrane and (ii) recognize and bind Ca-DPA2,6. Although their amino acid sequences are well conserved throughout evolution (25), as noted above SpoVA proteins except for SpoVAF exhibit no significant sequence homology to proteins of known function, and there also has been no specific functional or structural information about any of these proteins. However, recently the atomic structures of the SpoVAD proteins from B. subtilis (2.5 Å; Protein Data Bank [PDB] code 3LM6) and Bacillus licheniformis (2.0 Å; PDB code 3LMA) have been determined, and their structural coordinates were deposited in the RCSB Protein Data Bank (http://www.rcsb.org/pdb/). A search in the structural database revealed that these two structures exhibit significant homology to those of β-ketoacyl synthases and polyketide synthases (see Table S1 in the supplemental material). These enzymes all share a thiolase-like fold and are involved in reactions using coenzyme A (CoA) thioesters as substrates in the synthesis of fatty acids, flavonoids, polyketides, and a variety of other natural products (2, 3, 39). Strikingly, the locations and conformations of the substrate-binding sites of these enzymes are also conserved in the SpoVAD protein, including an active-site cysteine, even though there is no evidence that any SpoVA protein is an enzyme. It is, however, formally possible that these proteins are directly involved in DPA transport, a process that might well require catalytic activity in order to provide energy input into the transport process. In order to probe possible roles of this potential binding pocket in SpoVAD in DPA transport, we have used molecular docking to unambiguously place DPA2,6 in this SpoVAD pocket. We further show that the SpoVAD protein binds specifically to DPA2,6 and Ca-DPA2,6 but much more weakly to other DPA isomers that are not normally found in spores (9, 10). A site-directed mutational analysis of the putative binding site residues in SpoVAD provides evidence that conserved amino acid side chains in the putative DPA2,6-binding pocket are essential for DPA2,6 recognition in vitro and DPA2,6 uptake in B. subtilis sporulation. Taken together, our results delineate a critical role for SpoVAD in sporulation and provide direct evidence that SpoVA proteins are involved directly in DPA2,6 movement across the spore's inner membrane during both spore formation and spore germination.
MATERIALS AND METHODS
BLAST search and sequence conservation analysis.
The B. subtilis SpoVAD amino acid sequence (GenBank accession number BAA12659.1) was used as a query sequence for a BLAST search of spore-forming members of the Bacillales and Clostridiales orders on the NCBI genomic BLAST server (http://www.ncbi.nlm.nih.gov/sutils/genom_table.cgi). A single spoVAD gene was found for each species. The ClustalW alignment of SpoVAD homologs was performed using the MegAlign program from DNASTAR Lasergene suite 8 (DNASTAR Inc., Madison, WI).
Docking of DPA2,6 to SpoVAD.
Three-dimensional coordinates of DPA2,6 were adapted from the Hetero-Compound Information Centre (HIC-Up) database (15). Docking of DPA2,6 into the crystal structure of SpoVAD (PDB code 3LM6) was performed using the iterated Local Search Global Optimization algorithm provided by AutoDock Vina (34). The PDBQT format files (required as input) for both DPA2,6 and SpoVAD were generated using the AutoDock Tools package provided by AutoDock 4 (40). DPA2,6 was docked as a rigid body, and the entire surface of the SpoVAD structure was searched for possible binding sites without bias. A cubic box was built around the protein with 52 by 52 by 60 points as x, y, and z sizes. A spacing of 1.0 Å between the grid points was used, making the center of the protein the center of the cube, that is, x, y, and z centers at 18.661, −0.728, and 11.719, respectively. All other parameters were set as default as defined by AutoDock Vina. We have repeated the docking search using the FexX package (28).
Protein expression and purification.
The B. subtilis spoVAD variants were cloned into a pET22b plasmid (EMD Chemicals, Rockland, MA) containing a C-terminal His6 tag. The B. subtilis spoVAEa, gerD, and gerBC genes and the gene encoding the N-terminal hydrophilic domain of the A subunit of the gerA germinant receptor (GerAANTD; residues 1 to 239) were cloned into the pET15b vector (EMD Chemicals, Rockland, MA) containing an N-terminal His6 tag. All the proteins were expressed in Escherichia coli and purified to homogeneity by Ni2+-nitrilotriacetic acid (Ni2+-NTA) affinity, cation-exchange (for SpoVAD, GerBC, and GerAANTD), or anion-exchange (for SpoVAEa and GerD) and gel filtration (SD200; GE Healthcare, Piscataway, NJ) chromatography. The His6 tag was removed from the SpoVAD proteins by TEV protease cleavage.
Surface plasmon resonance (SPR) analysis.
The Biacore T100 (GE Healthcare, Piscataway, NJ) optical biosensor was used to measure the binding affinity of DPA/Ca-DPA to SpoVAD. The purified wild-type and mutant SpoVAD variants were covalently coupled to a carboxymethyl-dextran CM5 sensor chip to ∼5,000 resonance units. DPA2,6 and its isomers (DPA2,3, DPA2,5, and DPA3,5) in the absence and presence of equimolar CaCl2 were injected at various concentrations (0 to 60 mM, pH 7.4) over the immobilized SpoVAD protein at a flow rate of 30 μl/min for 4 min in buffer containing 10 mM HEPES (pH 7.5), 150 mM NaCl, and 0.05% (vol/vol) surfactant P20. The dissociation phase involved the injection of buffer alone for an additional 3 min followed by a regeneration step in which the bound DPA was removed by injection of 10 mM NaOH at 50 μl/min for 30 s. The control channel was treated in the identical way, omitting the immobilization of the protein. All these experiments were performed at 25°C. The titration data were analyzed by nonlinear curve fitting to a steady-state model using Biacore T100 analysis software (GE Healthcare, Piscataway, NJ) and OriginPro 7.5 (OriginLab Corporation, Northampton, MA).
B. subtilis strains used and spore preparation and purification.
The B. subtilis strains used in this work were PS832, a laboratory 168 strain, and two isogenic derivatives: FB112, in which the gene encoding SleB was largely deleted and replaced with a spectinomycin resistance (100 μg/ml) cassette (22), and FB122 (23), which lacks both SleB and the spoVF gene, encoding DPA2,6 synthetase (4). Spores of strain FB112 and FB112 derivatives in which the normal spoVAD gene in the spoVA operon was replaced with a cassette carrying either a wild-type or mutant spoVAD gene (see below) were prepared at 37°C on 2× SG agar plates without antibiotics (21, 23). Spores of strain FB122 were prepared at 37°C in liquid 2× SG medium (21) with or without various DPA isomers (1 mM). All spores were purified and stored as previously described (21, 23) and were free (>95%) of sporulating cells or germinated spores as determined by phase-contrast microscopy.
Site-directed mutagenesis.
Mutations were introduced into the spoVAD gene (1,018 bp) using an overlap PCR method (12). The PCR products were then cloned into a modified pBluescript II KS(−) vector containing a chloramphenicol resistance cassette separated by two inserts from the spoVA operon region. The first insert extends 300 bp inside the upstream spoVAC gene to the C-terminal region of the downstream spoVAEa gene. Downstream from this insert is the chloramphenicol resistance cassette, followed by 500 bp into the N-terminal region of the spoVAF gene in order to provide homology for a double crossover with the chromosomal DNA. The current study and previous studies (5) suggest that mutations that inactivate the spoVAF gene have no specific effect on sporulation and/or spore germination. The mutation sites in the plasmids were confirmed by DNA sequencing. The mutagenized plasmids were used to transform PS832 and FB112 competent cells, with selection for chloramphenicol resistance. Transformants in which the mutagenized spoVAD gene had integrated into the chromosome with replacement of the wild-type spoVAD gene were identified by PCR, and the PCR-amplified regions were sequenced to confirm the presence of the mutation.
DPA2,6 assays.
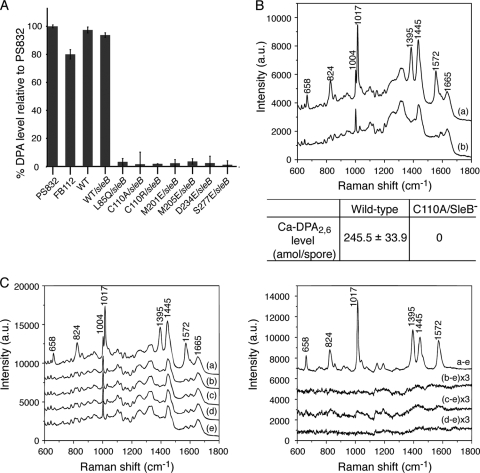
DPA2,6 levels in the spores were measured by two methods. In one, ∼1 ml of spore suspensions (optical density at 600 nm [OD600] of 5) were boiled in water for 30 min, cooled on ice for ≥15 min, and centrifuged for ∼5 min at 13,200 rpm. DPA2,6 in the supernatant fluid was measured by addition of aliquots of supernatant fluid to 50 μM TbCl3 in 25 mM HEPES buffer (pH 7.4). Tb+3-DPA2,6 levels were then determined fluorometrically as described previously (41). Each reaction mixture was tested in quadruplicate, and the fluorescence of the reaction mixture in the absence of TbCl3 or spore extract was subtracted from the averaged fluorescence reading of each reaction. The value for the averaged relative fluorescence units of the PS832 spore contents was considered to be 100%, and the percentage of the DPA level of spores of other strains was calculated against this maximum number. Ca-DPA2,6 levels were also measured in intact individual spores by Raman microspectroscopy of spores captured in an optical trap as described previously (14, 16). The content of Ca-DPA2,6 in individual spores was determined from the intensity of the Ca-DPA2,6-specific Raman peak at 1,017 cm−1 with reference to the intensity of Ca-DPA2,6 standards. Determination of levels of other DPA isomers in FB122 spores was also done by Raman microspectroscopy of individual spores as described above and with reference to the intensities of DPA isomer-specific Raman peaks given by the various pure DPA isomers.
Western blot analysis of the spore inner membrane fractions.
Inner membranes of the various spores were prepared as described previously (18). Equal amounts of the purified membrane fractions (prepared from ∼1 mg [dry weight] of decoated spores) were run on SDS-PAGE, and the SpoVAD protein was detected by immunoblotting using a polyclonal antiserum against SpoVAD (37).
Analytical ultracentrifugation analysis.
Sedimentation equilibrium measurements were carried out on a Beckman XL-A analytical ultracentrifuge (Beckman Coulter Inc., Brea, CA) using an An-60 Ti rotor (Beckman Coulter Inc., Brea, CA). Protein samples were dialyzed overnight against 15 mM Tris-HCl (pH 7.6), 100 mM NaCl, and 2 mM dithiothreitol, loaded at initial concentrations of 20, 60, and 180 μM, and analyzed at rotor speeds of 16,000 and 19,000 rpm at 20°C. Data were acquired at two wavelengths per rotor speed setting and processed globally for the best fit to a single-species model of absorbance versus radial distance by using the Origin software package provided by the manufacturer. Solvent density and protein partial specific volume were calculated according to solvent and protein compositions, respectively (17). Apparent molecular masses were within 10% of those calculated for an ideal monomer, with no systematic deviation of the residuals.
Ni2+-NTA affinity pulldown assays.
The indicated proteins (14 μM each) were incubated at room temperature for 10 min in 30 μl of binding buffer consisting of 50 mM Tris-HCl (pH 8.0), 200 mM NaCl, and 10 mM imidazole prior to addition of 28 μl of Ni2+-NTA resin (GE Healthcare, Piscataway, NJ). After 10 min, the resin was spun down, and ∼25 μl of supernatant, marked as the unbound (U) fraction in the figures, was removed. The resin was then washed three times with 0.6 ml of binding buffer, and the Ni2+-bound proteins were eluted with 30 μl of 50 mM Tris-HCl (pH 8.0), 200 mM NaCl, and 600 mM imidazole. One-third of each of the supernatant and eluted fractions (marked as the bound [B] fractions in the figures) was analyzed by SDS-PAGE and Coomassie blue staining.
RESULTS
Overall structure of SpoVAD.
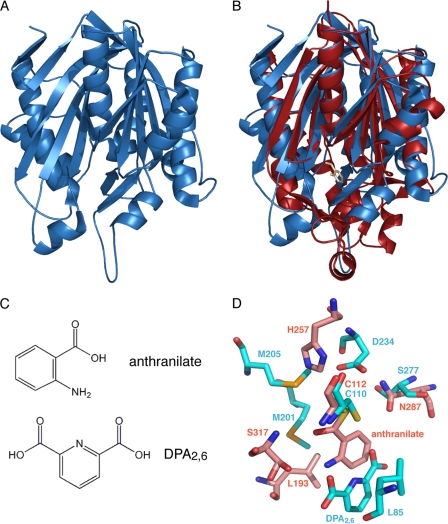
The crystal structures of the full-length B. subtilis (338 residues; PDB code 3LM6) and B. licheniformis (339 residues; PDB code 3LMA) SpoVAD proteins were recently determined by the Northeast Structural Genomics Consortium. B. subtilis SpoVAD adopts a compact, globular α/β mixed fold featuring two intertwined motifs related by pseudodyad duplication (Fig. 1A). The two motifs have similar topologies, with a five (S3 to S6 and S8)- or seven (S1, S2, S9 to S11, S13, and S14)-stranded mixed β-sheet surrounded by one helix on the inside (H5 or H10) and two helices (H3/H4 or H7/H8) on the outside, whereas each β-sheet consists of one antiparallel and four or six parallel β-strands (Fig. 1A; see Fig. 2A). The two substructures are related by a pseudo-2-fold axis, bringing the H5 and H10 helices together at the center of the molecule with the H3/H4 and H7/H8 pairs at the periphery. Assembly of these two domains thus creates a five-layer structure: the two core helices (H5 and H10) in the center are sandwiched between the two mixed sheets, while the two-helix pairs (H3/H4 and H7/H8) form the two outer layers. The three extended loops in the lower part of the molecule contain additional elements of secondary structure (H1/H2, H6/S7, and H9/S12) that are outside the repeated αβα domains. The overall fold of B. licheniformis SpoVAD is essentially the same as that of the B. subtilis protein. The two structures can be superimposed with a root mean square deviation (RMSD) of 1.1 Å for the 311 Cα atoms.
Fig 1.
The structure of the SpoVAD protein is similar to those of thiolase-fold containing proteins. (A) Ribbon diagram of the B. subtilis SpoVAD monomer (PDB code 3LM6). (B) Superimposition of SpoVAD (blue) and the covalent PqsD-anthranilate complex (red) (PDB code 3H77). The covalently bound anthranilate molecule in PqsD is shown as a ball-and-stick model and highlighted in pink. (C) Chemical structures of anthranilate and DPA2,6. (D) Superimposition of the active site of the PqsD-anthranilate complex (all carbon atoms in pink) and the presumed DPA2,6-binding pocket of SpoVAD (all carbon atoms in cyan). One of the nine top-ranked poses of DPA2,6 determined by AutoDock Vina (34) is shown as a ball-and-stick model (see Fig. S3 in the supplemental material for the conformations of the other eight poses for DPA2,6).
Fig 2.
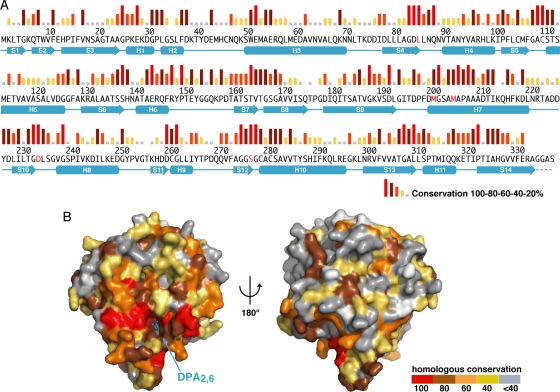
The presumed DPA2,6-binding site of SpoVAD is conserved. (A) Amino acid sequence of the B. subtilis SpoVAD protein. Sequence conservation is shown as a bar graph, with red bars indicating identity among the 80 SpoVAD orthologs in the Bacillales and Clostridiales species (see Fig. S4 in the supplemental material for a full alignment of the orthologous sequences). Secondary-structure assignments of SpoVAD are shown as cyan cylinders (α-helices) and arrows (β-strands), while disordered regions are shown as dashed lines. The specific amino acid residues that are mutated in this work are shown in red. (B) Molecular surface of SpoVAD, colored according to orthologous conservation. The DPA2,6 molecule in the same pose as in Fig. 1D is colored in cyan and shown as a ball-and-stick model.
Structural relatives of SpoVAD.
Although primary amino acid sequence analysis failed to identify any known protein motif in SpoVAD, a structure homology search of the Protein Data Bank using the Dali server (13) revealed unequivocal structural resemblance of SpoVAD with more than 400 thiolase-fold-containing enzymes (2, 39) (see Table S1 in the supplemental material). This superfamily of enzymes consists of many functionally diverse members, including β-ketoacyl-acyl carrier protein synthases and thiolases of fatty acid metabolism, the chalcone synthase/stilbene synthase superfamily of bacterial and plant polyketide synthases, and enzymes involved in bacterial natural product biosynthesis. In addition to catalysis of condensation reactions in biosynthetic pathways using groups in thioester linkage to CoA, some of these enzymes also carry out cyclization reactions leading to the formation of a variety of aromatic molecules. Each of these enzymes can be superimposed onto B. subtilis SpoVAD with an RMSD of ∼3 Å in the positions of over 250 Cα atoms of equivalent residues. Indeed, all these enzymes or their related subdomains have a common topology consisting of two mixed five-stranded β-sheets sandwiched between three helix pairs, a so-called αβαβα fold (2, 39). This overall fold closely resembles the determined SpoVAD structures.
All the thiolase-fold enzymes possess buried active sites in which an absolutely conserved catalytic cysteine is situated at the bottom edge of one of a pair of the central α-helices. The loops that extend from the conserved αβαβα core form a narrow CoA-binding tunnel to connect the catalytic cavity to the outside aqueous milieu. Importantly, the SpoVAD protein has these two common features: a highly conserved Cys-110 residue in B. subtilis SpoVAD lies at the N-terminal cap of the central helix H5, facing a deep buried pocket created principally by the extended loop regions (Fig. 1) (see below). The above structural analyses strongly suggest that SpoVAD belongs to the group of thiolase-fold enzymes capable of binding small-molecule substrates.
SpoVAD is a monomer in solution.
One of the characteristic features of the thiolase-fold-containing enzymes is that they all exist as a dimer in solution (2). Although the two monomers have virtually identical conformations in these enzymes, it has been suggested that reaction of a substrate molecule with one subunit of the dimer might induce a conformational change in the other subunit to reduce its binding affinity to the second ligand (1). Interestingly, SpoVAD also forms a dimer in the same fashion as those enzymes in the crystal structure (see Fig. S1 in the supplemental material). The dimer buries 2,400 Å2 of surface area, and two monomers can be superimposed with a 0.2-Å RMSD in Cα positions. The primary feature of the SpoVAD dimer interface is that the five-stranded β-sheet in one monomer aligns antiparallel with the corresponding sheet of another monomer to form a continuous ten-strand β-sheet that traverses the center of the entire molecule. Nonetheless, equilibrium ultracentrifugation experiments showed that SpoVAD forms a clean monomer in solution (see Fig. S2 in the supplemental material). These results are very similar to those for another germination protein, the C subunit of the B. subtilis GerB germinant receptor (GerBC), which is also a dimer in its crystal structure but monomeric in solution (19).
Molecular docking of DPA2,6 into SpoVAD.
Based on structural similarity, the B. subtilis and B. licheniformis SpoVAD proteins most closely resemble the Pseudomonas aeruginosa quinolone signal biosynthetic enzyme PqsD (3) (see Table S1 in the supplemental material). The PqsD and B. subtilis SpoVAD structures can be superimposed with an RMSD of 3.2 Å for 261 equivalent Cα atoms (Fig. 1B), although the two proteins have only ∼15% sequence identity. PqsD catalyzes a key chemical step in biosynthesis of the alkyl quinolones, the condensation and cyclization of anthraniloyl and malonyl thioesters, forming 2,4-dihydroxyquinoline (3). The PqsD active site contains the Cys-His-Asn catalytic triad, a typical feature of the β-ketoacyl-acyl carrier protein synthases and the chalcone synthase-like polyketide synthases (Fig. 1D). In the anthranilate-PqsD complex structure, Cys-112 of PqsD is covalently bound to anthranilate, a PqsD substrate that has a molecular size and chemical structure similar to those of DPA2,6 (Fig. 1C and D). Interestingly, the corresponding region in SpoVAD preserves the general structure of the PqsD active site, leading us to surmise that SpoVAD might bind DPA2,6 and/or Ca-DPA2,6.
As our attempts to crystallize the SpoVAD-DPA2,6 complex have failed, we decided to use molecular docking to identify a possible binding site on SpoVAD that could accommodate DPA2,6. The DPA2,6 model was docked as a rigid body to the B. subtilis SpoVAD crystal structure using AutoDock Vina (34), and the entire structural surface was searched for possible binding sites without bias. As expected, all nine top-ranked (i.e., lowest-binding-energy) docked conformations for DPA2,6 were predicted to bind to SpoVAD in the pocket at which anthranilate of PqsD binds, although the exact positions and orientations of these DPA2,6 molecules differ (Fig. 1D; see Fig. S3 in the supplemental material). For instance, in the most favorable binding conformation (Fig. 1D), the pyridine ring of DPA2,6 is oriented parallel to the benzene ring of anthranilate in the PqsD structure, while the whole molecule is shifted further away from the core Cys-110 and in close contact with the conserved Leu-85. As a positive control, we performed a similar binding site search to dock anthranilate to PqsD. Indeed, all nine top conformations of anthranilate were found in the same binding site revealed by the crystal structure of the anthranilate-PqsD complex. The docking search using the FexX package (28) also predicted the same DPA2,6-binding site, with six out of the top nine solutions in this site. Thus, we conclude that the Cys-110-containing pocket of SpoVAD is likely to be a binding site for DPA2,6.
SpoVAD contains an atypical substrate-binding site.
Despite the high degree of overall structural similarity, the putative DPA2,6-binding site of SpoVAD differs markedly from the active site of the thiolase-fold enzymes. Except for the Cys-110 residue, SpoVAD lacks the two conserved His-Asn residues that make up the catalytic triad of the enzymes discussed above. Although the corresponding Asp-234 and Ser-277 residues in B. subtilis SpoVAD are located in a position similar to that of the His-Asn pair, their large distances from Cys-110 (4.1 Å and 6.1 Å, respectively) do not support their role in deprotonating Cys-110. Moreover, the protein surface surrounding the entrance to the putative DPA2,6-binding site in SpoVAD is largely lined with hydrophobic residues (Leu-85, Phe-199, Met-201, Met-205, Leu-235, and Val-238) (Fig. 1D), while the corresponding arginine-rich protein surface in thiolase-fold enzymes promotes their interaction with CoA (2, 39). Thus, it seems that SpoVAD contains an atypical substrate-binding site that lacks the potential to effectively bind CoA or catalyze a condensation reaction.
The putative DPA2,6-binding site in SpoVAD is highly conserved among its homologs.
A broad BLAST search using the B. subtilis SpoVAD protein as the query sequence has identified 80 homologs of SpoVAD in spore-forming Bacillales and Clostridiales genomes (see Fig. S4 in the supplemental material). Sequence alignments of these SpoVAD homologs show that they share ∼41 to 90% pairwise sequence identities (Fig. 2A; see Fig. S4 in the supplemental material). As shown in Fig. 2A, those highly conserved residues (with red or brown bars above the sequence) are scattered throughout the sequences, and their functional significance remains elusive. Strikingly, mapping the conservation of all 80 homologs of SpoVAD onto the surface of its three-dimensional structure revealed that the region around the putative DPA2,6-binding site has a significantly higher conservation than the rest of the structure (Fig. 2B; note red and brown residues around the bound cyan DPA2,6). This supports our hypothesis that this site may play an important role in mediating the general function of SpoVAD and that SpoVAD is likely involved in DPA2,6 movement into and out of spores.
SpoVAD binds DPA2,6 and Ca-DPA2,6 in vitro.
To directly test the hypothesis noted above, we used SPR to measure the binding of various DPA isomers to the purified SpoVAD protein in the presence and absence of Ca2+. As expected, the injection of 2 mM DPA2,6 or Ca-DPA2,6 over immobilized SpoVAD displayed clear association signals (Fig. 3). In contrast, injection of other DPA isomers (DPA2,3, DPA2,5, and DPA3,5) resulted in much lower SPR association signals, suggesting that SpoVAD can specifically recognize and bind to both DPA2,6 and Ca-DPA2,6. Next, multiple DPA concentrations were titrated to determine their equilibrium dissociation constants (KD) with SpoVAD (Table 1; see Fig. S5A in the supplemental material). Following a steady-state fitting model, SpoVAD binds both DPA2,6 and its Ca chelate with a KD of ∼0.8 mM. As expected, titration of other DPA isomers, including DPA2,3, DPA2,5, and DPA3,5 with or without Ca2+, to SpoVAD gave much weaker or no SPR association signal even at the highest titration concentration (120 mM) (data not shown). Indeed, we were able to determine the binding affinity to SpoVAD of only one of the other DPA isomers, Ca-DPA2,3, and this affinity was 22-fold lower than that of Ca-DPA2,6 (Table 1). Together, the SPR data demonstrated that SpoVAD specifically binds DPA2,6 and Ca-DPA2,6 and with similar binding affinity.
Fig 3.
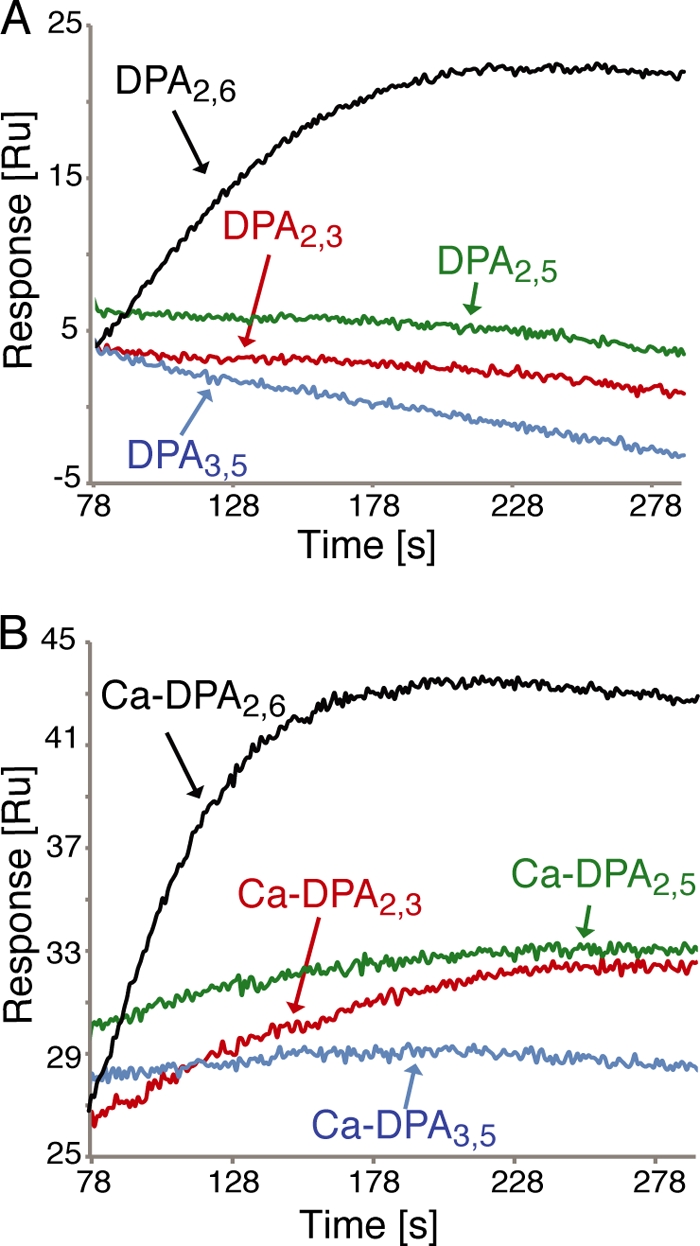
Binding of DPA and Ca-DPA to SpoVAD. (A) Overlay of the representative Biacore SPR sensorgrams displaying the association curves of 2 mM DPA2,6 and its isoforms (DPA2,3, DPA2,5, and DPA3,5) with the wild-type SpoVAD protein. (B) Overlay of the representative SPR sensorgrams displaying the association curves of 2 mM Ca-DPA2,6 and its isoforms (Ca-DPA2,3, Ca-DPA2,5, and Ca-DPA3,5) with the wild-type SpoVAD protein.
Table 1.
Binding of DPA and Ca-DPA isomers to SpoVAD proteins
| Protein |
KD, mM (mean ± SD), witha: |
||
|---|---|---|---|
| DPA2,6 | Ca-DPA2,6 | Ca-DPA2,3 | |
| SpoVADWT | 0.8 ± 0.4 | 0.8 ± 0.3 | 18 ± 3 |
| SpoVADC110A | 36 ± 6 | 5.3 ± 1.5 | ≥120b |
| SpoVADD234F | 13 ± 2 | 1.3 ± 0.5 | 13 ± 1 |
| SpoVADS277E | 2.8 ± 0.9 | 3.3 ± 0.7 | 21 ± 1 |
The binding affinities of the various purified SpoVAD proteins were determined as described in Materials and Methods. The KD values for the binding of DPA2,3, DPA2,5, DPA3,5, and the Ca chelates of the DPA2,5 and DPA3,5 isomers to the four SpoVAD proteins were too weak to be measured by SPR under our settings (Fig. 3 and data not shown; see Fig. S3 and S4 in the supplemental material).
The Ca-DPA2,3 binding to SpoVADC110A was too weak for the exact KD value to be measured even using the highest titration concentration (120 mM) (data not shown).
Mutations in SpoVAD affect DPA2,6 binding.
To further dissect the substrate specificity determinants of SpoVAD, we investigated the effects of mutating those conserved amino acid residues in the predicted DPA2,6/Ca-DPA2,6-binding site. We decided first to replace the three presumed core residues (Cys-110, Asp-234, and Ser-277) involved in substrate binding (Fig. 1D and 2A). Three SpoVAD mutants, C110A, D234F, and S277E, were expressed and purified. Mutation to alanine was selected because of its small size, while mutations to phenylalanine and glutamic acid were selected because an aromatic or acidic side chain at these positions might mimic the structural features of the substrate-binding sites in thiolase-fold-containing enzymes. The purified mutant proteins were eluted at essentially the same retention volume as wild-type SpoVAD on a gel filtration column (data not shown), suggesting that they fold properly. We then quantitated their binding for DPA/Ca-DPA using SPR. All three mutations in SpoVAD resulted in markedly reduced binding affinity to DPA2,6 and Ca-DPA2,6, confirming their identification as important elements of the substrate-binding site (Table 1; see Fig. S5B to D and S6 in the supplemental material). In particular, replacing the highly conserved Cys-110 with alanine decreased the substrate-binding affinity by ∼6.6- to 45-fold (DPA2,6, KD = 36 ± 6 mM; Ca-DPA2,6, KD = 5.3 ± 1.5 mM), indicating that this residue is absolutely required for normal DPA2,6-SpoVAD binding. Defects in the D234F and S277E mutants were less pronounced, with the affinities decreased by 1.6- to 16-fold. It is possible that their aromatic and/or acidic side chains might still be able to interact with DPA2,6 partially. We also found that the C110A mutation severely disrupted SpoVAD binding to the Ca-DPA2,3 isomer (KD ≥ 120 mM), while the D234F and S277E mutations had a minor effect on Ca-DPA2,3 binding, with KD values of 13 ± 1 mM and 21 ± 1 mM, respectively, suggesting that Asp-234 and Ser-277 might be specifically positioned to recognize the carboxylic side chains of DPA2,6 but not those of other DPA isomers. Together with the sequence conservation and docking analyses, the results indicated that SpoVAD likely interacts with DPA2,6 and Ca-DPA2,6 through the predicted substrate-binding site.
Mutations in SpoVAD affect sporulation and DPA2,6 uptake.
To determine whether alteration of the conserved SpoVAD amino acid residues in the DPA2,6-binding site affects sporulation and DPA2,6 uptake in B. subtilis, three core residues (Cys-110, Asp-234, and Ser-277) and three additional conserved hydrophobic amino acids surrounding the docked DPA2,6 molecule (Leu-85, Met-201, and Met-205) were chosen for our functional analyses (Fig. 1D and 2A). For those hydrophobic residues as well as Cys-110, a polar side chain was introduced to test whether hydrophobicity is important for SpoVAD to maintain its function. Seven spoVAD mutant genes carrying a single mutation (L85Q, C110A, C110R, M201E, M205E, D234F, and S277E) were constructed and introduced into the B. subtilis chromosome, replacing the wild-type spoVAD gene but in such a way that the downstream spoVAEb and spoVAEa genes were retained (see Materials and Methods). We note that the final spoVA cistron, spoVAF, was not retained under the control of the spoVA promoter in these constructs; our data (below) and previous studies (5) have suggested that this gene is not essential for DPA2,6 uptake in sporulation or DPA2,6 release in spore germination. Analysis of the sporulation of wild-type and mutant strains by phase-contrast microscopy indicated that the construct in which a wild-type spoVAD gene was inserted in the same manner as in the spoVAD mutant derivatives sporulated identically to the wild-type strain (PS832) and released phase-bright spores (data not shown). In striking contrast, while the strains with single point mutations in spoVAD appeared to begin sporulation normally and the spores in the sporangium were initially somewhat phase bright, they rapidly became phase dark and appeared to germinate within the sporangium and then lyse (data not shown). The same phenotype was observed in a spoVA null mutant strain, as seen in this work and previously (33).
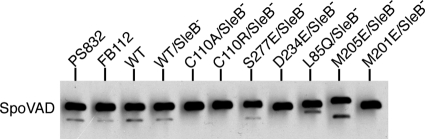
To determine the extent to which the above spoVAD mutations might cause complete loss of spore function, we moved the mutations into a sleB strain (FB112). Normally, spores that do not accumulate DPA2,6 are extremely unstable, most likely because in the absence of DPA2,6, the SleB protein, one of the two redundant cortex-lytic enzymes (CLEs) that degrade the Bacillus spore's peptidoglycan cortex during germination, becomes activated in the developing spore (23). Consequently, DPA2,6-less spores of Bacillus species are normally almost impossible to isolate. However, in the absence of SleB, such spores can be isolated, as the spore's other redundant CLE, CwlJ, is probably activated directly by Ca-DPA2,6 (20, 29). sleB strains carrying the spoVAD mutations sporulated relatively normally, although the spores were not as phase bright as the sleB spores with a wild-type spoVA operon (data not shown). The observed lower spore core refractive index likely resulted from the absence of DPA2,6 from the spore core. Indeed, analysis of the DPA2,6 levels in the spores of the various sleB strains either by bulk fluorescence assays of DPA2,6 in spore populations or by Raman spectroscopy of multiple individual spores showed that the spoVAD mutant spores accumulated either no DPA2,6 or ≤1% of the DPA2,6 level of the sleB spores with a wild-type spoVA operon (Fig. 4A and B). Since the spores of these sleB derivatives were stable, we then analyzed the levels of the SpoVAD protein in the spore's inner membrane by Western blot analysis (Fig. 5). This analysis showed clearly that the levels of the wild-type and variant SpoVAD proteins in the spore's inner membrane were relatively similar, indicating that these mutations do not affect protein assembly and localization. These results strongly suggest that the predicted substrate-binding site of SpoVAD is directly involved in DPA2,6 uptake during sporulation, consistent with the above structural and biochemical data.
Fig 4.
DPA/Ca-DPA contents of wild-type and mutant B. subtilis spores. (A) DPA2,6 levels of wild-type and spoVAD sleB mutant spores. The percent DPA2,6 level of spores of each strain was normalized against that of PS832 spores as described in Materials and Methods. The spoVAD mutation sites are shown below the x axis. The fluorescence measurements of the reaction mixtures in the absence of TbCl3 or spore extract were used as the negative control. Data represent means ± standard deviations (SD) for at least three independent measurements. (B) Average Raman spectra of wild-type (curve a) and mutant (spoVADC111A sleB) (curve b) spores. The spectra are the averages of the signals from 100 individual spores. The laser power used was 30 mW, and the integration time for Raman measurement was 20 s. Bottom, mean values of the Ca-DPA2,6 levels of spores from different preparations. (C) Average Raman spectra of FB122 spores prepared with various DPA isomers. In the left panel, curves a to e are the spectra for spores prepared with DPA2,6, DPA2,3, DPA2,5, and DPA3,5 and in the absence of DPA, respectively (see also Fig. S7 in the supplemental material). The spectra were averaged over the signals from 100 individual spores. The laser power used was 30 mW, and the integration time for Raman measurement was 20 s. The right panel shows the subtractions of curve e from curves a to d shown in the left panel. The subtractions of curve e from curves b to d were amplified by a factor of 3 for display.
Fig 5.
Levels of wild-type and mutant SpoVAD proteins in the inner membrane of spores. Inner membranes extracted from ∼1 mg dry decoated spores were run on SDS-PAGE and analyzed by Western blotting with the polyclonal anti-SpoVAD antiserum.
As each DPA isomer exhibits a unique Raman peak (see Fig. S7 in the supplemental material), we also examined the ability of spores to take up DPA2,6 and its isomers by Raman spectroscopy. The sleB spoVF strain (FB122) lacks sleB and spoVF, encoding DPA2,6 synthetase, and spores formed from this strain normally contain no DPA2,6 (4, 22). However, it has been shown that if DPA2,6 is provided during sporulation, it is taken up and the resultant DPA2,6 levels in the spores can reach those in wild-type spores (9, 10, 20, 23, 29). Indeed, when DPA2,6 and its three isomers were provided in the medium during sporulation, Raman spectroscopy of multiple individual spores showed that only DPA2,6, and not its isomers, were taken up by the spores (Fig. 4C). Together, these data reinforce the notion that DPA uptake is a specific process and that the SpoVAD-DPA2,6 interaction likely plays an important role in this process.
SpoVAD does not interact with several other germination proteins.
As all seven SpoVA proteins are encoded by a heptacistronic spoVA operon, it is possible that those proteins can interact to associate with each other. There is also evidence to suggest that germinant receptors can interact with some SpoVA proteins (35) and that the action of enzymes that hydrolyze the spore cortex during spore germination can influence rates of Ca-DPA2,6 release during germination (27). Hence, we employed an Ni2+-NTA affinity pulldown assay to assess the association of SpoVAD with its possible interaction partners that are currently available in purified form: (i) the N-terminal hydrophilic domain of the A subunit of the GerA germinant receptor (GerAANTD); (ii) GerBC; (iii) the auxiliary germination protein GerD, which is needed for optimal germination via triggering of the germinant receptors (26); and (iv) the SpoVAEa protein. We should point out that all four purified proteins are likely properly folded as demonstrated by gel filtration chromatography (for all four of them) (data not shown), analytical ultracentrifugation and circular dichroism spectroscopy (for GerD) (data not shown), crystallography (for GerBC) (19), and nuclear magnetic resonance (NMR) spectroscopy (for SpoVAEa) (data not shown). In the pulldown experiment, we incubated 14 μM purified SpoVAD with 14 μM purified His6-GerAANTD, His6-GerBC, His6-GerD, or His6-SpoVAEa, precipitated the mixture using Ni2+-NTA resin, eluted proteins from the resin with imidazole, and analyzed the eluates by SDS-PAGE and Coomassie blue staining. Figure S8 in the supplemental material shows that untagged SpoVAD does not elute with any of these His6-tagged proteins (lanes 8, 10, 16, and 18), suggesting that SpoVAD does not directly interact with these proteins in vitro. However, it is certainly possible that SpoVAD interacts with another SpoVA protein or proteins, but these are currently not available in purified form, nor are antisera against the other five SpoVA proteins available.
DISCUSSION
The molecular basis for DPA uptake and release in the sporulation and spore germination of Bacillus and Clostridium species has been the subject of intensive research for the past 3 decades. A number of studies have attempted to define or identify proteins that are involved in these processes. Although there is significant circumstantial evidence indicating that the SpoVA proteins are involved in both DPA2,6 uptake by the developing forespore and its release in germination, the exact mechanism of these proteins' action is unknown. The results in this communication provide the first direct evidence that at least one SpoVA protein, SpoVAD, is directly involved in DPA2,6 movement, at least in sporulation. We have identified novel conserved features of SpoVAD and demonstrated that SpoVAD specifically binds DPA2,6 and that mutations in the likely DPA2,6-binding site weaken DPA2,6 binding and eliminate DPA2,6 uptake by developing spores.
SpoVAD shows clear structural homology to thiolase-fold-containing enzymes. The monomers that make up these enzymes all contain the repeated core motif in an essentially identical conformation. Outside the conserved core motif, these enzymes exhibit considerable differences in the loop regions at the bottom of the molecule, which have evolved to recognize different substrates and carry out catalysis. The corresponding loop regions in SpoVAD share much higher conservation among all SpoVAD homologs than the rest of the molecule. Moreover, a number of conserved residues in these regions form a putative substrate-binding pocket, including a critical cysteine that plays a catalytic role in the enzymes that are structurally homologous to SpoVAD. Molecular docking analysis unambiguously locates DPA2,6 at this substrate-binding site. As two other critical catalytic residues in the thiolase-fold enzymes are missing in SpoVAD, we have not been able to identify any catalytic activity of SpoVAD based on the structural analysis, and indeed there may not be any such activity. However, our mutagenesis data have confirmed that this cysteine residue as well as several other conserved residues in the binding pocket are clearly essential for DPA2,6 binding and for SpoVAD function in DPA uptake in sporulation.
We show that both DPA2,6 and Ca-DPA2,6 can bind to SpoVAD, with KD values in the millimolar range. Importantly, the affinity of SpoVAD for DPA2,6 is clearly significantly higher than that for the other DPA isomers. This substrate specificity could explain the minimal if any uptake of DPA isomers other than DPA2,6 into spores (Fig. 4C) (9, 10). Nevertheless, we note that SpoVAD has relatively low affinity even for DPA2,6. We speculate that this low affinity is sufficient given the huge amount of DPA2,6 that must be synthesized by the mother cell in order for there to be sufficient DPA2,6 uptake to ∼12% of spore dry weight. Consistent with this notion, forespores' uptake of DPA2,6 made in the mother cell does not achieve 100% efficiency, as large amounts of DPA2,6 are released into the sporulation medium (6). In addition, all SpoVA proteins, not just SpoVAD, are thought to associate to form a complex in and on the forespores' inner membrane, and a number of SpoVA proteins are almost certainly integral membrane proteins. If such a complex does indeed exist, it is possible that interactions of SpoVAD with other SpoVA proteins could significantly increase SpoVAD's affinity for DPA2,6. Perhaps even more importantly, it also seems likely that the uptake of DPA2,6 from the mother cell to high levels in the forespore will be an energy-dependent process, since this DPA uptake will ultimately be against a concentration gradient, and again this energy dependence could increase SpoVAD's affinity for DPA2,6.
While the relatively low affinity of SpoVAD for DPA2,6 might be problematic to some degree for DPA2,6 uptake during sporulation, this should facilitate DPA2,6 release in spore germination. In this case, there is no energy requirement, since DPA2,6 will flow down a concentration gradient, and indeed there is no evidence that there is any energy generation that takes place in spore germination, whether this be ATP or a membrane potential (20, 26). Nevertheless, not all small molecules are released from the spores in the first minutes of spore germination, but only DPA2,6 and associated cations and some free amino acids (29, 30). Thus, the channel for DPA exit during spore germination must exhibit some specificity for DPA2,6 or Ca-DPA2,6, but the affinity can be low.
Like all the enzymes with the thiolase fold, SpoVAD forms a dimer in the crystalline state, and extensive interactions exist across the dimer interface through the formation of a joint 10-stranded β-sheet connecting two monomers. However, sedimentation equilibrium experiments revealed a monomeric SpoVAD in solution. While the dimer interface in the SpoVAD crystal is unlikely to affect DPA binding, it is possible that those solvent-accessible residues in the interface could participate in forming transient multiprotein complexes with other SpoVA proteins or proteins involved in sporulation and spore germination. This is consistent with our Western blotting results that show that a large amount of SpoVAD is present in the spore inner membrane, although SpoVAD by itself is certainly a soluble protein. Nevertheless, the affinity pulldown experiments show that SpoVAD does not interact directly with SpoVAEa and several other proteins involved in spore germination. As all the other SpoVA proteins and many other germinant proteins are either membrane-associated proteins or insoluble in the in vitro overexpression systems, we cannot yet test their interaction with SpoVAD in a purified system. Perhaps the determination of the exact function of SpoVAD and other SpoVA proteins in DPA2,6 transport may require the elucidation of the structure of an entire SpoVA protein complex.
Supplementary Material
ACKNOWLEDGMENTS
We are grateful to Min Lu of the Public Health Research Institute Center of UMDNJ-New Jersey Medical School for sedimentation analysis.
This work was supported by a Department of Defense Multi-University Research Initiative award through the U.S. Army Research Laboratory and the U.S. Army Research Office under contract number W911NF-09-1-0286.
Footnotes
Published ahead of print 10 February 2012
Supplemental material for this article may be found at http://jb.asm.org/.
REFERENCES
- 1. Alhamadsheh MM, et al. 2007. Alkyl-CoA disulfides as inhibitors and mechanistic probes for FabH enzymes. Chem. Biol. 14: 513– 524 [DOI] [PubMed] [Google Scholar]
- 2. Austin MB, Noel JP. 2003. The chalcone synthase superfamily of type III polyketide synthases. Nat. Prod. Rep. 20: 79– 110 [DOI] [PubMed] [Google Scholar]
- 3. Bera AK, et al. 2009. Structure of PqsD, a Pseudomonas quinolone signal biosynthetic enzyme, in complex with anthranilate. Biochemistry 48: 8644– 8655 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Daniel RA, Errington J. 1993. Cloning, DNA sequence, functional analysis and transcriptional regulation of the genes encoding dipicolinic acid synthetase required for sporulation in Bacillus subtilis. J. Mol. Biol. 232: 468– 483 [DOI] [PubMed] [Google Scholar]
- 5. Errington J. 1993. Bacillus subtilis sporulation: regulation of gene expression and control of morphogenesis. Microbiol. Rev. 57: 1– 33 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Errington J, Cutting SM, Mandelstam J. 1988. Branched pattern of regulatory interactions between late sporulation genes in Bacillus subtilis. J. Bacteriol. 170: 796– 801 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Errington J, Mandelstam J. 1986. Use of a lacZ gene fusion to determine the dependence pattern and the spore compartment expression of sporulation operon spoVA in spo mutants of Bacillus subtilis. J. Gen. Microbiol. 132: 2977– 2985 [DOI] [PubMed] [Google Scholar]
- 8. Fort P, Errington J. 1985. Nucleotide sequence and complementation analysis of a polycistronic sporulation operon, spoVA, in Bacillus subtilis. J. Gen. Microbiol. 131: 1091– 1105 [DOI] [PubMed] [Google Scholar]
- 9. Fukuda A, Gilvarg C. 1968. The relationship of dipicolinate and lysine biosynthesis in Bacillus megaterium. J. Biol. Chem. 243: 3871– 3876 [PubMed] [Google Scholar]
- 10. Fukuda A, Gilvarg C, Lewis JC. 1969. 4H-pyran-2,6-dicarboxylate as a substitute for dipicolinate in the spolation of Bacillus megaterium. J. Biol. Chem. 244: 5636– 5643 [PubMed] [Google Scholar]
- 11. Griffiths KK, Zhang J, Cowan AE, Yu J, Setlow P. 2011. Germination proteins in the inner membrane of dormant Bacillus subtilis spores colocalize in a discrete cluster. Mol. Microbiol. 81: 1061– 1077 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Heckman KL, Pease LR. 2007. Gene splicing and mutagenesis by PCR-driven overlap extension. Nat. Protoc. 2: 924– 932 [DOI] [PubMed] [Google Scholar]
- 13. Holm L, Kaariainen S, Rosenstrom P, Schenkel A. 2008. Searching protein structure databases with DaliLite v. 3. Bioinformatics 24: 2780– 2781 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Huang SS, et al. 2007. Levels of Ca2+-dipicolinic acid in individual Bacillus spores determined using microfluidic Raman tweezers. J. Bacteriol. 189: 4681– 4687 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Kleywegt GJ. 2007. Crystallographic refinement of ligand complexes. Acta Crystallogr. D Biol. Crystallogr. 63: 94– 100 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Kong L, Zhang P, Setlow P, Li Y. 2011. Multifocus confocal Raman microspectoscopy for rapid single-particle analysis. J. Biomed. Optics 16: 120503. [DOI] [PubMed] [Google Scholar]
- 17. Laue TM, Shah BD, Ridgeway TM, Pelletier SL. 1992. Computer-aided interpretation of analytical sedimentation data for proteins, p 90– 125 In Harding S, Rowe A, Horton J. (ed), Analytical ultracentrifugation in biochemistry and polymer science. Royal Society of Chemistry, Cambridge, United Kingdom [Google Scholar]
- 18. Li Y, et al. 2011. Structure-based functional studies of the effects of amino acid substitutions in GerBC, the C subunit of the Bacillus subtilis GerB spore germinant receptor. J. Bacteriol. 193: 4143– 4152 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Li Y, Setlow B, Setlow P, Hao B. 2010. Crystal structure of the GerBC component of a Bacillus subtilis spore germinant receptor. J. Mol. Biol. 402: 8– 16 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Magge A, et al. 2008. Role of dipicolinic acid in the germination, stability, and viability of spores of Bacillus subtilis. J. Bacteriol. 190: 4798– 4807 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Nicholson WL, Setlow P. 1990. Sporulation, germination and outgrowth, p 391– 450 In Harwood CR, Cutting SM. (ed), Molecular biological methods for Bacillus. John Wiley and Sons, Chichester, United Kingdom [Google Scholar]
- 22. Paidhungat M, Ragkousi K, Setlow P. 2001. Genetic requirements for induction of germination of spores of Bacillus subtilis by Ca2+-dipicolinate. J. Bacteriol. 183: 4886– 4893 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Paidhungat M, Setlow B, Driks A, Setlow P. 2000. Characterization of spores of Bacillus subtilis which lack dipicolinic acid. J. Bacteriol. 182: 5505– 5512 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Paredes-Sabja D, Setlow B, Setlow P, Sarker MR. 2008. Characterization of Clostridium perfringens spores that lack SpoVA proteins and dipicolinic acid. J. Bacteriol. 190: 4648– 4659 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Paredes-Sabja D, Setlow P, Sarker MR. 2011. Germination of spores of Bacillales and Clostridiales species: mechanisms and proteins involved. Trends Microbiol. 19: 85– 94 [DOI] [PubMed] [Google Scholar]
- 26. Pelczar PL, Igarashi T, Setlow B, Setlow P. 2007. Role of GerD in germination of Bacillus subtilis spores. J. Bacteriol. 189: 1090– 1098 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Peng L, Chen D, Setlow P, Li YQ. 2009. Elastic and inelastic light scattering from single bacterial spores in an optical trap allows the monitoring of spore germination dynamics. Anal. Chem. 81: 4035– 4042 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Rarey M, Kramer B, Lengauer T, Klebe G. 1996. A fast flexible docking method using an incremental construction algorithm. J. Mol. Biol. 261: 470– 489 [DOI] [PubMed] [Google Scholar]
- 29. Setlow B, Atluri S, Kitchel R, Koziol-Dube K, Setlow P. 2006. Role of dipicolinic acid in resistance and stability of spores of Bacillus subtilis with or without DNA-protective alpha/beta-type small acid-soluble proteins. J. Bacteriol. 188: 3740– 3747 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Setlow P. 2003. Spore germination. Curr. Opin. Microbiol. 6: 550– 556 [DOI] [PubMed] [Google Scholar]
- 31. Setlow P. 2006. Spores of Bacillus subtilis: their resistance to and killing by radiation, heat and chemicals. J. Appl. Microbiol. 101: 514– 525 [DOI] [PubMed] [Google Scholar]
- 32. Setlow P, Johnson EA. 2011. Spores and their significance, p 35– 67 In Doyle MP, Buchanan R. (ed), Food microbiology: fundamentals and frontiers. ASM Press, Washington, DC [Google Scholar]
- 33. Tovar-Rojo F, Chander M, Setlow B, Setlow P. 2002. The products of the spoVA operon are involved in dipicolinic acid uptake into developing spores of Bacillus subtilis. J. Bacteriol. 184: 584– 587 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Trott O, Olson AJ. 2010. AutoDock Vina: improving the speed and accuracy of docking with a new scoring function, efficient optimization, and multithreading. J. Comput. Chem. 31: 455– 461 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Vepachedu VR, Setlow P. 2007. Analysis of interactions between nutrient germinant receptors and SpoVA proteins of Bacillus subtilis spores. FEMS Microbiol. Lett. 274: 42– 47 [DOI] [PubMed] [Google Scholar]
- 36. Vepachedu VR, Setlow P. 2004. Analysis of the germination of spores of Bacillus subtilis with temperature sensitive spo mutations in the spoVA operon. FEMS Microbiol. Lett. 239: 71– 77 [DOI] [PubMed] [Google Scholar]
- 37. Vepachedu VR, Setlow P. 2005. Localization of SpoVAD to the inner membrane of spores of Bacillus subtilis. J. Bacteriol. 187: 5677– 5682 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Vepachedu VR, Setlow P. 2007. Role of SpoVA proteins in release of dipicolinic acid during germination of Bacillus subtilis spores triggered by dodecylamine or lysozyme. J. Bacteriol. 189: 1565– 1572 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. White SW, Zheng J, Zhang YM, Rock CO. 2005. The structural biology of type II fatty acid biosynthesis. Annu. Rev. Biochem. 74: 791– 831 [DOI] [PubMed] [Google Scholar]
- 40. Yanamala N, Tirupula KC, Klein-Seetharaman J. 2008. Preferential binding of allosteric modulators to active and inactive conformational states of metabotropic glutamate receptors. BMC Bioinformatics 9( Suppl. 1): S16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Yi X, Setlow P. 2010. Studies of the commitment step in the germination of spores of Bacillus species. J. Bacteriol. 192: 3424– 3433 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.