Abstract
Swarming is a mode of translocation dependent on flagellar activity that allows bacteria to move rapidly across surfaces. In several bacteria, swarming is a phenotype regulated by quorum sensing. It has been reported that the swarming ability of the soil bacterium Sinorhizobium meliloti Rm2011 requires a functional ExpR/Sin quorum-sensing system. However, our previous published results demonstrate that strains Rm1021 and Rm2011, both known to have a disrupted copy of expR, are able to swarm on semisolid minimal medium. In order to clarify these contradictory results, the role played by the LuxR-type regulator ExpR has been reexamined. Results obtained in this work revealed that S. meliloti can move over semisolid surfaces using at least two different types of motility. One type is flagellum-independent surface spreading or sliding, which is positively influenced by a functional expR gene mainly through the production of exopolysaccharide II (EPS II). To a lesser extent, EPS II-deficient strains can also slide on surfaces by a mechanism that is at least dependent on the siderophore rhizobactin 1021. The second type of surface translocation shown by S. meliloti is swarming, which is greatly dependent on flagella and rhizobactin 1021 but does not require ExpR. We have extended our study to demonstrate that the production of normal amounts of succinoglycan (EPS I) does not play a relevant role in surface translocation but that its overproduction facilitates both swarming and sliding motilities.
INTRODUCTION
Bacteria can move using different types of translocation. Swimming is a flagellum-driven motility that takes place in liquid environments. Bacterial translocation over surfaces can occur by twitching, gliding, sliding, and swarming (18, 19). Twitching is a slow cell movement on surfaces that is mediated by the extension and retraction of type IV pili. Gliding, a surface translocation extensively studied in myxobacteria, does not require flagella or pili but involves focal adhesion complexes, cell surface-associated complexes that anchor the bacterium to a substrate and might act as a motor. Sliding or spreading by expansion has been described as a passive surface translocation that is powered by the outward pressure of bacterial growth and facilitated by compounds that reduce friction between cells and surfaces. Swarming is a mode of surface translocation dependent on rotating flagella characterized by the rapid and coordinated movement of multicellular groups of bacteria. It is considered the fastest known type of bacterial motility on surfaces, with speeds of translocation very similar to the swimmer's speeds (up to 40 μm/s) (18). This allows swarmer cells to rapidly colonize different environments. An additional and distinguishing feature of swarming is that it can involve a complex process of morphological and physiological differentiation. Cells usually (but not always) become hyperflagellated and elongated, and substantial alterations in metabolic pathways and gene expression have been observed (24, 33, 46). This process is known to be triggered upon integration of several chemical and physical signals (12, 23, 45). Swarming has been described as a quorum-sensing-regulated phenotype in several bacteria (8). Quorum-sensing systems have been reported to be involved in the production of biosurfactants that act as wetting agents which reduce the surface tension during surface migration and in swarmer cell differentiation.
Swarming motility is not well characterized in the soil bacteria collectively known as rhizobia that are able to establish nitrogen-fixing symbiosis with legume plants. To date, within rhizobia, this surface motility has been described in Sinorhizobium meliloti, Rhizobium etli, and Rhizobium leguminosarum biovar viciae (7, 42, 44). R. etli has been demonstrated to have a quorum-sensing-regulated swarming motility: mutations affecting the cinIR quorum-sensing system abolish surface translocation in this bacterium. Moreover, it has been shown that N-acyl-homoserine lactones (AHLs) carrying a long-chain fatty acid moiety have a dual role in swarming of R. etli: as quorum-sensing signals and as biosurfactants which promote surface translocation (7).
S. meliloti possesses the ExpR/Sin quorum-sensing system, which is composed of two transcriptional regulators, ExpR and SinR, and the autoinducer synthase SinI, which is responsible for the synthesis of several AHLs (26). The sin AHLs, together with ExpR, control the expression of a large number of genes involved in several free-living and symbiotic cell functions, such as the production of the exopolysaccharides (EPS) succinoglycan (EPS I) and galactoglucan (EPS II) or motility (13, 16, 21, 22). In S. meliloti, the expression of motility genes is downregulated at high population densities. This control is exerted by the ExpR/Sin system via the visNR operon, which codes for the master regulator of flagellar, motility, and chemotaxis genes. At low cell densities, ExpR is required for the activation of motility-related genes, whereas at high population densities, ExpR, in conjunction with AHLs, inhibits transcription of the visNR operon, resulting in the repression of genes belonging to the flagellar regulon (16).
It has been reported that swarming of S. meliloti depends on the presence of a functional ExpR/Sin quorum-sensing system (2, 13). Two different laboratories have reported that only strains carrying a functional expR locus were able to swarm. However, our recent data are in disagreement with these findings. We have reported that the commonly used S. meliloti laboratory strain Rm1021 and the closely related strain Rm2011, both known to have a disrupted copy of expR, are able to swarm on semisolid minimal medium (32). To solve the discrepancies between these reports, in this work, we have reexamined the role played by the expR gene in swarming of S. meliloti. In addition, we have extended our studies to investigate the role of exopolysaccharides EPS I and EPS II in the surface motility of this bacterium.
MATERIALS AND METHODS
Bacterial strains, plasmids, and growth conditions.
Bacterial strains and plasmids used in this work and their relevant characteristics are listed in Table 1. Escherichia coli strains were grown in Luria-Bertani (LB) medium (38) at 37°C; S. meliloti strains were grown at 30°C either in complex tryptone yeast (TY) medium (4), in Bromfield medium (BM) (0.04% tryptone, 0.01% yeast extract, and 0.01% CaCl2 · 2H2O), or in minimal medium (MM) containing glutamate (6.5 mM), mannitol (55 mM), mineral salts (1.3 mM K2HPO4, 2.2 mM KH2PO4 · 3H2O, 0.6 mM MgSO4 · 7H2O, 0.34 mM CaCl2 · 2H2O, 0.022 mM FeCl3 · 6H2O, 0.86 mM NaCl), and vitamins (0.2 mg/liter biotin, 0.1 mg/liter calcium pantothenate) (37). To detect overproduction of EPS I, calcofluor white M2R (fluorescent brightener 28; Sigma) was added to TY or MM plates at a final concentration of 0.02%. When required, antibiotics were added at final concentrations of 50 μg ml−1 streptomycin, 100 μg ml−1 spectinomycin, and 50 μg ml−1 kanamycin for E. coli and 10 μg ml−1 nalidixic acid, 200 μg ml−1 streptomycin, 100 μg ml−1 spectinomycin, 100 μg ml−1 rifampin, 200 μg ml−1 kanamycin, 120 μg ml−1 neomycin, 75 to 100 μg ml−1 hygromycin, and 0.75 μg ml−1 oxytetracycline for S. meliloti. To improve reproducibility, all liquid cultures of S. meliloti were routinely initiated from glycerol stocks. The ability of the different strains to grow in liquid TY, BM, and MM was monitored every 2 h in a Bioscreen C apparatus (Oy Growth Curves Ab Ltd., Finland).
Table 1.
Bacterial strains and plasmids used in this study
| Strain or plasmid | Relevant characteristicsa | Reference or source |
|---|---|---|
| S. meliloti strains | ||
| Rm2011 | Wild type; Nalr Strr | 6 |
| Rm1021 | SU47 expR102::ISRm2011-1; Strr | 28 |
| Rm8530 | Rm1021 expR+; Strr | 14 |
| 2011m.E07 | 2011mTn5STM.1.03.E07, Rm2011 flgE::mini-Tn5; Nalr Strr Neor | 35 |
| 2011mTn5STM.4.06.G01 | Rm2011 wgeB::mini-Tn5; Nalr Strr Neor | 35 |
| Sm2B3001 | Rm2011 with a restored expR gene; Nalr Strr | 2 |
| Sm2B5005 | Sm2B3001 flgE::mini-Tn5; Nalr Strr Neor | 2 |
| Sm2B6005 | Sm2B3001 visN::Sptr; Nalr Strr Sptr | 2 |
| 2011R | Rm2011 with a restored expR gene; Nalr Strr | This study |
| 2011RFg | 2011R flgE::mini-Tn5; Nalr Strr Neor | This study |
| 2011rhbA62 | Rm2011 rhbA::Tn5lac; Strr Rifr Neor | 25 |
| QN1021 | Rm1021 with a fully deleted expR locus; Strr | This study |
| 1021F | Rm1021 flaA flaB; Strr Hygr | This study |
| 1021R | Rm1021 with a restored expR gene; Strr | This study |
| 1021rhbA | Rm1021 rhbA::Tn5lac; Strr Neor | This study |
| 1021Y | Rm1021 ΔexoY; Strr | This study |
| 1021YF | 1021Y flaA flaB; Strr Hygr | This study |
| 1021X | Rm1021 ΔexoX; Strr | This study |
| 1021XF | 1021X flaA flaB; Strr Hygr | This study |
| 1021XY | 1021X ΔexoY; Strr | This study |
| Rm11601 | Rm8530 flaA flaB; Strr Hygr | 16 |
| 8530Vis | Rm8530 with full deletion of visN visR; Strr | B. Scharf |
| 8530Fg | Rm8530 flgE::mini-Tn5; Strr Neor | This study |
| Rm9020 | Rm8530 exoY::Tn5-132; Strr Otcr | 15 |
| 11601Y | Rm11601 ΔexoY; Strr Hygr | This study |
| 8530W | Rm8530 wgeB::mini-Tn5; Strr Neor | This study |
| 11601W | Rm11601 wgeB::mini-Tn5; Strr Neor | This study |
| E. coli strains | ||
| DH5α | supE44 ΔlacU169 ϕ80d lacZΔM15 recA1endA1gyrA96thi-1relA1hsdR171 | Bethesda Research Lab |
| S17-1 | thiprorecAhsdRhsdM; Rp4Tc::Mu, Km::Tn7; Tmpr Strr Sptr | 41 |
| Plasmids | ||
| pCR-XL-TOPO | Cloning vector; Kanr | Invitrogen |
| pK18mobsacB | Suicide plasmid; Kanr | 39 |
| pK18-ΔexpR | pK18mobsacB carrying the deleted version of the expR locus; Kanr | This study |
| pK18-expR | pK18mobsacB carrying the expR gene from Rm8530; Kanr | This study |
| pK18-ΔexoY | pK18mobsacB carrying the deleted version of the exoY locus; Kanr | This study |
| pK18-ΔexoX | pK18mobsacB carrying the deleted version of the exoX locus; Kanr | This study |
Nalr, Strr, Neor, Sptr, Rifr, Hygr, Otcr, Tmpr, and Kanr indicate nalidixic acid, streptomycin, neomycin, spectinomycin, rifampin, hygromycin, oxytetracycline, trimethoprim, and kanamycin resistance, respectively.
Construction of S. meliloti strains.
For the construction of expR+ derivatives of Rm1021 (1021R) and Rm2011 (2011R), the functional expR gene of Rm8530 was PCR amplified using primers Rmpyc and SmndvA2 (Table 2), cloned into pCR-XL-TOPO, and sequenced. This construct was digested with EcoRI, and the 1,550-bp fragment containing the functional expR gene was isolated and subcloned into pK18mobsacB to yield plasmid pK18-expR. This plasmid was introduced into Rm1021 and Rm2011 via conjugation with E. coli strain S17-1, and allele replacement events were selected as described previously (39). In this case, clones in which allelic exchange occurred were easily identified, as they showed a very noticeable mucoid phenotype. S. meliloti strain QN1021 (expR) was obtained by replacing the disrupted expR locus of Rm1021 comprising the insertion sequence (IS) ISRm2011-1 and the IS-flanking loci smc03896 and smc03899 (http://iant.toulouse.inra.fr/bacteria/annotation/cgi/rhime.cgi) (34) with an unmarked deleted version. The expR deletion was generated in vitro by overlap extension PCR (20) using primers ExpR.1 to ExpR.4, listed in Table 2. The resulting fusion product, in which a deletion of 1,943 bp was created, was cloned into pCR-XL-TOPO and sequenced. Using the HindIII and BamHI restriction sites included in the outside primers, the insert was subcloned into vector pK18mobsacB, yielding plasmid pK18-ΔexpR. This construction was introduced into Rm1021 via conjugation with S17-1, and allele replacement events were selected as described previously (39). Likewise, S. meliloti mutant strains with deletion-containing versions of exoX and exoY were obtained by allelic replacement using the same methodology. The exoX and exoY mutant alleles harboring in-frame deletions of 274 and 501 bp, respectively, were generated in vitro by overlap extension PCR using primers listed in Table 2. The resulting PCR products were cloned into pCR-XL-TOPO, sequenced, and, by using the restriction sites included in the outside primers, subcloned into vector pK18mobsacB to yield plasmids pK18-ΔexoX and pK18-ΔexoY. pK18-ΔexoX was introduced into Rm1021, and after selection of allele replacement, the EPS I-overproducer 1021X strain was obtained. pK18-ΔexoY was introduced into Rm1021, Rm11601, and 1021X to yield the corresponding mutant strains defective in EPS I (1021Y, 11601Y, and 1021XY, respectively). Phage ΦM12 transduction (10) was employed to transfer mutations among strains in the following manners. (i) The flaA flaB mutants 1021F (expR flaA flaB), 1021YF (expR exoY flaA flaB), and 1021XF (expR exoX flaA flaB) were obtained by transferring the ΔflaA flaB::Hyg mutation from strain Rm11601 (expR+ flaA flaB) to strains Rm1021 (expR), 1021Y (expR exoY), and 1021X (expR exoX), respectively. (ii) The flgE mutants 2011RFg (expR+ flgE) and 8530Fg (expR+ flgE) were obtained by transferring the flgE::mini-Tn5 mutation from strain 2011mTn5STM.1.03.E07 to 2011R (expR+) and Rm8530 (expR+), respectively. (iii) The rhbA mutant 1021rhbA (expR rhbA) was obtained by transferring the rhbA::Tn5lac mutation from strain 2011rhbA62 to Rm1021 (expR). (iv) Likewise, the wgeB mutants 8530W (expR+ wgeB) and 11601W (expR+ flaA flaB wgeB) were obtained by transferring the mini-Tn5 disrupted locus wgeB from strain 2011mTn5STM.4.06.G01 to strains Rm8530 and Rm11601, respectively. All mutants constructed in this work were checked by PCR and Southern hybridization with specific probes.
Table 2.
Primers used in this study
| Primer | Sequence (5′ to 3′)a | Use |
|---|---|---|
| Rmpyc | AGAGTGGCGTGAACATTCGG | expR restoration |
| SmndvA2 | TCCTTCTGTGACGAGATCG | expR restoration |
| ExpR.1 | AAAAAGCTTGCTTTTCGAGATAGACCTCG (HindIII) | expR deletion |
| ExpR.2 | CGTACAGTTTCTGGCTGGTACATGAACG | expR deletion |
| ExpR.3 | CGTTCATGTACCAGCCAGAAACTGTACGAGC | expR deletion |
| ExpR.4 | AAAGGATCCCGTGAACTTCTTCAGTTCGC (BamHI) | expR deletion |
| delexoY.1 | AAAGGATCCACCTCATAAGAGTTGTTGCC (BamHI) | exoY deletion |
| delexoY.2 | GGACATATTGCGTGTTTGCCATACCTCC | exoY deletion |
| delexoY.3 | GGAGGTATGGCAAACACGCAATATGTCC | exoY deletion |
| delexoY.4 | AAAGGATCC AATACCGTCAAATTGGGAGC (BamHI) | exoY deletion |
| exoX1 | AATAAGCTTGGACTTCATAGAGGTGACTC (HindIII) | exoX deletion |
| exoX2 | GCTCAGGAATTGAGGGTGCGAACATGGC | exoX deletion |
| exoX3 | GCCATGTTCGCACCCTCAATTCCTGAGCGGC | exoX deletion |
| exoX4 | AATGGATCCGAGCGTAGAGATCGTAATC (BamHI) | exoX deletion |
Restriction sites used for cloning (underlined) are given in parentheses after the sequence.
Motility assays.
Swimming was examined on plates prepared with BM containing 0.3% Bacto agar and inoculated with 3-μl droplets of rhizobial cultures grown in TY (optical density at 600 nm = 1). Surface motility was analyzed using two different methodologies: (i) the motility assay described by Bahlawane et al. (2) in which 3 μl of overnight TY rhizobial cultures was inoculated onto the surface of BM containing 0.6% Bacto agar and (ii) the motility test described in our previous work (32, 42) in which 2 μl of washed, 10-fold-concentrated of cultures grown in TY broth to the late exponential phase was inoculated onto semisolid MM plates. For swimming and surface motility tests performed on BM, the migration zone was determined as the colony diameter in millimeters. In the case of surface motility tests performed on semisolid MM, in which fractal patterns with characteristic tendrils were formed, migration zones were calculated as the average length of the two sides of a rectangle able to exactly frame each colony.
CAS siderophore assay.
The determination of siderophores in liquid cultures was performed using the chrome azurol S (CAS) assay solution described by Schwyn and Neilands (40). Supernatants of S. meliloti cultures were mixed 1:1 with the CAS assay solution. After reaching equilibrium, the absorbance was measured at 630 nm.
RESULTS
ExpR promotes flagellum-independent surface spreading of S. meliloti.
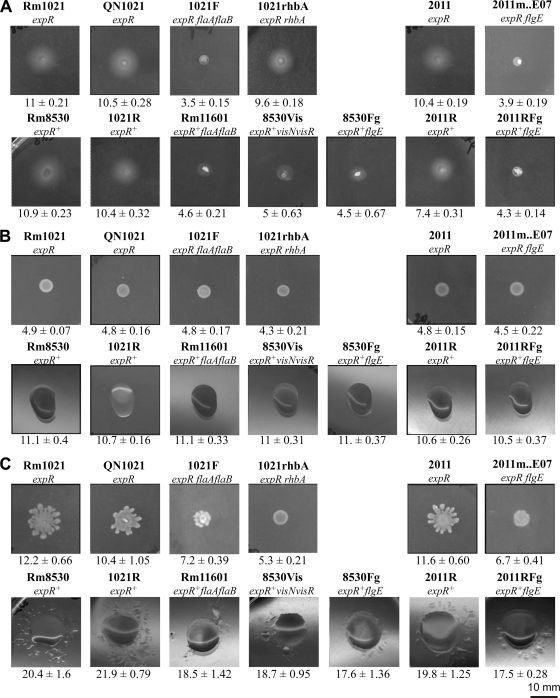
Swimming motility tests performed with Rm1021 (expR) and Rm8530 (expR+) revealed the abilities of these strains to swim without significant differences between them (Fig. 1A), thereby confirming previously published results (2). The same strains were assayed for surface motility on 0.6% agar BM highlighting different phenotypes (Fig. 1B). Whereas macrocolonies formed by Rm1021 were dry and did not show signs of significant surface expansion after 3 days of incubation, those formed by Rm8530 were highly mucoid and clearly covered a larger surface area. However, neither the macroscopic appearance nor the slow translocation over the surface of BM shown by Rm8530 (ca. 0.04 μm/s) was indicative of swarming motility. The two new Rm1021 derivative strains constructed in this study, QN1021 (expR) and 1021R (expR+), showed the same behavior as Rm1021 (expR) and Rm8530 (expR+), respectively. Bahlawane and coworkers described the surface expansion shown by their S. meliloti expR+ strains as swarming based on the fact that Rm2011 expR+ derivative strains defective in flagellum production (flgE and visN mutants) were nonmotile on semisolid BM (2). We tested the motility phenotype of three mutant derivative strains of Rm8530: Rm11601 (expR+ flaA flaB), lacking functional flagellar filaments, 8530Vis (expR+ visN visR), lacking the master regulator of flagellar, motility, and chemotaxis genes, and 8530Fg (expR+ flgE), affected in the gene putatively coding for the flagellar hook protein. These three mutant strains were devoid of flagella (16) (data not shown) and consequently were incapable of swimming (Fig. 1A). On the contrary, on 0.6% agar BM, all three strains showed the same behavior as the flagellated parental strain Rm8530 (Fig. 1B). These results were in disagreement with the data presented by Bahlawane et al. in which nonflagellated expR+ derivatives of the closely related strain Rm2011 were shown to be nonmotile under the same conditions. To investigate if the differences were due to strain-specific effects, the motility of Rm2011 (expR) and that of its derivatives Sm2B3001 (expR+), Sm2B5005 (expR+ flgE), and Sm2B6005 (expR+ visN) were tested on 0.6% agar BM (not shown in Fig. 1). The behavior of Rm2011 and the visN derivative mutant strain Sm2B6005 was reproducible in all our assays and in agreement with our results: colonies formed by the ExpR-deficient strain Rm2011 were dry, whereas those formed by the nonflagellated expR+ derivative strain Sm2B6005 were highly mucoid and spread over the surface of semisolid BM significantly more than colonies formed by Rm2011 (9.9 ± 0.2 mm versus 5.4 ± 0.2 mm, respectively). On the contrary, an unstable mucoid phenotype was observed for strains 2B3001 (expR+) and 2B5005 (expR+ flgE), leading to unreliable results. Therefore, we decided to construct two new Rm2011 derivative strains: 2011R (expR+) and 2011RFg (expR+ flgE). As shown in Fig. 1B, the new strains behaved like Rm8530 (expR+) and 8530Fg (expR+ flgE), respectively. Altogether, these data demonstrate that S. meliloti strains harboring a functional expR gene are able to spread on the surface of semisolid BM but that the mechanism used is not dependent on flagella and therefore cannot be described as swarming motility.
Fig 1.
Role of ExpR in motility of S. meliloti. (A) Swimming test in Bromfield medium (0.3% agar). (B) Surface motility on semisolid Bromfield medium (0.6% agar). (C) Surface motility on semisolid MM (0.6% agar). The relevant genotype is indicated under the name of each strain. Pictures were taken 2 days (A), 3 days (B), or 20 h (C) after inoculation. Under each image, the mean and standard deviation of the migration zones (given in millimeters and measured as described in the text) obtained from at least nine measurements are indicated.
ExpR is not required for swarming motility of S. meliloti.
When surface motility assays were performed on semisolid MM plates, surface translocation with characteristic tendril formation could be observed for the ExpR-deficient strains Rm1021, QN1021, and Rm2011 by 14 to 20 h after inoculation (Fig. 1C). To corroborate that the surface motility shown by S. meliloti ExpR-deficient strains on semisolid MM was dependent on flagella, the motility phenotypes of 1021F (expR flaA flaB) and 2011m.E07 (expR flgE) were assayed and compared to those of their corresponding parental strains Rm1021 and Rm2011, respectively. As expected for nonflagellated bacteria, 1021F and 2011m.E07 were nonmotile in swimming assays performed in BM containing 0.3% agar (Fig. 1A). Likewise, as expected for ExpR-deficient strains, they did not spread on 0.6% BM (Fig. 1B). On semisolid MM, surface translocation of these two strains was severely affected compared to that of their parental strains although not completely abolished, as is the case for 2011rhbA62 and 1021rhbA, mutant strains unable to produce the siderophore rhizobactin 1021 that are derived from Rm2011 and Rm1021, respectively (Fig. 1C) (32). This result demonstrates that ExpR-deficient S. meliloti strains are able to show flagellum-driven surface translocation (i.e., swarming motility) on semisolid MM, and therefore, we can conclude that ExpR is not required for swarming. The minor surface spreading shown by nonflagellated ExpR-deficient strains reveals the existence of a second type of surface motility which is not dependent on flagellar activity. This ExpR- and flagellum-independent surface motility seems to be regulated by the nutrient composition of the medium since it is manifested only on semisolid MM and not in semisolid BM. The fact that gene mutations (rhb) and environmental conditions (high iron) which block the synthesis of the siderophore rhizobactin 1021 render S. meliloti Rm1021/Rm2011 completely nonmotile on semisolid MM (Fig. 1C, 1021rhbA) (32) suggests that rhizobactin 1021 plays a role in both the flagellum-driven and the flagellum-independent surface motilities shown by ExpR-deficient S. meliloti strains. CAS assays performed with supernatants of Rm1021/Rm2011 cultures grown in BM revealed a lack of siderophore production (data not shown), which could explain the absence of surface motility on semisolid BM by these ExpR-deficient strains.
Surface motility assays on semisolid MM were also performed for S. meliloti strains harboring a functional expR gene (Fig. 1C). In contrast to the behavior shown by Rm1021/QN1021/Rm2011, colonies formed by 1021R/Rm8530/2011R (expR+ strains) were highly mucoid and showed smooth borders, although some tendrils could also be observed. Notably, the expR+ strains spread extensively over the surface, covering an area which was almost twice as large as the area colonized by ExpR-deficient strains, suggesting that ExpR promotes surface translocation not only on semisolid BM but also on semisolid MM. However, in contrast to expR mutant strains, the surface spreading displayed by Rm8530 was not significantly reduced in the absence of flagella, as revealed by the phenotypes exhibited by Rm11601 (expR+ flaA flaB), 8530Vis (expR+ visN visR), and 8530Fg (expR+ flgE) (Fig. 1C). Similar behavior was observed for 2011R and its nonflagellated derivative strain 2011RFg. These results indicate that, as on semisolid BM, the surface translocation shown by expR+ strains of S. meliloti on semisolid MM is not dependent on flagellar activity and therefore cannot be described as swarming.
EPS II is not required for swarming and promotes sliding motility in S. meliloti.
The observed correlation between mucoidy and flagellum-independent surface motility that was shown by expR+ strains, together with the role assigned to ExpR in EPS I and EPS II synthesis (21), prompted us to investigate the function of these exopolysaccharides in the surface spreading exhibited by S. meliloti.
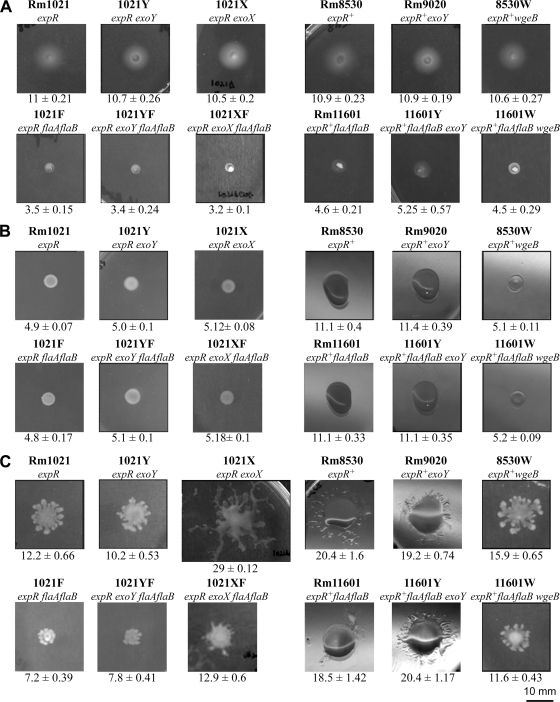
It is known that ExpR-deficient strains of S. meliloti, such as Rm1021, do not produce EPS II at detectable levels unless they are grown under low-phosphate conditions (29). Nevertheless, they are able to show swarming motility, as we have demonstrated in this and previous works (32), indicating that EPS II is not required for this flagellum-driven surface translocation. To investigate the role of EPS II in the motility of S. meliloti expR+ strains, we constructed wgeB (formerly expE2) mutants impaired in a glycosyl transferase involved in EPS II synthesis (3). Strain 8530W (expR+ wgeB) and nonflagellated 11601W (expR+ flaA flaB wgeB) showed a nonmucoid phenotype in different media, as expected for S. meliloti strains unable to synthesize EPS II. Moreover, no relevant differences in swimming rings were detected between these strains and their corresponding parental strains (Rm8530 and Rm11601, respectively) (Fig. 2A), suggesting that EPS II plays no role in swimming motility. However, in contrast to their parental strains and all the expR+ strains tested in this work, 8530W and 11601W did not spread over the surface of 0.6% BM, displaying the same phenotype as ExpR-deficient strains (Fig. 2B). 8530W and 1161W did not show defects in growth in liquid BM (data not shown). Thus, these results clearly demonstrate that the flagellum-independent surface translocation shown by ExpR strains of S. meliloti on semisolid BM is absolutely dependent on the production of EPS II. Therefore, this mode of translocation is most akin to sliding motility, whereby the production of EPS II promotes passive movement of cells across the agar surface.
Fig 2.
Role of exopolysaccharides EPS I and EPS II in motility of S. meliloti. (A) Swimming test in Bromfield medium (0.3% agar). (B) Surface motility on semisolid Bromfield medium (0.6% agar). (C) Surface motility on semisolid MM (0.6% agar). The relevant genotype is indicated under the name of each strain. Pictures were taken 2 days (A), 3 days (B), or 20 h (C) after inoculation. Under each image, the mean and standard deviation of the migration zones (given in millimeters and measured as described in the text) obtained from at least nine measurements are indicated.
The wgeB mutation also led to a significant reduction (23% in the case of flagellated Rm8530 and 37% for nonflagellated Rm11601) in the surface motility shown by expR+ strains of S. meliloti on semisolid MM (Fig. 2C). No differences in growth rates were detected in MM broth between the two wgeB mutants and their corresponding parental strains (data not shown). Therefore, EPS II also contributes to flagellum-independent surface translocation or sliding, which seems to be the predominant mode of translocation of expR+ strains, on MM. Interestingly, when EPS II production is blocked, these strains exhibit swarming motility, manifested by the ca. 4.25-mm difference in surface spreading displayed between 8530W (expR+ wgeB) (15.9 mm) and 11601W (expR+ flaA flaB wgeB) (11.6 mm) which, indeed, is very similar to the difference in surface spreading (5 mm) shown by Rm1021 (expR) (12.2 mm) and 1021F (expR, flaA flaB) (7.2 mm). These results indicate that as in expR mutants, EPS II is not essential for the swarming motility of expR+ strains. The sliding motility promoted by EPS II allows for a larger surface colonization than the swarming motility exhibited by the same strain when EPS II synthesis is blocked. This makes it difficult to determine if swarming and sliding coexist in expR+ strains or if, alternatively, EPS II production inhibits flagellum-driven surface motility. In either case, our data revealed that once EPS II production is impeded, ExpR does not significantly influence swarming motility in S. meliloti.
As is the case for the nonflagellated strain 1021F (expR flaA flaB), the ability of 11601W (expR+ flaA flaB wgeB) to move over the surface of semisolid MM was not abolished. Indeed, the flagellum-independent surface translocation shown by 11601W seemed to be enhanced compared to that shown by 1021F. This behavior could be the result of the better growth rate shown by 1161W in liquid MM compared to that of 1021F (data not shown). Regardless of the effect of growth on surface translocation, our results suggest that an ExpR-controlled factor might play a role in flagellum-independent surface translocation. It is tempting to speculate that this factor might be rhizobactin 1021 based on the role played by this siderophore in surface motility of ExpR-deficient strains and on the reported transcriptomic data which revealed higher expression of the rhrA gene (encoding the AraC-like regulator, which positively regulates the production and transport of rhizobactin 1021) in Rm8530 (expR+) than in Rm1021 (expR) (21). However, this hypothesis has not been investigated here.
Overproduction of EPS I promotes both sliding and swarming motilities in ExpR-deficient S. meliloti strains.
To investigate the role of EPS I in the different types of motility shown by S. meliloti, several exoY mutants lacking a sugar transferase essential in EPS I synthesis (30) were generated. As shown in Fig. 2, under the three conditions tested, the phenotypes exhibited by the exoY mutants 1021Y (expR exoY), 1021YF (expR exoY flaA flaB), Rm9020 (expR+ exoY), and 11601Y (expR+ flaA flaB exoY) were similar to those of their corresponding isogenic strains harboring a functional exoY locus (Rm1021, 1021F, Rm8530, and Rm11601, respectively). These results demonstrate that the production of normal amounts of EPS I does not play a significant role in either swimming, swarming (observed in ExpR-deficient strains on semisolid MM), or flagellum-independent surface spreading (shown by expR+ strains on both semisolid BM and MM and by ExpR-deficient strains on semisolid MM).
We decided to test if an increased production of EPS I could have an effect on motility similar to the effect caused by the large amount of EPS II produced by expR+ strains. The overproduction of EPS I was achieved by deleting most of the coding sequence of the exoX gene, whose disruption has been shown to cause overproduction of low-molecular-weight EPS I in S. meliloti (36). The gene deletion eliminates essential amino acids required for the inhibitory effect of ExoX on exopolysaccharide synthesis, an effect that it is thought to occur posttranslationally in a mechanism in which the stoichiometry with ExoY is important.
The exoX derivative mutant strains 1021X and 1021XF were more mucoid on MM plates than the corresponding parental strains Rm1021 and 1021F. Moreover, the higher fluorescence shown under long-wave UV light by 1021X and 1021XF, grown on TY plates supplemented with the fluorescent dye calcofluor white, compared to that by their parental strains confirmed EPS I overproduction (data not shown). Motility tests performed with these strains revealed no significant differences in swimming (Fig. 2A). In addition, no surface translocation associated with the exoX mutation could be observed on semisolid BM (Fig. 2B). Interestingly, on semisolid MM, 1021X (expR exoX), which showed a growth rate in liquid MM that was similar to that of the parental strain, exhibited the largest surface translocation of all the strains tested in this work, colonizing a surface area which was 2.4-fold wider than that of the parental strain Rm1021 (Fig. 2C). This movement was strongly diminished in the absence of flagella, as revealed by the behavior of 1021XF (expR exoX flaA flaB), demonstrating that 1021X shows swarming motility. Furthermore, the flagellum-promoted surface spreading exhibited by 1021X (expR exoX) (ca. 16 mm) was approximately 3-fold larger than the flagellum-driven surface translocation shown by Rm1021 (expR) (ca. 5 mm) (Fig. 2C), indicating that overproduction of EPS I promotes swarming motility. On the other hand, EPS I overproduction also promotes flagellum-independent surface translocation on MM, as revealed by the larger area colonized by 1021XF (expR exoX flaA flaB) compared to the surface area colonized by 1021F (expR flaA flaB) (Fig. 2C). Introducing an exoY mutation into the 1021X strain led to phenotypes (mucoidy, calcofluor brightness, and surface motility) that were similar to those shown by Rm1021 (expR) and 1021Y (expR exoY) (data not shown), demonstrating that the overproduction of EPS I was the only cause of the observed effects in exoX mutants.
DISCUSSION
This work was aimed at solving the existing discrepancies concerning the role of the LuxR-type regulator ExpR in the swarming motility of S. meliloti. Two different groups reported independently that swarming of S. meliloti depends on the presence of a functional expR locus (2, 13). However, we recently reported that strains Rm1021 and Rm2011, both known to have a disrupted copy of expR, are able to swarm on semisolid medium (32). We have reexamined the role played by ExpR by using different mutants in different genetic backgrounds and assaying their motility phenotypes under the experimental conditions described in the contradicting publications.
The new data showed that although ExpR-deficient strains do not display surface translocation on semisolid BM, as reported by Bahlawane et al. (2), they exhibit flagellum-driven surface translocation on semisolid MM. Therefore, we can conclude that ExpR is not essential for swarming motility. Moreover, it became clear that, as previously reported for a S. meliloti fadD mutant (42), the swarming motility of ExpR-deficient strains is greatly influenced by the nutrient composition of the medium. Besides flagella, the production of the siderophore rhizobactin 1021, which requires the presence of low iron concentrations in the medium, is the only factor known up to now to play an essential role in the swarming motility of ExpR-deficient strains.
In addition to demonstrating the dispensability of ExpR for swarming motility in S. meliloti, this study has unveiled the existence in S. meliloti of an additional mode of surface translocation which does not require flagellar activity. This type of movement was especially noticeable in strains harboring a functional expR locus in both semisolid BM and MM. By using up to 4 different nonflagellated derivative mutants (including those used in the study by Bahlawane et al.), we clearly demonstrated that the surface spreading shown by expR+ strains on semisolid medium was not significantly diminished by the absence of flagella and therefore cannot be considered swarming. However, when synthesis of galactoglucan (EPS II) was blocked by generating wgeB mutations, surface spreading of expR+ strains was completely abolished on BM and significantly reduced on MM. Considering these data, the surface translocation shown by expR+ strains of S. meliloti is most akin to sliding motility (18, 19), whereby the production of EPS II promotes passive movement of cells across the agar surface.
To the best of our knowledge, this work represents the first report on sliding motility in Rhizobium. Sliding or spreading by expansion has been described for a diverse group of bacteria, such as mycobacteria, Bacillus subtilis, Vibrio cholerae, Serratia marcescens, Pseudomonas aeruginosa, and Legionella pneumophila (1, 5, 9, 27, 31, 43), in which a strong correlation between sliding and the production of surfactants has been established. For example, the production of rhamnolipids in Pseudomonas, the lipopeptides surfactin and serrawettin in Bacillus and Serratia, respectively, or a surfactant-like material in Legionella facilitates flagellum-independent surface translocation in these bacteria. Most of these surfactants also play a crucial role in swarming motility (reviewed in references 8, 23, and 45). We are not aware of the possible surfactant properties of the galactoglucan produced by S. meliloti, and we can only speculate about its role in sliding motility. It is possible that the high levels of EPS II excreted by expR+ strains serve either as a hydrated milieu that gives sufficient moisture to facilitate the spreading of the colony or as a lubricant that reduces friction between cells and surfaces. In any case, and in contrast to surfactants such as rhamnolipids, surfactin, or serrawettin, EPS II is not essential for the swarming motility of S. meliloti, as indicated by the flagellum-dependent translocation shown by EPS II-defective strains, regardless of whether they have a functional ExpR regulator.
In addition to swarming and EPS II-promoted sliding motility, S. meliloti strains can also spread over surfaces, although to a lesser extent, using a flagellum- and EPS II-independent type of motility. In ExpR-deficient strains, this motility relies on the production of the rhizobactin 1021 siderophore, since abolishment of its synthesis renders Rm1021/Rm2011 strains completely nonmotile. Therefore, rhizobactin 1021 plays a crucial role in both swarming and flagellum-independent surface translocation shown by expR strains of S. meliloti. We recently observed that purified rhizobactin 1021 shows drop collapse activity (our unpublished results), a property probably conferred by the presence of the long-chain fatty acid (E)-2-decenoic acid in its chemical structure. Thus, it is very probable that, as reported for other surfactants which play roles in swarming and sliding motilities, rhizobactin 1021 contributes to the surface migration of S. meliloti by acting as a wetting agent. We have not demonstrated in this work if rhizobactin 1021 also accounts for the flagellum- and EPS II-independent surface translocation exhibited by expR+ strains on semisolid MM, although this possibility is very likely.
The results presented in this work also provide further insights into additional factors contributing to surface translocation in S. meliloti. The phenotype exhibited by exoX derivative mutants of Rm1021 demonstrates that the overproduction of EPS I, but not the production of normal amounts of this EPS, facilitates both sliding and swarming motilities. Extracellular polysaccharides have been involved in surface translocation in other bacteria. Thus, the acidic capsular polysaccharide produced by Proteus mirabilis that is known as colony migration factor (Cmf), which is an important component of the extracellular matrix that surrounds swarmer cells, plays a key role in swarming motility by reducing surface friction during translocation (17). A similar role could be attributed to EPS I but only at high levels of production.
This work has unveiled the unexpected complexity of surface translocation in S. meliloti, raising questions that require further investigation. It is clear that EPS II-dependent sliding is the most relevant type of surface translocation displayed by expR+ strains of S. meliloti, allowing these bacteria to colonize surfaces more efficiently than strains displaying only swarming motility. Whether swarming and sliding take place at the same time in expR+ strains or whether EPS II inhibits swarming motility still remains unclear. Furthermore, although we show here that ExpR is not required and does not significantly influence swarming motility in S. meliloti, we cannot rule out the possibility of population density regulation of swarming motility in this bacterium. Therefore, efforts should be continued to identify and characterize other regulators and components which play key roles in sliding and/or swarming. Another interesting question to be solved is the role these types of surface motilities play in the different lifestyles of Rhizobium spp. Whereas the role of swarming motility in the establishment of the Rhizobium-legume symbiosis is still unclear, sliding motility may allow S. meliloti to colonize surfaces under conditions where flagellar expression is downregulated, for instance, at high cell population densities and during the invasion process. In line with this, a collective sliding movement of bacteria toward the infection thread tip has been proposed to contribute to colonization of the thread (11). The biological significance of the ability to slide or swarm in Rhizobium-legume symbiosis remains to be elucidated.
ACKNOWLEDGMENTS
This work was supported by grants BIO2007-62988 and BIO2010-18005 from the Ministerio de Ciencia e Innovación (Spain), CVI 03541 from the Junta de Andalucía (Spain), and FEDER funds. J.N. and L.B.R. were supported by grants from the Consejería de Innovación, Ciencia y Empresa (Junta de Andalucía, Spain).
We are grateful to Juan E. González, Anke Becker, and Birgit Scharf for providing several S. meliloti strains.
Footnotes
Published ahead of print 10 February 2012
REFERENCES
- 1. Agustí G, Astola O, Rodríguez-Güell E, Julián E, Luquin M. 2008. Surface spreading motility shown by a group of phylogenetically related, rapidly growing pigmented mycobacteria suggests that motility is a common property of mycobacterial species but is restricted to smooth colonies. J. Bacteriol. 190: 6894– 6902 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Bahlawane C, McIntosh M, Krol E, Becker A. 2008. Sinorhizobium meliloti regulator MucR couples exopolysaccharide synthesis and motility. Mol. Plant Microbe Interact. 21: 1498– 1509 [DOI] [PubMed] [Google Scholar]
- 3. Becker A, et al. 1997. The 32-kilobase exp gene cluster of Rhizobium meliloti directing the biosynthesis of galactoglucan: genetic organization and properties of the encoded gene products. J. Bacteriol. 179: 1375– 1384 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Beringer JE. 1974. R factor transfer in Rhizobium leguminosarum. J. Gen. Microbiol. 84: 188– 198 [DOI] [PubMed] [Google Scholar]
- 5. Brown II, Häse CC. 2001. Flagellum-independent surface migration of Vibrio cholerae and Escherichia coli. J. Bacteriol. 183: 3784– 3790 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Casse F, Boucher C, Julliot JS, Michel M, Dénarié J. 1979. Identification and characterization of large plasmids in Rhizobium meliloti using agarose-gel electrophoresis. J. Gen. Microbiol. 113: 229– 242 [Google Scholar]
- 7. Daniels R, et al. 2006. Quorum signal molecules as biosurfactants affecting swarming in Rhizobium etli. Proc. Natl. Acad. Sci. U. S. A. 103: 14965– 14970 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Daniels R, Vanderleyden J, Michiels J. 2004. Quorum sensing and swarming migration in bacteria. FEMS Microbiol. Rev. 28: 261– 289 [DOI] [PubMed] [Google Scholar]
- 9. Fall R, Kearns DB, Nguyen T. 2006. A defined medium to investigate sliding motility in a Bacillus subtilis flagella-less mutant. BMC Microbiol. 6: 31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Finan TM, et al. 1984. General transduction in Rhizobium meliloti. J. Bacteriol. 159: 120– 124 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Fournier J, et al. 2008. Mechanism of infection thread elongation in root hairs of Medicago truncatula and dynamic interplay with associated rhizobial colonization. Plant Physiol. 148: 1985– 1995 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Fraser GM, Hughes C. 1999. Swarming motility. Curr. Opin. Microbiol. 2: 630– 635 [DOI] [PubMed] [Google Scholar]
- 13. Gao M, et al. 2005. sinI- and expR-dependent quorum sensing in Sinorhizobium meliloti. J. Bacteriol. 187: 7931– 7944 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Glazebrook J, Walker GC. 1989. A novel exopolysaccharide can function in place of the calcofluor-binding exopolysaccharide in nodulation of alfalfa by Rhizobium meliloti. Cell 56: 661– 672 [DOI] [PubMed] [Google Scholar]
- 15. Glazebrook J, Walker GC. 1991. Genetic techniques in Rhizobium meliloti. Methods Enzymol. 204: 398– 418 [DOI] [PubMed] [Google Scholar]
- 16. Gurich N, González JE. 2009. Role of quorum sensing in Sinorhizobium meliloti-alfalfa symbiosis. J. Bacteriol. 191: 4372– 4382 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Gygi D, et al. 1995. A cell-surface polysaccharide that facilitates rapid population migration by differentiated swarm cells of Proteus mirabilis. Mol. Microbiol. 17: 1167– 1175 [DOI] [PubMed] [Google Scholar]
- 18. Harshey RM. 2003. Bacterial motility on a surface: many ways to a common goal. Annu. Rev. Microbiol. 57: 249– 273 [DOI] [PubMed] [Google Scholar]
- 19. Henrichsen J. 1972. Bacterial surface translocation: a survey and a classification. Bacteriol. Rev. 36: 478– 503 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Ho SN, Hunt HD, Horton RM, Pullen JK, Pease LR. 1989. Site-directed mutagenesis by overlap extension using the polymerase chain reaction. Gene 77: 51– 59 [DOI] [PubMed] [Google Scholar]
- 21. Hoang HH, Becker A, González JE. 2004. The LuxR homolog ExpR, in combination with the Sin quorum sensing system, plays a central role in Sinorhizobium meliloti gene expression. J. Bacteriol. 186: 5460– 5472 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Hoang HH, Gurich N, González JE. 2008. Regulation of motility by the ExpR/Sin quorum-sensing system in Sinorhizobium meliloti. J. Bacteriol. 190: 861– 871 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Kearns DB. 2010. A field guide to bacterial swarming motility. Nat. Rev. Microbiol. 8: 634– 644 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Kim W, Surette MG. 2004. Metabolic differentiation in actively swarming Salmonella. Mol. Microbiol. 54: 702– 714 [DOI] [PubMed] [Google Scholar]
- 25. Lynch D, et al. 2001. Genetic organization of the region encoding regulation, biosynthesis, and transport of rhizobactin 1021, a siderophore produced by Sinorhizobium meliloti. J. Bacteriol. 183: 2576– 2585 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Marketon MM, Gronquist MR, Eberhard A, González JE. 2002. Characterization of the Sinorhizobium meliloti sinR/sinI locus and the production of novel N-acyl homoserine lactones. J. Bacteriol. 184: 5686– 5695 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Matsuyama T, Bhasin A, Harshey RM. 1995. Mutational analysis of flagellum-independent surface spreading of Serratia marcescens 274 on a low-agar medium. J. Bacteriol. 177: 987– 991 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Meade HM, Signer ER. 1977. Genetic mapping of Rhizobium meliloti. Proc. Natl. Acad. Sci. U. S. A. 74: 2076– 2078 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Mendrygal KE, González JE. 2000. Environmental regulation of exopolysaccharide production in Sinorhizobium meliloti. J. Bacteriol. 182: 599– 606 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Müller P, et al. 1993. Genetic analysis of the Rhizobium meliloti exoYFQ operon: ExoY is homologous to sugar transferases and ExoQ represents a transmembrane protein. Mol. Plant Microbe Interact. 6: 55– 65 [DOI] [PubMed] [Google Scholar]
- 31. Murray TS, Kazmierczak BI. 2008. Pseudomonas aeruginosa exhibits sliding motility in the absence of type IV pili and flagella. J. Bacteriol. 190: 2700– 2708 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Nogales J, et al. 2010. Transcriptome profiling of a Sinorhizobium meliloti fadD mutant reveals the role of rhizobactin 1021 biosynthesis and regulation genes in the control of swarming. BMC Genomics 11: 157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Overhage J, Bains M, Brazas MD, Hancock RE. 2008. Swarming of Pseudomonas aeruginosa is a complex adaptation leading to increased production of virulence factors and antibiotic resistance. J. Bacteriol. 190: 2671– 2679 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Pellock BJ, Teplitski M, Boinay RP, Bauer WD, Walker GC. 2002. A LuxR homolog controls production of symbiotically active extracellular polysaccharide II by Sinorhizobium meliloti. J. Bacteriol. 184: 5067– 5076 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Pobigaylo N, et al. 2006. Construction of a large signature-tagged mini-Tn5 transposon library and its application to mutagenesis of Sinorhizobium meliloti. Appl. Environ. Microbiol. 72: 4329– 4337 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Reed JW, Capage M, Walker GC. 1991. Rhizobium meliloti exoG and exoJ mutations affect the ExoX-ExoY system for modulation of exopolysaccharide production. J. Bacteriol. 173: 3776– 3788 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Robertsen BK, Aiman P, Darwill AG, Mcneil M, Albersheim P. 1981. The structure of acidic extracellular polysaccharides secreted by Rhizobium leguminosarum and Rhizobium trifolii. Plant Physiol. 67: 389– 400 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Sambrook J, Russell DW. 2001. Molecular cloning: a laboratory manual, 3rd ed Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY [Google Scholar]
- 39. Schäfer A, et al. 1994. Small mobilizable multi-purpose cloning vectors derived from the Escherichia coli plasmids pK18 and pK19: selection of defined deletions in the chromosome of Corynebacterium glutamicum. Gene 145: 69– 73 [DOI] [PubMed] [Google Scholar]
- 40. Schwyn B, Neilands JB. 1987. Universal chemical assay for the detection and determination of siderophores. Anal. Biochem. 160: 47– 56 [DOI] [PubMed] [Google Scholar]
- 41. Simon R, Priefer U, Pühler A. 1983. A broad host range mobilization system for in vivo genetic engineering: transposon mutagenesis in Gram-negative bacteria. Nat. Biotechnol. 1: 784– 791 [Google Scholar]
- 42. Soto MJ, Fernández-Pascual M, Sanjuán J, Olivares J. 2002. A fadD mutant of Sinorhizobium meliloti shows multicellular swarming migration and is impaired in nodulation efficiency on alfalfa roots. Mol. Microbiol. 43: 371– 382 [DOI] [PubMed] [Google Scholar]
- 43. Stewart CR, Rossier O, Cianciotto NP. 2009. Surface translocation by Legionella pneumophila: a form of sliding motility that is dependent upon type II protein secretion. J. Bacteriol. 191: 1537– 1546 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Tambalo DD, Yost CK, Hynes MF. 2010. Characterization of swarming motility in Rhizobium leguminosarum bv. viciae. FEMS Microbiol. Lett. 307: 165– 174 [DOI] [PubMed] [Google Scholar]
- 45. Verstraeten N, et al. 2008. Living on a surface: swarming and biofilm formation. Trends Microbiol. 16: 496– 506 [DOI] [PubMed] [Google Scholar]
- 46. Wang Q, Frye JG, McClelland M, Harshey RM. 2004. Gene expression patterns during swarming in Salmonella typhimurium: genes specific to surface growth and putative new motility and pathogenicity genes. Mol. Microbiol. 52: 169– 187 [DOI] [PubMed] [Google Scholar]