Abstract
The hydrothermal vent gammaproteobacterium Thiomicrospira crunogena inhabits an unstable environment and must endure dramatic changes in habitat chemistry. This sulfur chemolithoautotroph responds to changes in dissolved inorganic carbon (DIC) (DIC = CO2 + HCO3− + CO3−2) availability with a carbon-concentrating mechanism (CCM) in which whole-cell affinity for DIC, as well as the intracellular DIC concentration, increases substantially under DIC limitation. To determine whether this CCM is regulated at the level of transcription, we resuspended cells that were cultivated under high-DIC conditions in chemostats in growth medium with low concentrations of DIC and tracked CCM development in the presence and absence of the RNA polymerase inhibitor rifampin. Induction of the CCM, as measured by silicone oil centrifugation, was hindered in the presence of rifampin. Similar results were observed for carboxysome gene transcription and assembly, as assayed by quantitative reverse transcription-PCR (qRT-PCR) and transmission electron microscopy, respectively. Genome-wide transcription patterns for cells grown under DIC limitation and those grown under ammonia limitation were assayed via microarrays and compared. In addition to carboxysome genes, two novel genes (Tcr_1019 and Tcr_1315) present in other organisms, including chemolithoautotrophs, but whose function(s) has not been elucidated in any organism were found to be upregulated under low-DIC conditions. Likewise, under ammonia limitation, in addition to the expected enhancement of ammonia transporter and PII gene transcription, the transcription of two novel genes (Tcr_0466 and Tcr_2018) was measurably enhanced. Upregulation of all four genes (Tcr_1019, 4-fold; Tcr_131, ∼7-fold; Tcr_0466, >200-fold; Tcr_2018, 7-fold), which suggests that novel components are part of the response to nutrient limitation by this organism, was verified via qRT-PCR.
INTRODUCTION
Hydrothermal fluid emitted from cracks in the earth's crust provides reduced chemicals (e.g., H2S, H2, CH4, and Fe+2) utilized by chemolithoautotrophic microbes to fuel carbon fixation (6, 12, 22). The deep-sea hydrothermal vent environment, while being an extremely productive ecosystem (15, 20), presents challenges to which vent organisms must adapt. In this habitat, turbulent eddies of dilute hydrothermal fluid (30°C), which have a low pH and carry H2S, mix with bottom seawater (2°C), which has alkaline pH and carries O2, causing wide fluctuations in habitat chemistry over time (16). Concentrations of dissolved inorganic carbon (DIC) (DIC = CO2 + HCO3− + CO3−2; 2 mM to 7 mM) and pH values (∼5 to 8) vary considerably, presenting very divergent concentrations of CO2 (20 to 2,000 μM) and HCO3− to the autotrophs growing there (13, 17), which may necessitate adaptations to maintain a steady supply of DIC.
One such adaptation is a carbon-concentrating mechanism (CCM) (4). Ribulose 1,5-bisphosphate carboxylase/oxygenase (RubisCO), the carboxylase of the Calvin-Benson-Bassham cycle, has a low affinity for CO2 and can use both CO2 and O2 as substrates. Two components of CCMs compensate for these catalytic limitations. First, active HCO3− transport generates a high intracellular concentration of this compound (26). Next, intracellular HCO3− enters carboxysomes, which are protein-bound inclusions where RubisCO is sequestered, with a trace of carbonic anhydrase activity. The carbonic anhydrase catalyzes the conversion of HCO3− to CO2, which is then fixed by RubisCO before it can escape the carboxysome (7). CCMs have been well studied in cyanobacteria and facilitate the growth of these cells under low-CO2 conditions, and they vary among species (4). Thus far, three evolutionarily distinct HCO3− transporters have been discovered: BCT1, an ABC transporter which is induced under carbon limitation (23); a Na+-dependent transporter (29); and SulP, which is evolutionarily related to sulfate transporters (24). Regulation of DIC uptake occurs both at the level of transcription and posttranslationally (26).
The hydrothermal vent gammaproteobacterial chemolithoautotroph Thiomicrospira crunogena is the first chemolithoautotroph in which elevated intracellular DIC concentrations, consistent with the presence of a CCM, which may enable it to grow steadily despite environmental CO2 fluctuations, have been measured (10). T. crunogena utilizes the Calvin-Benson-Bassham cycle for CO2 fixation and grows rapidly even when the culture DIC is below 20 μM. T. crunogena cells cultivated under low-DIC conditions are capable of generating intracellular DIC concentrations 100-fold higher than extracellular concentrations (10) and can utilize extracellular CO2 and HCO3− for carbon fixation. Whole-cell affinities for DIC respond to the DIC concentration (KDIC) present during growth; when DIC concentrations are low, cell affinities for this substrate are substantially higher (KDIC = 26 μM) than when cells are cultivated under elevated DIC concentrations (KDIC = 660 μM) (10).
Some components of a typical CCM are apparent in the genome of T. crunogena, but key components are not. A carboxysome operon, which encodes the shell proteins, carbonic anhydrase (csoSCA), and carboxysomal form I RubisCO, is present. Elsewhere in the genome, two other RubisCOs are encoded (one form I and one form II), as well as an alpha carbonic anhydrase (α-CA) and two β-CA genes (including csoSCA). However, CsoSCA is the only carbonic anhydrase that appears to play a role in DIC uptake and fixation (9), and no orthologs to the bicarbonate transporters found in cyanobacteria are apparent in the T. crunogena genome (28).
The objectives of this study were to determine whether CCM induction in T. crunogena occurs at the level of transcription and to compare the transcriptomes of T. crunogena cultivated under low- and high-DIC conditions to identify the genes whose expression is stimulated by growth under low-DIC conditions as a step in resolving all of the components of this proteobacterial CCM.
MATERIALS AND METHODS
Cultivation.
To prepare cells for experiments to monitor changes in DIC uptake and fixation in response to exposure to low-DIC conditions and to determine whether these changes were dependent on transcription, we cultivated T. crunogena XCL-2 cells (DSM 25203) (1) overnight under ammonia-limited conditions in two chemostats (dilution rate = 0.1 h−1) in artificial seawater (ASW) supplemented with thiosulfate (40 mM) and a high concentration of DIC (50 mM, as quantified with a gas chromatograph) (9). The pH (8) and oxygen concentration (∼20 to 100 μM) were monitored with electrodes and maintained with 10 N potassium hydroxide (KOH) and sparging with 5% CO2, balance O2 (9). The following day, cells were aseptically harvested via centrifugation (10,000 × g, 10 min, 4°C) and resuspended to their original volume (2 × 1 liter; optical density at 600 nm [OD600] of ∼0.1) in thiosulfate-supplemented ASW with a low concentration (1 mM) of DIC. Rifampin was added to one culture at a concentration that was found to be growth inhibitory in solid culture (50 mg/liter). Both cultures were then run as bioreactors (dilution rate = 0) and periodically sparged with pure O2 as directed by the O2 electrode to maintain the concentration of this dissolved gas. Samples from both cultures (2 × 30 ml, 1 × 60 ml) were harvested via centrifugation at 30-min intervals over a 3-h time course. At their initial optical density (0.1), cells were in early exponential phase growth and continued to consume oxygen readily over the entire time course of the experiment.
To identify genes with more abundant transcripts during cultivation under DIC limitation conditions, we cultivated cells in chemostats as described above under conditions of DIC limitation [low-DIC cells; reservoir nutrient concentration = 2 mM DIC, 13 mM (NH4)2SO4, 3.1 mM PO4−x; growth chamber DIC concentration of ∼0.1 mM as quantified via gas chromatography; growth chamber was sparged with pure O2 as described above], ammonia limitation [high-DIC cells; reservoir nutrient concentration = 50 mM DIC, 0.8 mM (NH4)2SO4, 3.1 mM PO4−x; sparged with 5% CO2; balance O2; growth chamber DIC concentration of ∼50 mM DIC], or phosphate limitation [low-PO4−x; 50 mM DIC, 13 mM (NH4)2SO4, 0.025 mM PO4−x; sparged with 5% CO2; balance O2; growth chamber DIC concentration of ∼50 mM DIC]. Cells were maintained at a steady state until harvesting (dilution rate = 0.1 h−1). After harvesting by centrifugation (10,000 × g, 5 min, 4°C), T. crunogena cells were flash frozen in liquid nitrogen and stored at −80°C.
DIC uptake and fixation.
To assay changes in DIC uptake and fixation after resuspension in low-DIC growth medium, we washed the cell pellets from the 60-ml samples taken from both cultures (rifampin and rifampin-free) once and resuspended them in 1 ml of DIC-free ASW. DIC fixation (conversion of 14C-DIC to acid-stable 14C) and uptake (fixation plus intracellular 14C-DIC) were measured immediately after resuspension via silicone oil centrifugation (10).
Carboxysome quantification.
To track changes in carboxysome abundance over time after resuspension in low-DIC growth medium, we resuspended the cell pellets from one 30-ml sample of each culture in ASW supplemented with glutaraldehyde (2.5%, vol/vol) and stored them overnight at 4°C. The following day, cells in these samples were harvested via centrifugation and washed three times with ASW (4°C) before treatment for 1 h with 1% osmium tetroxide in ASW. Cells were harvested and rinsed three times with distilled water, after which they were embedded in 3% agar. After trimming to 1-mm3 pieces, agar blocks were dehydrated with an ethanol series (50%, 70%, 95%, and 100% twice) and ethanol/propylene oxide (50:50, 0:100, twice). Spurr's resin was infused into the samples (propylene oxide/Spurr's, 50:50, 25:75, 0:100, four times; first incubation was overnight; all incubations were agitated with a rotator). Spurr's resin was polymerized overnight at 70°C, and 80-nm sections were cut and stained for 10 min with 8% aqueous uranyl acetate, followed by a 5-min staining with Reynold's lead citrate stain. Electron micrographs were produced with an FEI Morgagni 268D transmission electron microscope at 60 kV, and images were captured with an Olympus SIS MegaView III side-mounted camera with 1.4 megapixel image capture. Approximately 50 images were collected from each time point, and carboxysomes were tallied from at least 150 cell cross-sections per sample. Cell cross-section areas were estimated by measuring their long (L) and short (S) axes in Photoshop (CS3; Adobe), modeling them as rectangles with a half circle on each end, and calculating areas as π × (S2/2) · (SL − S2).
csoSCA transcription quantification.
In preparation for RNA extraction, cell pellets from one 30-ml sample from both cultures were flash frozen with liquid nitrogen and stored at −80°C. RNA was purified and reverse transcribed using the RiboPure (Ambion) and ImProm-II RT (Promega) systems. Transcription of csoSCA (locus tag Tcr_0841) was tracked via quantitative reverse transcription-PCR (qRT-PCR) using TaqMan primers and probes (ABI) as described in reference 9.
Transcriptional profiling.
Oligonucleotide arrays were fabricated with probes designed to represent all genes within the T. crunogena genome, with two or three probes per gene and a probe length of 35 nucleotides (CombiMatrix, Mukilteo, WA). RNA was isolated from cells grown in six chemostats (three low-DIC and three high-DIC; ammonia-limited) as described in reference 9. RNA was purified further with an RNeasy MinElute cleanup kit (Qiagen, Germantown, MD), which also served to remove EDTA remaining from the RiboPure system, and was eluted in RNase-free water. One microgram of total RNA was directly labeled for 1 h with a LabelIT Cy5 labeling reaction (Mirus Bio, Madison, WI). After labeling, the RNA underwent ethanol (EtOH) precipitation (5 min at −80°C and 30 min centrifugation). The RNA pellet was resuspended in 16 μl of water and 4 μl of fragmentation buffer (final concentration, 40 mM Tris-acetate [pH 8.1], 100 mM potassium acetate [KOAc], 30 mM MgOAc).
Prior to hybridization, arrays were rehydrated at 65°C with water for 10 min and then incubated at 45°C for 2 h in prehybridization solution (final concentration, 6× SSPE [1× SSPE is 0.18 M NaCl, 10 mM NaH2PO4, and 1 mM EDTA {pH 7.7}; Lonza Accugene], 0.05% Tween 20, 20 mM EDTA, 5× Denhardt's solution, 100 ng/μl of herring sperm, 0.05% SDS). Labeled RNA was then added to hybridization solution (final concentration, 6× SSPE, 0.05% Tween 20, 20 mM EDTA, 25% distilled formamide,100 ng/μl of herring sperm DNA, 0.04% SDS), and the array was incubated overnight at 45°C. Arrays were washed and imaged per the hybridization and imaging protocol for the CombiMatrix CustomArray 12K microarray. All arrays used were stripped and rehybridized a maximum of three times each, as subsequent stripping and hybridization resulted in substantial loss of signal (data not shown). Global normalizing was used for all six arrays (three low-DIC and three high-DIC) to compensate for between-array variation in overall signal strength. For normalization, the average signal strength for each array was calculated using the fluorescence intensities for all spots on each array. Spot intensities on all arrays were normalized so that each array would have the same average signal. For a particular probe, fold changes in transcription were calculated by dividing the average spot intensity for microarrays hybridized with labeled mRNA purified from low-DIC cells by the spot intensity for those hybridized with labeled mRNA from high-DIC cells (low-DIC/high-DIC) or vice-versa (high-DIC/low-DIC). The Comparative Marker Selection module of GenePattern (14, 27) was used to generate asymptotic P values associated with gene expression levels. Asymptotic P values were calculated from P values from a standard independent two-sample Student t test. Rather than create an empirical distribution of the scores, we computed the P values with the assumption that test statistic scores follow Student's t test distribution. Changes in transcription levels for genes were considered noteworthy if average spot intensities for more than one probe per gene were enhanced at least 2-fold.
Quantification of gene transcription via qRT-PCR.
RNA was isolated from low- and high-DIC cells (as well as low-PO4−x cells) as described above; the primers used for cDNA synthesis are listed in Table 1. Verification of amplification efficiencies was carried out with primer/probe sets directed against 16S (calibrator) and target genes (Tcr_1019, Tcr_1315, Tcr_466, and Tcr_2018) as described in reference 9. Fold enhancement of transcription in low-DIC cells relative to that in high-DIC cells was calculated according to the 2−ΔΔCT method, where ΔΔCT = [(CT target − CT calibrator) − (CT targetRef − CT calibratorRef)], CT target and CT calibrator are the threshold cycle (CT) values for target and 16S gene amplification in low-DIC cells, and CT targetRef − CT calibratorRef is the corresponding value from high-DIC cells (19). Fold enhancement in high-DIC cells was calculated by using low-DIC cells as the reference.
Table 1.
Primers and probes used to target T. crunogena conserved hypothetical genes
| Locus tag | Purpose | Function | Sequence (5′–3′)a | Location on geneb (nt) |
|---|---|---|---|---|
| Tcr_1019 | cDNA synthesis | R primer | ACGCGGTTAGATCCCATTG | 206 |
| qRT-PCR | F primer | AGAAAGCCGGCCGCTAAAA | 130 | |
| R primer | CCGGTTCTTCTTTTTCAGGTTGTTT | 175 | ||
| Probe | FAM CCGGTTGCCAAACAG NFQ | 150 | ||
| Tcr_1315 | cDNA synthesis | R primer | GGTATACGCCAGGTCATTGG | 653 |
| qRT-PCR | F primer | CCGTCGGGATTTTGAATGAAACC | 427 | |
| R primer | GGGTTACGCTCAACGCCATAA | 468 | ||
| Probe | FAM ACGAACCACCAACTTT NFQ | 452 | ||
| Tcr_0466 | cDNA synthesis | R primer | ACGGCACCATCTTTTGTTTC | 541 |
| qRT-PCR | F primer | GGGTTTGACCGCATGTATAACGA | 343 | |
| R primer | GATCGTAACGCCACCGAAAC | 382 | ||
| Probe | FAM CTGCCGACAGATTTA NFQ | 368 | ||
| Tcr_2018 | cDNA synthesis | R primer | ATATCGGCTTTTGGACAACG | 1423 |
| qRT-PCR | F primer | GCCGATATCGCTGGTGTAGAATTT | 1075 | |
| R Primer | GTTTTGCCAGGTTGAAGTCGTATT | 1117 | ||
| Probe | FAM CCAAGCGCCATATTC NFQ | 1099 | ||
| 16S (Tcr_R0053) | cDNA synthesis | R primer | TTTATGAGATTCGCGCACTG | 1206 |
| qRT-PCR | F primer | CGAATATGCTCTACGGAGTAAAGGT | 110 | |
| R primer | CGCGGGCTCATCCTTTAG | 157 | ||
| Probe | VIC CCCTCTCCTTGGAAGGT NFQ | 136 |
FAM and VIC refer to the fluorescence tags, while NFQ refers to the major groove binder and nonfluorescent quencher (ABI, Carlsbad, CA).
Number of nucleotides at the 3′ end of the start codon of the gene.
Taxonomic distribution and phylogenetic analysis of homologs to novel differentially transcribed genes.
Apparent orthologs (bidirectional BLAST best hits) to Tcr_1019 and Tcr_1315 were collected from the Integrated Microbial Genomes (IMG) database (21). For Tcr_1315, hits whose pairwise alignments with Tcr_1315 covered at least 50% of both genes were selected for further analysis. A multiple alignment was then generated via MUSCLE (11) as implemented in MEGA5 (32). Genes with less than 50% sequence identity to Tcr_1315 aligned poorly and were eliminated from the alignment. The remaining genes were realigned, and this alignment was trimmed using relaxed criteria in gBlocks (31) as implemented in the Phylogeny software (http://www.phylogeny.fr) (8). The trimmed alignment of 72 residues was used to construct a maximum likelihood tree in MEGA5 (18, 32) using the JTT substitution model, uniform rates among sites, complete deletion of missing data, and the nearest neighbor interchange heuristic method. For Tcr_1019, too few orthologs were present in the database to undertake these analyses.
Microarray data accession number.
Microarray data are available at Gene Expression Omnibus (GEO) under accession number GSE36254.
RESULTS
DIC uptake and fixation.
After resuspension and cultivation for 1 h in growth medium containing a low concentration of DIC (1 mM), T. crunogena cells expressed an ability to generate elevated intracellular DIC concentrations in short-term (30-s) incubations in the presence of 0.1 mM DIC (Fig. 1A). Carbon fixation lagged behind intracellular DIC accumulation, being substantially elevated after ∼1.5 h of cultivation (Fig. 1B). Cells incubated in the presence of rifampin did not develop an ability to accumulate intracellular DIC or to fix carbon under low-DIC conditions (Fig. 1).
Fig 1.
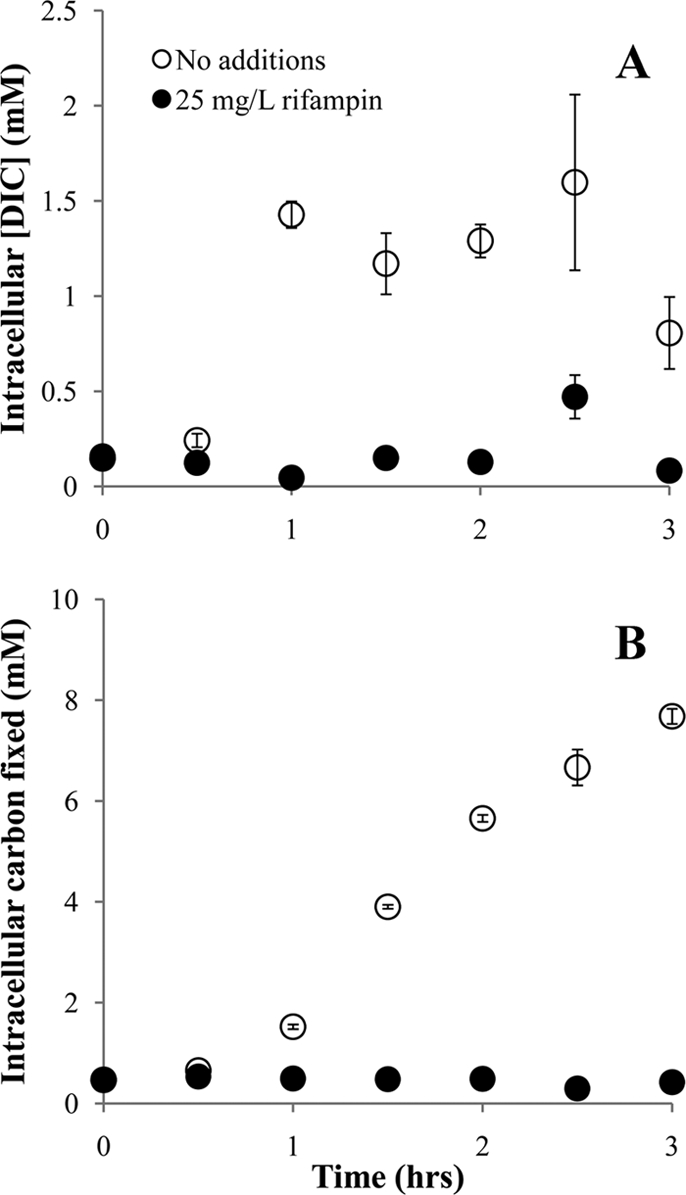
Dissolved inorganic carbon uptake (A) and fixation (B) by T. crunogena cells after resuspension and cultivation in growth medium containing 1 mM DIC. Cells were initially cultivated in the presence of 50 mM DIC, harvested, washed, and resuspended in growth medium containing 1 mM DIC and either dimethyl sulfoxide (DMSO; 0.1% [vol/vol]) or rifampin dissolved in DMSO. Cells were harvested at the indicated times and incubated in the presence of 0.1 mM DIC for the short-term (30-s) incubations whose results are depicted here. Error bars indicate the standard errors of the measurements.
Carboxysome presence and csoSCA transcription quantification.
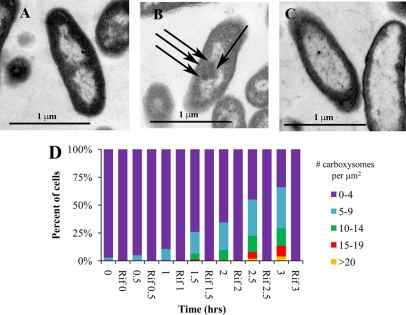
During cultivation under low-DIC conditions, carboxysomes became increasingly abundant in cells over the time course sampled here (Fig. 2), paralleling carbon fixation rates (Fig. 1). Cultures to which rifampin had been added did not have any visible carboxysomes 3 h after resuspension in low-DIC growth medium (Fig. 2). Carboxysome gene transcript abundance (csoSCA) was somewhat elevated in cells resuspended in low-DIC growth medium containing rifampin after 30 min and 1 h, which may indicate that the action of this antibiotic on RNA polymerase is not instantaneous. However, levels in cells that had not been exposed to rifampin were always substantially elevated relative to those in cells cultivated in the presence of this compound. (Fig. 3).
Fig 2.
Accumulation of carboxysomes in cells over time, after transfer to growth medium with 1 mM DIC. Cells were initially cultivated in the presence of 50 mM DIC, harvested, washed, and resuspended in medium with 1 mM DIC. Depicted in transmission electron micrographs are cells cultivated in 50 mM DIC (A) and cells 3 h after resuspension in the absence (B) or presence (C) of 25 mg/liter of rifampin. For one cell, carboxysomes are indicated with arrows. (D) Frequencies of carboxysomes per cell area in electron micrographs. Cultures to which rifampin have been added are indicated as “Rif.”
Fig 3.
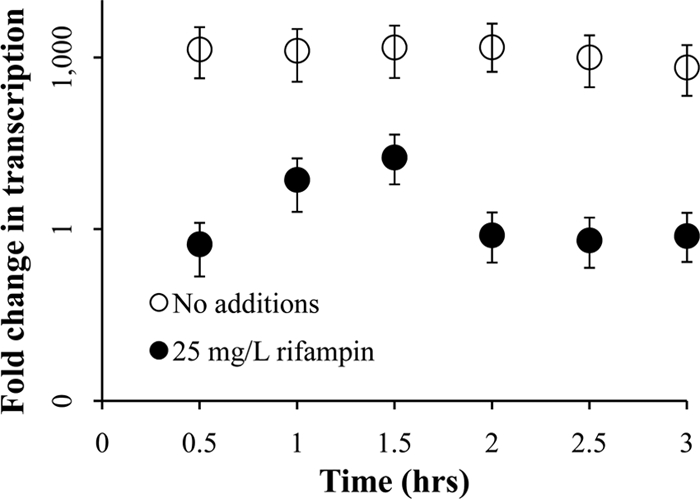
Transcription of the carboxysomal carbonic anhydrase gene (csoSCA) over time, after resuspension in growth medium with 1 mM DIC. Cells were initially cultivated in the presence of 50 mM DIC and then were harvested and washed before resuspension. Fold changes in csoSCA transcription relative to time zero were measured via qRT-PCR targeting csoSCA transcripts and are presented with error bars indicating the 95% confidence interval of the measurement. The y axis is logarithmic.
Transcriptional profiling.
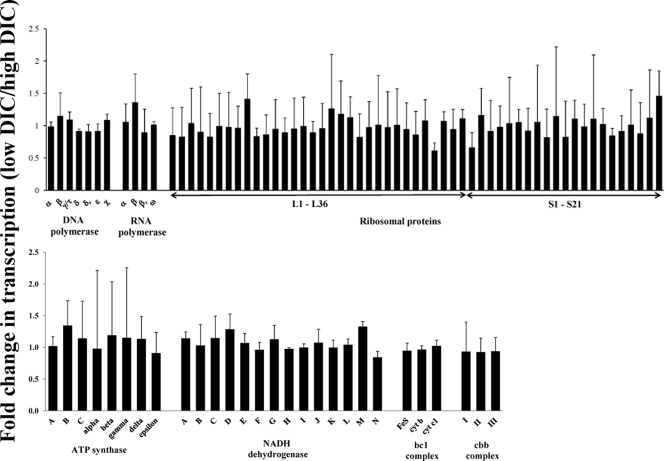
Housekeeping genes, including those encoding RNA and DNA polymerase, ribosomal proteins, ATP synthase, and components of the electron transport chain (NADH dehydrogenase, bc1 complex, and the cbb3 complex), had similar transcript levels in low- and high-DIC cells (Fig. 4).
Fig 4.
Housekeeping genes that do not have a measureable change in transcription under low- versus high-DIC conditions, as determined with microarrays. Shown here listed by subunits are genes involved in DNA, RNA, and protein synthesis, as well as ATP synthesis and electron transport (NADH dehydrogenase, bc1 complex, cbb complex). Two or three probes were designed to target each gene, and results from all are averaged per gene. Error bars represent standard deviations as propagated from normalized spot intensities from three low-DIC and three high-DIC chemostats.
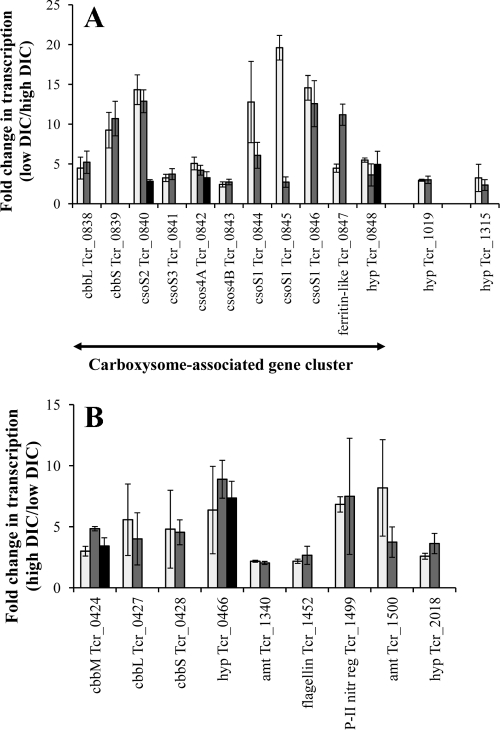
When cells were cultivated under low-DIC conditions, transcripts from the gene cluster encoding the carboxysomal shell proteins and enzymes (RubisCO and carbonic anhydrase; Tcr_0838 to Tcr_0848) were more abundant (Fig. 5). Transcripts from two other genes (Tcr_1019 and Tcr_1315, also verified by qRT-PCR) (Table 2) were more abundant under low-DIC conditions and may be novel components of the CCM of T. crunogena. Confirming previous results, transcripts from the α-CA gene (Tcr_1545) and the noncarboxysomal β-CA gene (Tcr_0421) were similar in abundance under both low- and high-DIC conditions (9).
Fig 5.
Enhanced transcription of genes under low-DIC (A) or high-DIC (B) conditions, as determined with microarrays. Genes are presented with locus tags from the genome annotation. (A) cbbL and cbbS encode a carboxysomal form IA RubisCO, while csoS1 to csc4 encode carboxysome shell proteins. “hyp” indicates that the gene encodes a hypothetical protein. (B) cbbM encodes a form II RubisCO, while cbbL and cbbS encode the subunits of a form IA RubisCO with ∼80% amino acid sequence identity with the carboxysomal RubisCO. Amount (amt) genes encode Amt family proteins and are likely to be ammonium transporters, while “P-II nitr reg” encodes the PII protein, which is involved in regulating the Ntr regulon for nitrogen assimilation. Two or three probes were designed to target each gene and are illustrated with individual bars on the graphs. Only those genes for which more than one probe showed more than 2-fold enhancement of transcription are presented here. Error bars represent standard deviations as propagated from normalized spot intensities from three low-DIC and three high-DIC chemostats.
Table 2.
Response of T. crunogena hypothetical gene transcription to changes in growth conditions
| Locus tag | Response to low DIC |
Response to low NH4+ |
Response to low PO4−x |
|||
|---|---|---|---|---|---|---|
| Fold increasea,b | ΔΔCT ± SD | Fold increasea,c | ΔΔCT ± SD | Fold increasea,d | ΔΔCT ± SD | |
| Tcr_1019 | 4.4 | −2.1 ± 0.2 | ||||
| Tcr_1315 | 6.7 | −2.8 ± 0.3 | ||||
| Tcr_0466 | 202 | −7.6 ± 1.0 | 0.77 | 0.3 ± 2.3 | ||
| Tcr_2018 | 7.0 | −2.8 ± 1.2 | 0.69 | 0.6 ± 1.4 | ||
qRT-PCR was carried out on RNA extracted from T. crunogena cells using 16S transcripts as the calibrator.
Fold increase is the frequency of transcription in low-DIC (high-NH4+, high-PO4−x) cells divided by the frequency in high-DIC (low-NH4+, high-PO4−x) cells.
Fold increase is the frequency of transcription in low-NH4+ (high-DIC, high-PO4) cells divided by the frequency in high-NH4+ (low-DIC, high-PO4−x) cells.
Fold increase is the frequency of transcription in low-PO4−x (high-DIC, high-NH4+) cells divided by the frequency in high-PO4−x (high-DIC, high-NH4+) cells.
Under low-NH4+ (high-DIC) growth conditions, genes associated with a nitrogen starvation response had increased transcript levels (Fig. 5). This included two Amt family ammonium transporters and the regulatory protein PII, which stimulates transcription of the gene encoding glutamine synthetase as well as posttranslational modification of this enzyme via adenylation/deadenylation (3). Genes encoding two conserved hypothetical proteins, Tcr_0466, and Tcr_2018, were found to have increased transcription under low-NH4+ conditions (Fig. 5, confirmed by qRT-PCR; Table 2). Transcription levels of these genes were not enhanced under high-DIC, low-phosphate conditions, consistent with a response to ammonia limitation, not to high-DIC conditions.
Taxonomic distribution and phylogenetic analysis of homologs to Tcr_1019 and Tcr_1315.
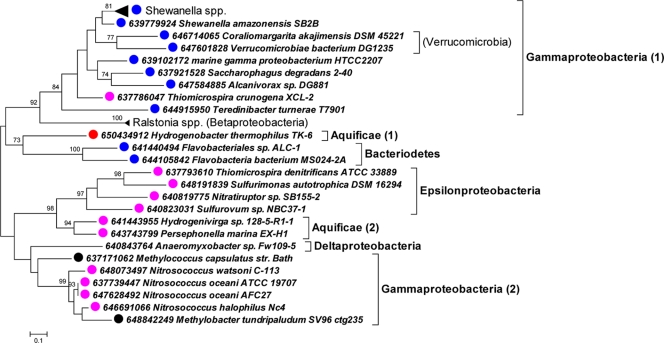
For Tcr_1019, only two genes were returned from BLAST queries of the IMG database (Table 3). The organisms from which these two genes derive are both capable of growing chemolithoautotrophically and phylogenetically disparate: Thioalkalivibrio sp. K90, a member of the Gammaproteobacteria, and Nitrobacter winogradskyi, an alphaproteobacterium. Many apparent orthologs of Tcr_1315 are present in the IMG database and have substantial sequence similarity (Fig. 6). Tcr_1315 falls within a well-supported clade with homologs from other marine proteobacteria.
Table 3.
Orthologs of Tcr_1019
| Organism | Locus taga | Length of amino acid sequenceb | Portion aligning to Tcr_1019c | % similarity of portion aligning | E value |
|---|---|---|---|---|---|
| T. crunogena | Tcr_1019 | 75 | 1–75 | 100 | N/A |
| N. winogradskyi | Nwi_1977 | 75 | 7–75 | 55 | 3e−08 |
| Thioalkalivibrio sp. strain K90 | TK90_0870 | 140 | 48–140 | 47 | 7e−07 |
Sequences were collected from the Integrated Microbial Genomes system (http://img.jgi.doe.gov/cgi-bin/w/main.cgi [21]).
Predicted from the nucleic acid sequence.
Based on results from the “Align two sequences” feature at the National Center for Biotechnology Information (http://www.ncbi.nlm.nih.gov/) website, which uses BLASTp (2).
Fig 6.
Phylogenetic analysis of amino acid sequences predicted from genes orthologous to Tcr_1315. The maximum likelihood method was implemented in MEGA5 to create an unrooted tree (32). Bootstrap values of >70% are displayed above the branches. Taxa are labeled with dots as follows: blue, marine; red, autotroph; purple, marine autotroph; black, methylotroph or methanotroph.
DISCUSSION
Given the novel nature of a CCM in a member of the Proteobacteria (10), we were interested in determining the levels at which its components are induced in T. crunogena in response to low-DIC conditions. Unlike cyanobacterial DIC uptake, which is regulated both at the level of transcription and at posttranslational modification (26), it appears that DIC uptake by T. crunogena is regulated at the level of transcription, as the RNA polymerase inhibitor rifampin prevented development of the ability to generate elevated intracellular concentrations of DIC upon exposure to low-DIC conditions (Fig. 1). Like that of the betaproteobacterium Halothiobacillus neapolitanus (5), carboxysome synthesis is regulated at the level of transcription (Fig. 2 and 3). Interestingly, it appears that the lag in DIC uptake ability (∼1 h) (Fig. 1A) is shorter than the lag in carbon fixation/carboxysome development (∼1.5 to 2 h) (Fig. 1B and 2). Given that carboxysome genes and transporter genes are not cotranscribed (DIC transporter genes are not apparent in the carboxysome gene cluster), differences in timing for the development of these two components of the CCM might be expected. Furthermore, given the size and structural complexity of carboxysomes, it seems feasible that their synthesis and assembly would take more time than insertion of structurally simpler transporter proteins into the cell membrane. However, the observation that csoSCA transcript abundance is high after 30 min suggests that CsoSCA protein should be more abundant as well. If CsoSCA is active before carboxysome assembly, it should be reflected in lower intracellular DIC concentrations, as free cytoplasmic carbonic anhydrase should convert DIC to CO2, which would diffuse from the cell (25). The high intracellular DIC concentration at 30 min despite elevated levels of csosCA transcripts is puzzling. Perhaps translation of csoSCA transcripts is delayed or CsoSCA protein is somehow less active until packaged into a carboxysome.
As this is the first genome-wide assay of transcriptional response to DIC and ammonia abundance in an obligate chemolithoautotroph, we anticipated that novel genes/patterns of transcription would be apparent. Indeed, transcripts from two conserved hypothetical genes (Tcr_1019 and Tcr_1315) were more abundant under low-DIC conditions (Fig. 5A; Table 2). The proteins encoded by both genes are predicted to have an amino-terminal signal peptide without downstream transmembrane alpha helices. The protein encoded by Tcr_1019 is likely to be localized in the periplasm, while the protein encoded by Tcr_1315 falls within protein family IPR011250 (InterPro), whose members typically have a beta-barrel structure, which indicates that it may be present in the outer membrane.
Intriguingly, orthologs to both genes are present in other chemolithoautotrophs (Table 3; Fig. 6). While Tcr_1019 homologs are restricted to two other bacteria, Tcr_1315 homologs are present in many marine bacteria, many of which are also autotrophs. The function of this group of homologous genes in any organism has not been elucidated, and it will be of great interest to determine whether it has a uniform function (e.g., DIC acquisition or more generally facilitating growth at seawater pH and salinity) in all of these organisms. Gene disruption experiments to determine if either protein plays a role in the T. crunogena CCM are under way.
Novel genes (Tcr_0466 and Tcr_2018) were discovered in the ammonia-limited (high-DIC) cells as well (Fig. 5B; Table 2). The function of these conserved hypothetical proteins could not be discerned based on their amino acid sequences. However, both these postulated genes have predicted amino-terminal signal peptides. Both also lack downstream transmembrane helices, suggesting a periplasmic location. Genes homologous to Tcr_0466 and Tcr_1315 are widespread among the Proteobacteria; this is the first report of differential transcription of these genes and it will be of interest to see whether these proteins all play a role in nitrogen metabolism among their different host organisms.
These observations of novel gene transcription under DIC- and ammonia-limited conditions are strengthened both by consistency in the transcription of housekeeping genes (Fig. 4) and by transcription patterns consistent with previous work. For example, transcript levels from RubisCO genes varied with the DIC concentration available during growth; the carboxysomal form I had higher transcript levels under low-DIC conditions, while the other two (noncarboxysomal form I RubisCO and form II RubisCO) had increased transcript levels under high-DIC conditions (Fig. 5), which is consistent with what has been observed in the close relative Hydrogenovibrio marinus, a gammaproteobacterial hydrogen-oxidizing autotroph with similar RubisCO operon structure (33). Further, elevated transcript levels of Amt family ammonia transporters were observed in response to ammonium limitation, also consistent with observations of these proteins in other organisms (30).
It is surprising that relatively few genes appeared to have significant changes in transcript abundance under these growth conditions. This is likely the result of both the consistency of the growth conditions in chemostats, where only a single nutrient concentration differs between treatments and growth rates can be regulated very precisely, and the conservative nature of the criteria used for deciding which genes were represented with higher transcript levels. Still, it might be anticipated that more genes would have been highlighted; for example, it seems reasonable to expect that molecular chaperones might be more abundant when cells are grown under low-DIC conditions, due to greatly enhanced carboxysome assembly. The observation that these genes do not appear to respond this way suggests either that carboxysome assembly does not require these proteins or that the basal level present in the cell suffices.
A full understanding of how the CCM works in chemolithoautotrophs will be instrumental in gaining insight into how these organisms succeed in environments like the hydrothermal vents. Furthermore, identifying proteobacterial CCM components in T. crunogena will make it possible to identify them in other microorganisms. Until then, understanding of DIC uptake and fixation in the many noncyanobacterial autotrophs that fix carbon dioxide in their challenging habitats is quite limited.
ACKNOWLEDGMENTS
We are deeply thankful to the National Science Foundation for their support of this project (NSF-MCB-0643713 to K.M.S.).
We are also thankful to anonymous reviewers for suggestions that substantially improved the quality of the manuscript.
Footnotes
Published ahead of print 10 February 2012
REFERENCES
- 1. Ahmad A, Barry JP, Nelson DC. 1999. Phylogenetic affinity of a wide, vacuolate, nitrate-accumulating Beggiatoa sp. from Monterey Canyon, California, with Thioploca spp. Appl. Environ. Microbiol. 65: 270– 277 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Altschul SF, et al. 1997. Gapped BLAST and PSI-BLAST: a new generation of protein database search programs. Nucleic Acids Res. 25: 3389– 3402 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Arcondéguy T, Jack R, Merrick M. 2001. PII signal transduction proteins, pivotal players in microbial nitrogen control. Microbiol. Mol. Biol. Rev. 65: 80– 105 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Badger MR, Price GD, Long BM, Woodger FJ. 2006. The environmental plasticity and ecological genomics of the cyanobacterial CO2 concentrating mechanism. J. Exp. Bot. 57: 249– 265 [DOI] [PubMed] [Google Scholar]
- 5. Cai F, Heinhorst S, Shively JM, Cannon GC. 2008. Transcript analysis of the Halothiobacillus neapolitanus cso operon. Arch. Microbiol. 189: 141– 150 [DOI] [PubMed] [Google Scholar]
- 6. Campbell AC, et al. 1988. Chemistry of hot springs on the Mid-Atlantic Ridge. Nature 335: 514– 519 [Google Scholar]
- 7. Cannon GC, Heinhorst S, Kerfeld CA. 2010. Carboxysomal carbonic anhydrases: structure and role in microbial CO2 fixation. Biochim. Biophys. Acta 1804: 382– 392 [DOI] [PubMed] [Google Scholar]
- 8. Dereeper A, et al. 2008. Phylogeny.fr: robust phylogenetic analysis for the non-specialist. Nucleic Acids Res. 36: W465– W469 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Dobrinski KP, Boller AJ, Scott KM. 2010. Expression and function of four carbonic anhydrase homologs in the deep-sea chemolithoautotroph Thiomicrospira crunogena. Appl. Environ. Microbiol. 76: 3561– 3567 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Dobrinski KP, Longo DL, Scott KM. 2005. The carbon-concentrating mechanism of the hydrothermal vent chemolithoautotroph Thiomicrospira crunogena. J. Bacteriol. 187: 5761– 5766 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Edgar RC. 2004. MUSCLE: multiple sequence alignment with high accuracy and high throughput. Nucleic Acids Res. 32: 1792– 1797 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Edmond JM, Von Damm KL, McDuff RE, Measures CI. 1982. Chemistry of hot springs on the East Pacific Rise and their effluent dispersal. Nature 297: 187– 191 [Google Scholar]
- 13. Goffredi SK, et al. 1997. Inorganic carbon acquisition by the hydrothermal vent tubeworm Riftia pachyptila depends upon high external PCO2 and upon proton-equivalent ion transport by the worm. J. Exp. Biol. 200: 883– 896 [DOI] [PubMed] [Google Scholar]
- 14. Gould J, Getz G, Monti S, Reich M, Mesirov JP. 2006. Comparative gene marker selection suite. Bioinformatics 22: 1924– 1925 [DOI] [PubMed] [Google Scholar]
- 15. Govenar B, Freeman M, Bergquist DC, Johnson GA, Fisher CR. 2004. Composition of a one-year-old Riftia pachyptila community following a clearance experiment: insight to succession patterns at deep-sea hydrothermal vents. Biol. Bull. 207: 177– 182 [DOI] [PubMed] [Google Scholar]
- 16. Johnson KS, Childress JJ, Hessler RR, Sakamoto-Arnold CM, Beehler CL. 1988. Chemical and biological interactions in the Rose Garden hydrothermal vent field, Galapagos spreading center. Deep Sea Res. A Oceanogr. Res. Pap. 35: 1723– 1744 [Google Scholar]
- 17. Le Bris N, Govenar B, LeGall C, Fisher CR. 2006. Variability of physico-chemical conditions in 9°50′N EPR diffuse flow vent habitats. Mar. Chem. 98: 167– 182 [Google Scholar]
- 18. Lewis PO. 1998. Maximum likelihood as an alternative to parsimony for inferring phylogeny using nucleotide data, p 132–163. In Soltis DE, Soltis PS, Doyle JJ. (ed), Molecular systematics of plants II: DNA sequencing. Kluwer Academic Publishers, Norwell, MA. [Google Scholar]
- 19. Livak KJ, Schmittgen TD. 2001. Analysis of relative gene expression data using real-time quantitative PCR and the 2−ΔΔCT method. Methods 25: 402– 408 [DOI] [PubMed] [Google Scholar]
- 20. Lutz RA, et al. 1994. Rapid growth rates at deep-sea vents. Nature 371: 663– 664 [Google Scholar]
- 21. Markowitz VM, et al. 2010. The integrated microbial genomes system: an expanding comparative analysis resource. Nucleic Acids Res. 38: D382– D390 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Michard G, et al. 1984. Chemistry of solutions from the 13°N East Pacific Rise hydrothermal site. Earth Planet. Sci. Lett. 67: 297– 307 [Google Scholar]
- 23. Omata T, et al. 1999. Identification of an ATP-binding cassette transporter involved in bicarbonate uptake in the cyanobacterium Synechococcus sp. strain PCC 7942. Proc. Natl. Acad. Sci. U. S. A. 96: 13571– 13576 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Price GD, Woodger FJ, Badger MR, Howitt SM, Tucker L. 2004. Identification of a SulP-type bicarbonate transporter in marine cyanobacteria. Proc. Natl. Acad. Sci. U. S. A. 101: 18228– 18233 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Price GD, Badger MR. 1989. Expression of human carbonic anhydrase in the cyanobacterium Synechococcus PCC7942 creates a high CO2-requiring phenotype. Plant Physiol. 91: 505– 513 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Price GD, Badger MR, Woodger FJ, Long BM. 2008. Advances in understanding the cyanobacterial CO2-concentrating-mechanism (CCM): functional components, Ci transporters, diversity, genetic regulation and prospects for engineering into plants. J. Exp. Bot. 59: 1441– 1461 [DOI] [PubMed] [Google Scholar]
- 27. Reich M, et al. 2006. GenePattern 2.0. Nat. Genet. 38: 500– 501 [DOI] [PubMed] [Google Scholar]
- 28. Scott KM, et al. 2006. The genome of deep-sea vent chemolithoautotroph Thiomicrospira crunogena XCL-2. PLoS Biol. 4: e383. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Shibata M, et al. 2002. Genes essential to sodium-dependent bicarbonate transport in cyanobacteria: function and phylogenetic analysis. J. Biol. Chem. 277: 18658– 18664 [DOI] [PubMed] [Google Scholar]
- 30. Soupene E, He L, Yan D, Kustu S. 1998. Ammonia acquisition in enteric bacteria: physiological role of the ammonium/methylammonium transport B (AmtB) protein. Proc. Natl. Acad. Sci. U. S. A. 95: 7030– 7034 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Talavera G, Castresana J. 2007. Improvement of phylogenies after removing divergent and ambiguously aligned blocks from protein sequence alignments. Syst. Biol. 56: 564– 577 [DOI] [PubMed] [Google Scholar]
- 32. Tamura K, et al. 2011. MEGA5: molecular evolutionary genetics analysis using maximum likelihood, evolutionary distance, and maximum parsimony methods. Mol. Biol. Evol. 28: 2731– 2739 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Yoshizawa Y, Toyoda K, Arai H, Ishii M, Igarashi Y. 2004. CO2-responsive expression and gene organization of three ribulose-1,5-bisphosphate carboxylase/oxygenase enzymes and carboxysomes in Hydrogenovibrio marinus strain MH-110. J. Bacteriol. 186: 5685– 5691 [DOI] [PMC free article] [PubMed] [Google Scholar]