Abstract
Clinical urine specimens are usually considered to be sterile when they do not yield uropathogens using standard clinical cultivation procedures. Our aim was to test if the adult female bladder might contain bacteria that are not identified by these routine procedures. An additional aim was to identify and recommend the appropriate urine collection method for the study of bacterial communities in the female bladder. Consenting participants who were free of known urinary tract infection provided urine samples by voided, transurethral, and/or suprapubic collection methods. The presence of bacteria in these samples was assessed by bacterial culture, light microscopy, and 16S rRNA gene sequencing. Bacteria that are not or cannot be routinely cultivated (hereinafter called uncultivated bacteria) were common in voided urine, urine collected by transurethral catheter (TUC), and urine collected by suprapubic aspirate (SPA), regardless of whether the subjects had urinary symptoms. Voided urine samples contained mixtures of urinary and genital tract bacteria. Communities identified in parallel urine samples collected by TUC and SPA were similar. Uncultivated bacteria are clearly present in the bladders of some women. It remains unclear if these bacteria are viable and/or if their presence is relevant to idiopathic urinary tract conditions.
INTRODUCTION
Culture-dependent methods are typically used to test if clinical urine specimens contain uropathogens, and the results play a pivotal role in the diagnosis and treatment of urinary tract infection symptoms in women. Clinical urine cultures are considered positive when the colony count of a recognized uropathogen, such as Escherichia coli, Pseudomonas, Klebsiella, or group B Streptococcus reaches a predefined threshold (14). Bacterial urinary tract infections (UTI) caused by these typical uropathogens elicit symptoms that typically improve or resolve in response to appropriate antibiotic therapy. For other common urinary disorders, including overactive bladder, urinary incontinence, and a spectrum of pain disorders, e.g., painful bladder syndrome and interstitial cystitis, the clinical urine culture is negative and antibiotics are not given for clinical treatment. Under these conditions, the etiology is unknown, and research into these conditions so far has not incorporated culture-independent assessments of bladder infection, such as bacterial 16S rRNA PCR and metagenomic sequencing approaches.
Recently, a concerted international effort, known as the Human Microbiome Project (http://commonfund.nih.gov/hmp/), has begun to catalogue the core microbial composition of the healthy human body in order to determine if changes to the core microbial communities affect health. Sequence analysis of 16S rRNA, the workhorse of that effort, has been used to determine the microflora composition of healthy skin (12, 13), the gastrointestinal tract (7, 9, 36), the mouth (24, 25, 28), and the vagina (11, 16, 27, 39) and to correlate certain diseases with changes in this composition (9, 11, 27). A common theme from all of these studies is that our rather limited capacity to cultivate microorganisms has caused us to overlook much of the diversity of the bacterial communities that colonize the human body.
Based upon the results of culture-independent 16S rRNA sequencing of samples obtained by voided urine, Nelson and coworkers (26) reported that diverse bacteria colonize the adult male urogenital tract. Many of the taxa identified in this study either cannot be or are not routinely cultivated by clinical microbiology laboratories (hereinafter called uncultivated bacteria). A related study determined that first-catch urine and urethral swab samples collected from adult men contained highly similar bacterial communities (6). These and other reports suggest that uncultivated bacteria can colonize the male urogenital tract and might be relevant to male urinary and reproductive tract syndromes.
Colonization of the vagina with diverse populations of fastidious and uncultivated microorganisms is common and clearly associated with bacterial vaginosis (BV) and other idiopathic upper reproductive tract conditions (11, 16, 27, 39). In contrast, it is not known whether uncultivated bacteria in the female urinary tract could be relevant to female urinary tract disease.
The goals of this study were to test if the bladders of women who do not meet the clinical definition for UTI contain uncultivated bacteria. An additional aim was to identify and recommend the most suitable urine collection method for the culture-independent characterization of bacterial communities that specifically reside in the female bladder.
MATERIALS AND METHODS
Study design and patients.
Following Loyola institutional review board (IRB) approval for all phases of this project, participants gave verbal and written consent for the collection and analysis of their urine for research purposes. Eligibility required the absence of known UTI by clinical assessment (physician history and physical) and catheterized urine culture prior to surgery, using the clinical laboratory recommendation to interpret >104 CFU/ml of a uropathogen as clinical infection. Urinary symptoms were assessed for all participants with a validated symptom questionnaire, the Pelvic Floor Distress Inventory (PFDI) (3).
Urine was collected from two groups of women. The control group, composed of patients undergoing surgery for benign gynecologic conditions, reported no urinary symptoms. The comparison group included patients undergoing surgery for treatment of common urogynecologic conditions, including pelvic organ prolapse (POP) and/or urinary incontinence (UI), and who reported at least one PFDI symptom. Metadata, including patient age, race/ethnicity, and body mass index, were abstracted from the electronic medical record for descriptive purposes only.
Sample collection.
Clean-catch, midstream voided urine samples were obtained with a special emphasis on proper clinical collection procedures. The remaining samples were collected at the start of clinically indicated surgery prior to antibiotic administration. Vaginal mucosal swabs were obtained following induction of anesthesia but prior to standard surgical skin preparation. Urinary samples were collected following skin preparation and sterile surgical draping. Because suprapubic aspirate (SPA) specimen collection involves transcutaneous puncture of the abdomen, controls were included to assess skin microorganism contamination of the urine collected by this method. First, a swab of the suprapubic skin was collected. Second, a needle was used to puncture the suprapubic skin, but not the bladder, and then preserved. Third, a second needle was introduced into the retropubic bladder and the SPA urine collected. Finally, the transurethral catheter (TUC) sample was obtained.
Light microscopy.
Raw urine samples were centrifuged for 10 min at 3,000 rpm at 4°C, and the pellet was resuspended with 10 ml phosphate-buffered saline (PBS). The suspension was recentrifuged, and the pellet was resuspended in 1% methylene blue, which does not stain human cells and thus highlights bacteria among human cells. After 15 min of incubation at room temperature, the sample was screened by phase-contrast light microscopy. Some urine samples also were examined using a concentrated Gram stain technique as follows: two drops of well-mixed urine collected in a sterile container was cytocentrifuged at 1,200 rpm for 5 min onto a glass slide, heat fixed, and Gram stained using a standard protocol. Twenty to 40 representative fields were observed under oil immersion (100×) for the presence of microorganisms and white blood cells.
Microbial DNA isolation.
Within 4 h of collection, the urine samples were centrifuged at 5,000 × g for 10 min, and the resulting pellets were resuspended in DNA stabilization buffer (35). Swabs were washed with sterile PBS supplemented with DNA stabilization buffer. All samples were frozen at −80°C until used for microbial DNA isolation and sequence analysis. A laminar flow hood was used to isolate genomic DNA (gDNA) from the urine samples and from the reagent-only control samples. DNA was isolated from urine specimens using a Qiagen DNeasy tissue extraction kit with the Gram-positive bacteria protocol. The gDNA was stored at 4°C until 16S PCR amplification and quantified by fluorescent double-stranded DNA (dsDNA) assay.
Sequencing and analysis.
A combination of approaches was used to identify bacterial 16S rRNA genes in gDNA isolated from the experimental and reagent-only control specimens. Near-full-length 16S rRNA alleles were amplified from gDNA of some samples with degenerate eubacterial primers and were TA cloned and Sanger sequenced as we recently described (26). For pyrosequencing, barcoded degenerate primers targeting the hypervariable V1-V3 subregion of eubacterial 16S rRNA were employed, and the resulting amplicons were sequenced in multiplex by using the Roche 454 Titanium platform (6, 29, 31). Only those sequence reads that perfectly matched primer barcode sequences were decoded into individual samples. After the trimming of primer and barcode sequences, sequences of less than 200 bp in length and/or with average quality scores of less than 25 were discarded (6). These short and low-quality sequences represent PCR noise, which prevents confident taxonomic classification and leads to an inflated number of falsely unclassified sequence reads. Since the length and quality cutoffs were applied to all samples, this discarding of PCR noise did not bias the statistical analysis. All sequences were compared to the human genome using the Basic Local Alignment Search Tool (BLAST) (1), and sequences with significant similarity (E-value threshold of ≤10−10) were excluded from subsequent analyses. Taxonomic classifications were assigned using RDP Classifier v.2.2 (37) with four different RDP Classifier confidence cutoffs: 90%, 80%, 70%, and 60%. As a measure of distance, hierarchical clustering was performed using either Bray-Curtis or Spearman's rank correlation coefficient. To compare microbial distributions between TUC and SPA samples, after normalizing sequence read counts for each assigned taxon to total bacteria sequence reads in the sample, paired t test, McNemar's test, and the Kolmogorov-Smirnov test were implemented in the programming language R (6). To compare the beta distance of different microbial communities, principal coordinate analyses (PCA) of microbial communities were performed using the UniFrac algorithm (15, 20).
To determine if the female bladder can contain uncultivated bacteria, we first identified the urine collection method most suitable for DNA-based studies. To test if voided urine samples accurately surveyed female bladder microbiomes, we compared the bacteria present in pairs of voided and TUC urine samples from the same participant using light microscopy, cultivation, and 16S rRNA sequencing.
To test if TUC sample microbial communities were similar to bladder microbial communities, we collected TUC and SPA samples from 12 asymptomatic participants without clinical signs of UTI and 11 culture-negative participants with POP/UI. Because SPA sample collection involves transcutaneous puncture of the abdomen and thus could be contaminated with microorganisms from the superficial skin, we also collected swabs of the sample site and needles that punctured the skin but did not enter the bladder. V1-V3 16S rRNA PCR and pyrosequencing were used to identify bacteria in these samples.
RESULTS
Results of microscopy, cultivation, and PCR amplification indicated that the microbiomes in voided urine and TUC samples differed. The majority of voided samples (5 of 6) produced colonies upon cultivation in aerobic and/or anaerobic conditions, and 16S rRNA could be amplified from all 6 samples by PCR (termed PCR-positive samples). In contrast, none of the paired TUC samples produced colonies upon cultivation in either aerobic or anaerobic conditions (see Fig. S1 in the supplemental material; also data not shown), while half of the TUC samples (3 of 6) were PCR positive (data not shown). In the voided samples, bacteria could be easily identified by microscopy (for example, see Fig. S2A, C, and F in the supplemental material). In contrast, they were more difficult to find in TUC samples (data not shown). Bacteria were found both free in solution and specifically associated with human nonkeratinized squamous cells of the type often found in the bladder (see Fig. S3 and S2, respectively). In contrast, bacteria were not observed in culture-negative, PCR-negative TUC samples (for example, see Fig. S2D and E). Bacteria also were not observed in some culture-negative but PCR-positive TUC samples (data not shown). Importantly, however, both Gram-negative and -positive bacteria were observed for some culture-negative, but PCR-positive TUC samples (for example, see Fig. S2B and S3). These results confirmed that bacteria were present in at least a subpopulation of the culture-negative TUC samples.
To identify these bacteria, total gDNA was isolated from the samples and 16S rRNA alleles were PCR amplified using the broad-range eubacterial primers F11 and 926R (8, 33). These amplicons were cloned and sequenced, and taxonomy of the resulting sequences was assigned using BLAST and the RDP Classifier (Table 1). The PCR-positive TUC samples contained diverse bacterial genera, including those associated with healthy (e.g., Lactobacillus) and pathological (e.g., Prevotella) vaginal microbiomes (11).
Table 1.
Comparison of bacterial genera from five patients identified by Sanger sequencing
| Patient identifier | Sample source (no. of samples)a | Species | % of total sequences |
|---|---|---|---|
| 1 | Void (10) | Lactobacillus iners | 30 |
| Prevotella timonensis | 20 | ||
| Corynebacterium sp. | 10 | ||
| Anaerococcus sp. | 10 | ||
| Finegoldia magna | 30 | ||
| 1 | TUC (10) | Lactobacillus iners | 100 |
| 6 | Vag (10) | Prevotella timonensis | 10 |
| Uncultured bacterium BF0002B042 | 30 | ||
| Atopobium vaginae | 30 | ||
| Clostridiales genomosp. BVAB3 | 10 | ||
| Leptotrichia amnionii | 10 | ||
| Eggerthella sp. | 10 | ||
| 6 | Void (10) | Prevotella timonensis | 50 |
| Uncultured bacterium BF0002B042 | 10 | ||
| Atopobium vaginae | 20 | ||
| Prevotella bivia | 20 | ||
| 6 | TUC (10) | Prevotella timonensis | 30 |
| Uncultured bacterium BF0002B042 | 10 | ||
| Clostridiales genomosp. BVAB3 | 10 | ||
| Vibrio sp. | 10 | ||
| Streptococcus mitis | 10 | ||
| Lactobacillus iner | 20 | ||
| Corynebacterium aurimucosum | 10 | ||
| 7 | Vag (10) | Streptococcus agalactiae | 60 |
| Prevotella bergensis | 20 | ||
| Prevotella buccalis | 20 | ||
| 7 | Void (9) | Streptococcus agalactiae | 22 |
| Prevotella bergensis | 11 | ||
| Prevotella buccalis | 11 | ||
| Lactobacillus iners | 22 | ||
| Uncultured Prevotella sp. | 11 | ||
| Xanthomonas arboricola | 11 | ||
| Uncultured bacterium clone HRX_H16 | 11 | ||
| 8 | Vag (10) | Lactobacillus iners | 80 |
| Lactobacillus gasseri | 10 | ||
| Enterococcus faecalis | 10 | ||
| 8 | Void (10) | Lactobacillus iners | 80 |
| Dialister propionicifaciens | 20 | ||
| 8 | TUC (10) | Lactobacillus iners | 80 |
| Corynebacterium aurimucosum | 10 | ||
| Blautia hansenii | 10 | ||
| 9 | Vag (10) | Lactobacillus crispatus | 20 |
| Lactobacillus gasseri | 80 | ||
| 9 | Void (10) | Lactobacillus crispatus | 40 |
| Lactobacillus gasseri | 40 | ||
| Peptoniphilus sp. | 10 | ||
| Prevotella buccalis | 10 |
Void, voided urine sample; Vag, vaginal swab sample; TUC, transurethral catheter urine sample.
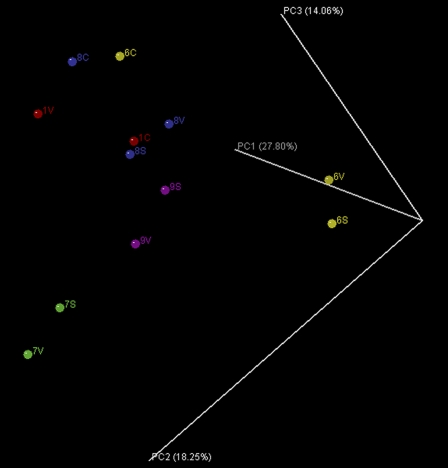
UniFrac principal component analysis was used to test if the microbial communities in voided and TUC samples from the same individual were more closely related to one another than were TUC samples collected from different participants (Fig. 1). On average, the UniFrac distance between the voided sample (V) and the vaginal swab (S) from the same participant was shorter than the distance between the voided and the TUC (C) samples from that same participant (participants 6 and 8). The average UniFrac distance separating TUC samples collected from different participants was also shorter than that of TUC and voided samples from the same participant. These results suggested that the voided samples were contaminated with vulvo-vaginal bacteria and that one could not generally assume that the microbial communities in these samples reflected that of TUC samples, which presumably predominately contained bacteria from the bladder and/or urethra.
Fig 1.
Unweighted UniFrac principal component analysis comparing vaginal swabs (S) to voided (V) and TUC (C) urine samples from patients 1 (red), 6 (yellow), 8, (blue), and 9 (magenta).
To test if TUC microbial communities were similar to bladder microbial communities, we collected TUC and SPA samples from 12 asymptomatic participants without clinical signs of UTI and 11 culture-negative POP/UI participants. V1-V3 16S rRNA PCR and pyrosequencing were used to identify bacteria in these samples.
Taxonomy of the sequences using TUC and SPA samples from 12 asymptomatic participants without clinical signs of UTI and 11 culture-negative POP/UI participants was assigned using RDP Classifier v.2.2 at 90% confidence. We obtained 372,983 high-quality bacterial 16S sequences for subsequent analysis: 206,271 from controls (45,096 from TUC, 48,588 from SPA, 56,838 from skin, and 55,749 from needle samples) and 166,712 from patient samples (29,146 from TUC, 30,083 from SPA, 51,523 from skin, and 55,960 from needle samples). In total, from all four types of samples, 392 genera from 22 bacterial phyla were identified. In the control participants, totals of 165, 226, 205, and 181 genera were detected in TUC, SPA, skin, and needle samples, respectively. In the samples from participants with POP/UI, totals of 150, 191, 185, and 179 genera were detected in TUC, SPA, skin, and needle samples, respectively.
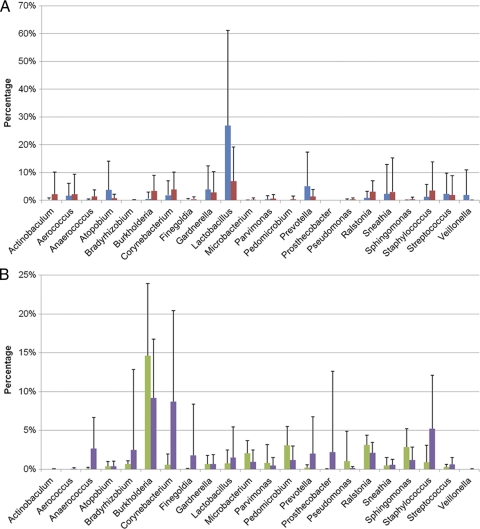
Fourteen genera out of 321 total (<5%) in samples obtained by TUC and SPA were present in different relative abundances (P < 0.05, Student's t test, no multiple correction). Of those 14 genera, only Lactobacillus represented greater than 1% of the total sequences present in either the TUC (26.96%) or SPA (6.94%) samples. The most abundant genera found for TUC samples were similar to those found for SPA urine samples (Fig. 2A), while the most abundant genera found for skin samples were similar to those found for needle samples (Fig. 2B). However, the genera detected for the skin and needle samples were dissimilar from those in the TUC and SPA samples.
Fig 2.
Comparison of the total bacterial communities in (A) TUC (blue) and SPA (red) samples and (B) in needle sticks (green) and skin swabs (purple) of culture-negative participants. Numbers of genera in each sample type were totaled, and the percentage of that total was calculated for each genus. Only the most abundant genera are shown. Error bars represent standard deviations. Note the difference in the scales of the y axes.
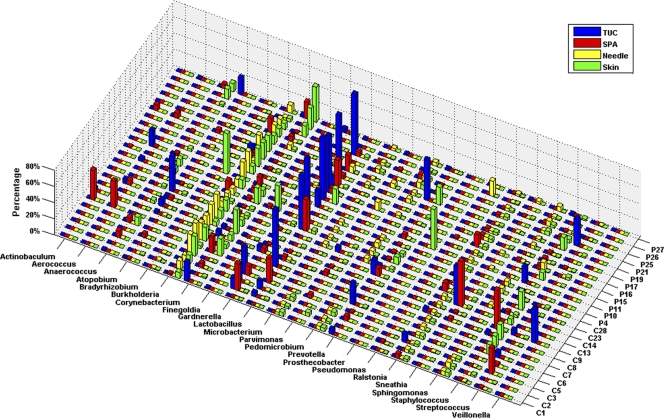
Bacterial 16S rRNA alleles corresponding to taxa from diverse phyla and genera were detected in most of the culture-negative samples from both the asymptomatic controls and the POP/IU participants (Fig. 3). Importantly, 21 of 23 SPA samples, which due to their route of collection were less likely to have been exposed to vulvo-vaginal contamination than the TUC samples, contained detectable levels of 16S rRNA. As noted above, the microbial communities in these samples also were clearly dissimilar to those in the corresponding swab and needle samples, indicating that superficial contamination with vaginal flora on pubic skin was unlikely to be the source of these organisms. However, 16S rRNA could not be amplified from 2 of 23 SPA samples, indicating that the bladders of some women may not contain bacteria.
Fig 3.
Comparison of bacterial communities from each culture-negative participant. The percentage of each genus was calculated for each sample. Only the most abundant genera are shown. TUC (blue), SPA (red), needle (yellow), and skin (green) samples are shown. C, control; P, patient.
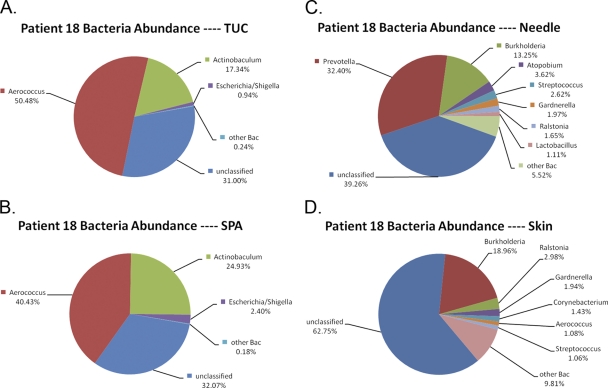
To control for the possibility that the organisms detected in SPA samples represented extremely low-abundance contaminants, we sequenced samples from a single culture-positive participant (>105 CFU/ml E. coli) (data not shown). We obtained 19,832 high-quality bacterial 16S sequences for subsequent analysis: 4,578 from TUC, 5,127 from SPA, 4,905 from skin, and 5,222 from needle samples. The sets of sequences for the TUC and for the SPA samples were similar (Fig. 4A and B). In both samples, the two most abundant genera were Aerococcus (50.48% in TUC and 40.43% in SPA) and Actinobaculum (17.34% in TUC and 24.93% in SPA); a much smaller fraction of sequences corresponded to Escherichia/Shigella (0.94% in TUC and 2.40% in SPA). The bacterial communities in skin and needle samples were highly dissimilar from those found in the TUC and SPA samples (Fig. 4C and D). Although all of the samples shared some low-abundance genera (Burkholderia, Ralstonia, Gardnerella, and Streptococcus), the major constituents were different (Prevotella, Atopobium, and Lactobacillus in needle samples and Corynebacterium and Aerococcus in skin samples). We conclude that at least some of the organisms detected in the SPA samples are not low-abundance contaminants.
Fig 4.
The most abundant genera in TUC (A), SPA (B), needle (C), and skin (D) samples from a culture-positive patient.
DISCUSSION
Bacterial community composition, as revealed by deep 16S sequence analyses, is argued to contribute to diverse human health and disease states (10). While the microbial community structure has been shown to influence susceptibility to infection in models of gastrointestinal disease (2, 4, 5, 34), the application of this concept to the female urinary tract has not been pursued. To define the existence and compositions of bladder bacterial communities in human females without the confounding factor of possible vulvo-vaginal contamination, we carefully sampled urine directly from female bladders using TUC and SPA. Deep 16S rRNA gene sequencing of these samples revealed that bacterial bladder communities of different types do exist in women, although not in all individuals. These data confirm and extend results of earlier studies (17, 21–23), clearly showing that urine specimens reported to clinicians as “culture-negative” or “insignificant growth” can contain varied bacterial communities that can be simple or extremely diverse and can be composed of typical uropathogens or of genera not identified by standard cultivation techniques.
While our data provide evidence for the presence of bacteria and of bacterial DNA in bladders of symptomatic and asymptomatic adult women, the viability of these bacteria remains unclear. To test the viability of certain organisms identified in the SPA samples (e.g., Actinobaculum and Lactobacillus spp.), modified clinical cultivation procedures could suffice. For many organisms detected in SPA, cultivation techniques do not currently exist, and thus viability must be assessed indirectly, such as by in situ measures of bacterial metabolic activity, e.g., shotgun transcriptomics and/or metabolomics. Unfortunately, neither method would suffice to differentiate between two distinct previously hypothesized scenarios: (i) that diverse populations of bacteria can exist in the adult female bladder and may influence urinary tract health (21) and (ii) that diverse populations of viable bacteria are episodically delivered to the bladder but that only known uropathogens proliferate in the bladder and are relevant there. Differentiating between these possibilities will likely require a combination of longitudinal culture-independent measurements of bladder microorganisms in the context of relevant clinical outcomes, such as unexplained pyuria, and the strong association of specific microorganisms with unexplained urinary tract syndromes. Confirmations that fastidious, anaerobic, and difficult-to-cultivate organisms, including Ureaplasma and Actinobaculum spp. (17–19, 30), are causal in some cases of urinary tract syndromes provide a precedent for these sorts of future studies.
For years, the urine culture has been the gold standard for assessment of UTI (32). The utility of this assay, however, is limited. Standard clinical microbiological procedures favor detection of fast-growing bacteria that grow in the presence of oxygen. They consistently undercount slow-growing bacteria and can detect neither anaerobic bacteria nor those whose preferred growth conditions remain unknown (38). This point is driven home by our observation that urine, obtained from participant 18 and determined by standard cultivation procedures to be positive for E. coli, was rich in DNA associated with the fastidious genera Aerococcus (∼40% and ∼50% in SPA and TUC, respectively) and Actinobaculum (∼25% and ∼17%) yet poor in DNA identified with E. coli (∼3% and ∼1%) (Fig. 4). The disparity between the culture and molecular assays raises a question: are this woman's clinical symptoms due to the relatively small number of the “typical” uropathogen E. coli, the numerically superior fastidious bacteria, or a combination of both?
Despite clinical differences between the control and POP/UI populations, we found bacterial communities in most subjects. To properly interpret these findings for subjects with urinary tract disorders, longitudinal studies that assess clinical correlates as well as variability within and among healthy individuals are essential. While DNA-dependent techniques provided evidence of a urinary microbial community in the majority of subjects, they failed to detect bacteria in a minority. The existence of these two states (detectable bacteria and no detectable bacteria) is interesting, and further research that links our findings to various human health conditions will provide guidance as to the “preferred” biological state. As with other areas of the body, one can easily envision “protective” communities or “vulnerable” communities.
For the purpose of microbiome research, the SPA sampling technique appears to be the best at minimizing contamination from nonurinary sites and thus obtaining the best view of the bacteria present within the bladder. The pragmatic aspects of this collection method, however, limit its utility for most research teams. For these teams, we recommend the use of the TUC sampling method. The bacterial communities assessed via TUC and SPA in the same person were highly similar, suggesting that TUC is a sufficient approximation of SPA. While voided samples may be currently sufficient for clinical care, special analytic approaches may be necessary to subtract likely vulvo-vaginal contaminants as part of subsequent microbiome analyses.
Scientific inquiry into the female urinary microbiome is in its infancy. At this early stage of investigation, insufficient information exists concerning variability of urinary communities in health to inform robust sample size calculations for group comparisons. As this field advances, important clinical questions can be addressed using optimal techniques and larger cohorts of appropriately selected subjects.
The likelihood that diverse bacteria can exist within the female urinary tract has important implications for urinary tract disorder researchers. These findings should stimulate work to advance our understanding of the roles played by these bacterial communities in the development of UTI and other urinary tract disorders that remain poorly understood. Such studies could make it possible to identify at-risk UTI populations and allow hypothesis-based research of improved targeted treatments and/or prevention efforts.
Supplementary Material
ACKNOWLEDGMENTS
We thank Sierra Molecular for the generous gift of DNA stabilization reagent, Kashi Revanna for providing computational support for the data analysis, Diane Jaworski for secretarial assistance with manuscript formatting and referencing, and Eva Wojcik for help interpreting cytology.
We gratefully acknowledge the following unrestricted research funding sources: Falk Foundation Research Award, Loyola University Chicago Stritch School of Medicine Research Funding Committee, and Doyle Family Philanthropic Support (to L. Brubaker and A. J. Wolfe) and NIH grant 1RC2HG005806-01, which provides computational support and data analysis (to Q. Dong and D. E. Nelson).
The following authors have no disclosures to report: A. J. Wolfe, E. Toh, N. Shibata, R. Rong, K. Kenton, M. FitzGerald, Q. Dong, D. E. Nelson, and L. Brubaker. The following authors have disclosures to report: E. R. Mueller, research grant from Allergan; P. Schreckenberger, independent contractor/speakers bureau for Cubist, Merck, Forest Laboratories, bioMérieux, Remel, and Hardy Diagnostics; consultant/advisory committees for Abbott Molecular, Elmhurst Hospital, Edward Hospital, Doctors Data Inc., and Forest Laboratories; and review panels for, boards of, or research grants from Cepheid, Abbott Molecular, Siemens, Becton-Dickinson, and bioMérieux.
The authors have full control and free access to all specimens and data relevant to this research without restriction from any research-funding source.
Footnotes
Published ahead of print 25 January 2012
Supplemental material for this article may be found at http://jcm.asm.org/.
REFERENCES
- 1. Altschul SF, et al. 1997. Gapped BLAST and PSI-BLAST: a new generation of protein database search programs. Nucleic Acids Res. 25:3389–3402 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Bailey MT, et al. 2010. Stressor exposure disrupts commensal microbial populations in the intestines and leads to increased colonization by Citrobacter rodentium. Infect. Immun. 78:1509–1519 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Barber MD, et al. 2009. The minimum important differences for the urinary scales of the Pelvic Floor Distress Inventory and Pelvic Floor Impact Questionnaire. Am. J. Obstet. Gynecol. 200:580.e1–580.e7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Bereswill S, et al. 2011. Novel murine infection models provide deep insights into the “ménage à trois” of Campylobacter jejuni, microbiota and host innate immunity. PLoS One 6:e20953. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Croswell A, Amir E, Teggatz P, Barman M, Salzman NH. 2009. Prolonged impact of antibiotics on intestinal microbial ecology and susceptibility to enteric Salmonella infection. Infect. Immun. 77:2741–2753 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Dong Q, et al. 2011. The microbial communities in male first catch urine are highly similar to those in paired urethral swab specimens. PLoS One 6:e19709. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Eckburg PB, et al. 2005. Diversity of the human intestinal microbial flora. Science 308:1635–1638 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Forsman M, Sandstrom G, Sjostedt A. 1994. Analysis of 16S ribosomal DNA sequences of Francisella strains and utilization for determination of the phylogeny of the genus and for identification of strains by PCR. Int. J. Syst. Bacteriol. 44:38–46 [DOI] [PubMed] [Google Scholar]
- 9. Frank DN, et al. 2007. Molecular-phylogenetic characterization of microbial community imbalances in human inflammatory bowel diseases. Proc. Natl. Acad. Sci. U. S. A. 104:13780–13785 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Frank DN, Zhu W, Sartor RB, Li E. 2011. Investigating the biological and clinical significance of human dysbioses. Trends Microbiol. 19:427–434 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Fredricks DN, Fiedler TL, Marrazzo JM. 2005. Molecular identification of bacteria associated with bacterial vaginosis. N. Engl. J. Med. 353:1899–1911 [DOI] [PubMed] [Google Scholar]
- 12. Gao Z, Tseng CH, Pei Z, Blaser MJ. 2007. Molecular analysis of human forearm superficial skin bacterial biota. Proc. Natl. Acad. Sci. U. S. A. 104:2927–2932 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Grice EA, et al. 2009. Topographical and temporal diversity of the human skin microbiome. Science 324:1190–1192 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Gupta K, et al. 2011. International clinical practice guidelines for the treatment of acute uncomplicated cystitis and pyelonephritis in women: a 2010 update by the Infectious Diseases Society of America and the European Society for Microbiology and Infectious Diseases. Clin. Infect. Dis. 52:e103–e120 [DOI] [PubMed] [Google Scholar]
- 15. Hamady M, Lozupone C, Knight R. 2010. Fast UniFrac: facilitating high-throughput phylogenetic analyses of microbial communities including analysis of pyrosequencing and PhyloChip data. ISME J. 4:17–27 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Hyman RW, et al. 2005. Microbes on the human vaginal epithelium. Proc. Natl. Acad. Sci. U. S. A. 102:7952–7957 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Imirzalioglu C, Hain T, Chakraborty T, Domann E. 2008. Hidden pathogens uncovered: metagenomic analysis of urinary tract infections. Andrologia 40:66–71 [DOI] [PubMed] [Google Scholar]
- 18. Lawson PA, Falsen E, Akervall E, Vandamme P, Collins MD. 1997. Characterization of some Actinomyces-like isolates from human clinical specimens: reclassification of Actinomyces suis (Soltys and Spratling) as Actinobaculum suis comb. nov. and description of Actinobaculum schaalii sp. nov. Int. J. Syst. Bacteriol. 47:899–903 [DOI] [PubMed] [Google Scholar]
- 19. Lee YS, et al. 2010. Prevalence and treatment efficacy of genitourinary mycoplasmas in women with overactive bladder symptoms. Korean J. Urol. 51:625–630 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Lozupone C, Hamady M, Knight R. 2006. UniFrac—an online tool for comparing microbial community diversity in a phylogenetic context. BMC Bioinformatics 7:371. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Maskell RM. 2010. The natural history of urinary tract infection in women. Med. Hypoth. 74:802–806 [DOI] [PubMed] [Google Scholar]
- 22. Meijer-Severs GJ, Aarnoudse JG, Mensink WF, Dankert J. 1979. The presence of antibody-coated anaerobic bacteria in asymptomatic bacteriuria during pregnancy. J. Infect. Dis. 140:653–658 [DOI] [PubMed] [Google Scholar]
- 23. Mohanty NK, Jolly BB. 1996. Incidence of anaerobic bacterial infection following transurethral instrumentation. Indian J. Pathol. Microbiol. 39:33–36 [PubMed] [Google Scholar]
- 24. Nasidze I, Li J, Quinque D, Tang K, Stoneking M. 2009. Global diversity in the human salivary microbiome. Genome Res. 19:636–643 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Nasidze I, et al. 2009. Comparative analysis of human saliva microbiome diversity by barcoded pyrosequencing and cloning approaches. Anal. Biochem. 391:64–68 [DOI] [PubMed] [Google Scholar]
- 26. Nelson DE, et al. 2010. Characteristic male urine microbiomes associate with asymptomatic sexually transmitted infection. PLoS One 5:e14116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Oakley BB, Fiedler TL, Marrazzo JM, Fredricks DN. 2008. Diversity of human vaginal bacterial communities and associations with clinically defined bacterial vaginosis. Appl. Environ. Microbiol. 74:4898–4909 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Paster BJ, et al. 2001. Bacterial diversity in human subgingival plaque. J. Bacteriol. 183:3770–3783 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Petrosino JF, Highlander S, Luna RA, Gibbs RA, Versalovic J. 2009. Metagenomic pyrosequencing and microbial identification. Clin. Chem. 55:856–866 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Reinhard M, et al. 2005. Ten cases of Actinobaculum schaalii infection: clinical relevance, bacterial identification, and antibiotic susceptibility. J. Clin. Microbiol. 43:5305–5308 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Ronaghi M. 2001. Pyrosequencing sheds light on DNA sequencing. Genome Res. 11:3–11 [DOI] [PubMed] [Google Scholar]
- 32. Sanford JP, Favour CB, Mao FH, Harrison JH. 1956. Evaluation of the positive urine culture; an approach to the differentiation of significant bacteria from contaminants. Am. J. Med. 20:88–93 [DOI] [PubMed] [Google Scholar]
- 33. Schwieger F, Tebbe CC. 1998. A new approach to utilize PCR-single-strand-confirmation polymorphism for 16S rRNA gene-based microbial community analysis. Appl. Environ. Microbiol. 64:4870–4876 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Sekirov I, et al. 2008. Antibiotic-induced perturbations of the intestinal microbiota alter host susceptibility to enteric infection. Infect. Immun. 76:4726–4736 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Stothard DR, Boguslawski G, Jones RB. 1998. Phylogenetic analysis of the Chlamydia trachomatis major outer membrane protein and examination of potential pathogenic determinants. Infect. Immun. 66:3618–3625 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Turnbaugh PJ, et al. 2009. A core gut microbiome in obese and lean twins. Nature 457:480–484 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Wang Q, Garrity GM, Tiedje JM, Cole JR. 2007. Naive Bayesian classifier for rapid assignment of rRNA sequences into the new bacterial taxonomy. Appl. Environ. Microbiol. 73:5261–5267 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Wilson ML, Gaido L. 2004. Laboratory diagnosis of urinary tract infections in adult patients. Clin. Infect. Dis. 38:1150–1158 [DOI] [PubMed] [Google Scholar]
- 39. Zhou X, et al. 2007. Differences in the composition of vaginal microbial communities found in healthy Caucasian and black women. ISME J. 1:121–133 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.