Abstract
Activation-induced deaminase (AID) is an enzyme required for class switch recombination (CSR) and somatic hypermutation (SHM), processes that ensure antibody maturation and expression of different immunoglobulin isotypes. AID function is tightly regulated by tissue- and stage-specific expression, nuclear localization, and protein stability. Transcription factor YY1 is crucial for early B cell development, but its function at late B cell stages is unknown. Here, we show that YY1 conditional knockout in activated splenic B cells interferes with CSR. Knockout of YY1 did not affect B cell proliferation, transcription of the AID and IgM genes, or levels of various switch region germ line transcripts. However, we show that YY1 physically interacts with AID and controls the accumulation of nuclear AID, at least in part, by increasing nuclear AID stability. We show for the first time that YY1 plays a novel role in CSR and controls nuclear AID protein levels.
INTRODUCTION
The immune system recognizes and dynamically responds to an immense variety of pathogens. The large repertoire of IgM surface receptors is created during early stages of B cell development through rearrangement of heavy chain and light chain immunoglobulin (Ig) variable, diversity, and joining (VDJ) gene segments (10, 19, 27). After exposure to antigen, B cells enter two possible pathways. First, a population of B cells differentiates into plasma cells that secrete original antibody of low affinity and IgM isotype. Second, other B cells enter germinal centers, where they undergo further antibody maturation and late-stage development. Two processes occur during the germinal center reaction: class switch recombination (CSR) and somatic hypermutation (SHM) (33). While SHM diversifies antigen binding sites through mutations in immunoglobulin variable regions, CSR rearranges constant regions of the Ig heavy chain, enabling antibodies to be distributed throughout the body and to carry out different effector functions.
Both CSR and SHM require the enzyme activation-induced cytidine deaminase (AID) (35, 36). AID knockout mice, and patients with autosomal recessive AID mutations, generate only low-affinity antibodies of IgM isotype and thus suffer from a severe immunodeficiency known as hyper-IgM syndrome type 2 (HIGM2) (52). CSR and SHM both require that AID deaminate cytidine to uracil, followed by either mutagenic processing by error-prone repair mechanisms (SHM) or double-strand breaks, leading to rearrangement (CSR) (33). AID function must be tightly regulated to avoid deleterious mutagenic activity because, in addition to diversifying the immune response, AID-catalyzed cytidine deamination is believed to be involved in generation of lymphomagenic chromosome translocations, and overexpression of AID in transgenic animals leads to T cell lymphomas and tumors in the lung epithelium (31, 39, 43, 63). An increasing number of non-Ig genes have also been revealed to be hypermutated by AID in wild-type B cells (31).
AID expression levels directly correlate with the frequency of AID-dependent DNA-remodeling events and the incidence of c-myc/IgH translocations (13, 15, 56, 63, 64). Therefore, limiting AID levels in the nucleus protects the B cell genome from mistargeted mutations, and this is regulated by multiple mechanisms. Upon stimulation of B cells, AID expression is dramatically upregulated in germinal center B cells (36). However, most AID is retained in the cytoplasm, and only a small fraction translocates to the nucleus to mediate CSR and SHM (5, 25, 34, 50). In addition, AID stability is greatly reduced in the nucleus compared to the cytoplasm (1).
Factors that interact with AID and potentially control AID targeting are only now being identified; they include the splicing factors CTNNBL1 and PTBP2, 14-3-3 adaptor proteins, Crm1 exportin protein, the translational elongation factor eEF1A, the DNA repair proteins UNG and Msh2-Msh6, the repressor proteins KAP1 and HP1, the transcriptional pausing protein Spt5, the calcium and integrin binding protein CIB1, RNA exosome proteins, and hsp90 (4, 11, 16, 24, 26, 38, 40, 44, 51, 71). Some of these AID partner proteins have recently been reviewed (57, 58), and it appears that some, such as CIB1 and CTNNBL1, are unlikely to be necessary for CSR (12, 23). As the nuclear levels of AID are clearly important for Ig gene diversification and disease processes, identifying the factors that regulate AID nuclear accumulation is crucial.
Transcription factor YY1 is a ubiquitously expressed GLI-Kruppel zinc finger transcription factor that can both activate and repress a large number of promoters (65). YY1 associates with Ig enhancer elements in both the Ig heavy chain (intron and 3′ enhancers) and the Ig kappa light chain (3′ enhancer) loci (21, 42). YY1 participates in numerous biological processes, including transcriptional activation, transcriptional repression, Polycomb group function, cell cycle regulation, X chromosome inactivation, imprinting, and oncogenesis (2, 14, 20, 28, 61). Cytoplasmic and nuclear localization of YY1 can also be regulated during development (17, 29, 41, 72), suggesting that YY1 might regulate the subcellular localization of interacting partner proteins. Conditional knockout of YY1 in the B cell lineage results in a developmental block at the pro-B cell stage characterized by reduced Ig locus contraction and rearrangement of distal Vh genes (30). YY1 also appears to be important for germinal center B cell development (22).
We show here for the first time that YY1 plays a critical role in regulating CSR. We find that conditional knockout of YY1 in splenic B cells activated ex vivo interferes with CSR to multiple isotypes. YY1 did not affect B cell proliferation, transcription of the Aicda or IgM gene, or transcription of various switch region germ line transcripts needed for efficient CSR. Instead, we found that YY1 physically interacts with AID and modulates the levels of AID in the nucleus, at least in part by increasing AID nuclear stability, thus regulating CSR. We show for the first time that YY1 has a novel role in CSR by controlling nuclear AID protein levels.
MATERIALS AND METHODS
DNA constructs.
The DNA construct glutathione S-transferase (GST)-PU.1, GST-PAX5, cytomegalovirus (CMV)-PAX5, GST-IRF4, CMV-PU.1, CMV-YY1, and Gal-YY1 deletion mutants were previously described (6, 7, 32, 37, 45, 49, 68). GST-Bcl6 was a gift from R. Dalla-Favera (Columbia University Medical Center), and GST-YY1 was supplied by F. Wilkinson (Philadelphia University). The mouse AID cDNA (supplied by Fred Alt, Harvard University) was cloned into the pcDNA3.1 vector at the BamHI-EcoRV sites with the Flag peptide inserted at the N terminus. The QuikChange Site-Directed Mutagenesis kit (Stratagene) was used to create point mutations in the AID sequence in Flag-pcDNA3.1. AID point mutations were cloned according to mutations described in HIGM2 patients: F11V, R24W, S43P, H56Y, L59F, and W80R (52, 62) or phosphorylation sites T27A and S38A.
Mice.
yy1flox/flox mice on a C57BL/6 background were a gift from Y. Shi (Harvard). yy1flox/flox mice or C57BL/6 controls between 8 and 12 weeks of age were used for experiments. The mice were bred and maintained under pathogen-free conditions. All animal studies were performed in compliance with the U.S. Department of Health and Human Services guidelines and were approved by the University of Pennsylvania Institutional Animal Care and Use Committee.
Purification of B cells, CFSE staining, TAT-CRE treatment, in vitro stimulation, and flow cytometry.
Follicular B cells were purified from mouse spleen with anti-CD23-biotin (eBioscience) and streptavidin microbeads (MACS; Miltenyi Biotec). In order to track cell proliferation, the cells were stained with carboxyfluorescein diacetate succinimidyl ester (CFSE) (Invitrogen) according to the manufacturer's instructions. Conditional YY1 knockout in splenic B cells was performed ex vivo using TAT-CRE enzyme purified from bacteria. The cells were washed three times with opti-MEM (Invitrogen) and then incubated with TAT-CRE for 45 min at 37°C. To inactivate TAT-CRE, fetal bovine serum was added to a final concentration of 10%. The cells were washed once with splenic B medium (RPMI 1640, 10% HyClone fetal bovine serum [FBS] [Thermo Scientific], 1× OPI-Media Supplement Hybri-max, 55 μM 2-mercaptoethanol [Sigma], 1× minimal essential medium [MEM] nonessential amino acids, 2 mM l-glutamine, 1% gentamicin [Invitrogen]) and then cultured at 37°C in a 5% CO2 atmosphere. The cells were activated ex vivo with 10 μg/ml lipopolysaccharide (LPS) (Sigma) to stimulate proliferation and switching to isotypes IgG3 and IgG2b, 20 ng/ml interleukin 4 (IL-4) (Peprotech) was added to switch to isotypes IgG1 and IgE, 1 ng/ml human transforming growth factor β1 (TGF-β1) (R&D Systems) was added to stimulate switching to isotype IgA, and 0.5 μg/ml gamma interferon (IFN-γ) (Peprotech) was added to stimulate switching to IgG2A. Additionally, cells were stimulated to switch to IgG3 with 20 μg/ml LPS (Sigma) and 3 ng/ml anti-IgD–dextran (Fina Biosolutions LLC) or to IgA with 20 μg/ml LPS, 20 ng/ml IL-4 (Peprotech), 1.5 ng/ml IL-5 (Peprotech), 1 ng/ml TGF-β (R&D Systems), and 3 ng/ml anti-IgD–dextran. Isotype switching was measured by flow cytometry. The cells were stained with anti-IgG3-fluorescein isothiocyanate (FITC) or IgA-FITC and anti-IgM–phycoerythrin (PE)-Cy7 antibodies (BD Pharmingen). Dead cells stained with 7-aminoactinomycin D (7-AAD) were excluded from analysis. Splenic B cells were stained with PE–anti-mouse IgG1 (BD Pharmingen) and 7-aminoactinomycin D (Invitrogen). Flow cytometry was performed on a FACS Canto II machine, and data were analyzed using FlowJo software. In order to analyze plasma cell differentiation, cells cultured in 5 μg/ml LPS and 20 ng/ml IL-4 for 3 days were stained with anti-CD138–allophycocyanin (APC), as well as anti-IgG1–PE (BD Pharmingen). [6-3H]thymidine (Perkin Elmer) with specific activity of >10 Ci/mmol was diluted in splenic B cell medium to a final concentration of 5 μCi/ml 6 h before harvest.
Splenocyte genotyping.
YY1flox/flox CD23-positive splenocytes were treated with TAT-CRE and subsequently stimulated in culture to switch to IgG1 with 20 μg/ml LPS and 20 ng/ml IL-4. Splenocytes were harvested 0, 18, 48, and 72 h after TAT-CRE or mock treatment. Genomic DNA was prepared using a DNeasy Blood and Tissue kit (Qiagen). PCR primers 1 and 2 (Fig. 1A) amplified the YY1 floxed allele, while primers 1 and 4 (Fig. 1A) amplified DNA after exon 1 deletion as described by Liu et al. (30). The products were normalized to the Cκ region of the immunoglobulin gene using forward primer CCAAGGACGAGTATGAACGACATAACAGCTATAC and reverse primer GTGTAATCTCACGGTATAGAGGTCCTTGAAG.
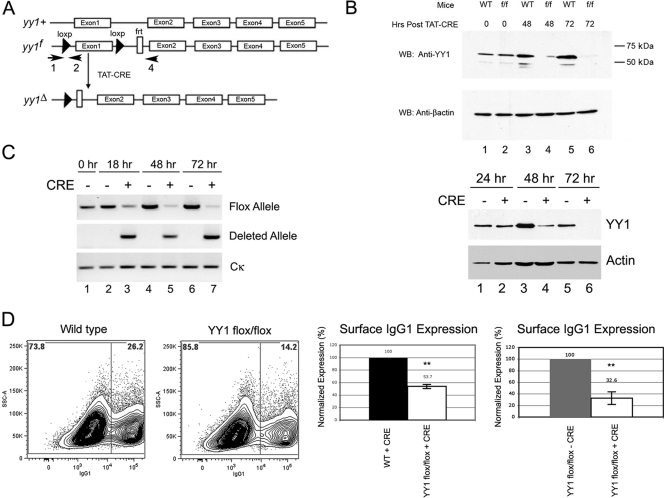
Fig 1.
Conditional YY1 knockout reduces CSR and surface IgG1 expression in activated mouse B cells. (A) Schematic representation of YY1 loci from wild-type (yy1+), yy1 floxed (yy1f), and yy1 CRE-deleted (yy1Δ) mice. The arrows show the locations of primers that detect floxed (primers 1 and 2) and deletion (primers 1 and 4) alleles (30). (B) Recombinant TAT-CRE protein efficiently deletes the floxed yy1 gene in primary splenic B cells ex vivo. Splenic B cells were isolated from yy1flox/flox mice and then treated with recombinant TAT-CRE protein. Cells were harvested at 0, 48, or 72 h (top) or at 18, 48, and 72 h (bottom) after TAT-CRE addition, and whole-cell lysates were assayed for the presence of YY1 by Western blotting (WB) with anti-YY1 antibody. YY1 levels were greatly reduced at 48 h and nearly undetectable at 72 h. Western blotting with anti-β-actin antibody served as a loading control. (C) Genotyping of yy1flox/flox splenic B cells treated with TAT-CRE. Cells were harvested at 0, 18, 48, and 72 h after TAT-CRE addition. Genomic DNA was subjected to PCR with primers that detect the yy1 floxed allele or the yy1-deleted allele. Primers to the Cκ constant region served as loading controls. (D) Splenic B cells were purified from wild-type (WT) or yy1flox/flox mice and treated with recombinant TAT-CRE protein. Levels of surface IgG1 were determined by FACS (left). A histogram of surface IgG1 expression in wild-type or yy1flox/flox B cells treated with TAT-CRE is shown (second from right). Alternatively, yy1flox/flox B cells were treated with TAT-CRE or were mock treated with vehicle control (far right). After a 3-day induction with LPS and IL-4, surface IgG1 expression was measured by FACS. Dead cells were excluded from analysis based on 7-AAD staining. Levels of surface IgG1 expression are shown from 6 or 14 independent experiments (second from right and far right, respectively). The error bars indicate standard deviations, and the asterisks indicate significance (P < 0.001).
RT-PCR.
RNA was isolated from splenic B cells stimulated ex vivo for 4 days using the RNeasy Plus minikit (Invitrogen). RNA was reverse transcribed with the SuperScript first-strand synthesis system (Gibco BRL, Rockville, MD). PCRs were performed for 29 cycles with primers published by Muramatsu et al. (35). IgE and IgG2b germ line transcripts were amplified in 32 and 33 cycles, respectively. AID primers were described by Wuerffel et al. (70). Gamma actin was amplified with primers Gamma actin F, CACGGCATTGTCACTAACT, and Gamma actin R, CAGCCAGGTCCAGACGCAAGAT.
Digestion-circularization PCR (DC-PCR).
Splenic B cells activated with LPS and IL-4 were cultured for 4 days, and then genomic DNA was purified from the live cell population. The DNA (2 μg) was digested overnight with 20 units EcoRI, extracted with phenol-chloroform, and concentrated to 50 μl by ethanol precipitation. Ten percent of the purified DNA was ligated overnight with 400 units of T4 DNA ligase in a 55-μl total volume. The μ-γ1 switch region was amplified by 32 cycles of PCR using primers and conditions described by Ballantyne et al. (3). The nicotinic acetylcholine receptor β-subunit gene was used as a ligation and loading control as described by Chu et al. (9).
GST pulldown.
GST proteins were prepared as previously described (37). AID and RPA70 proteins were labeled with [35S]methionine during in vitro translation using rabbit reticulocyte lysate (Promega, Madison, WI). Equivalent trichloroacetic acid (TCA)-precipitable counts of AID or RPA70 were incubated for 2 h at 4°C with similar amounts of GST or GST fusion proteins bound to glutathione-Sepharose in 100 mM NaCl–1 mM EDTA–20 mM Tris (pH 8.0)–0.5% NP-40 (NETN) supplemented with 1 mg/ml bovine serum albumin and 20 μg/ml ethidium bromide to prevent nonspecific protein-DNA interactions. Beads were washed 6 times with NETN. Bound proteins were eluted in 2× Laemmli buffer supplemented with 200 mM 2-mercaptoethanol, resolved on 10% SDS-polyacrylamide gels, dried, and subjected to autoradiography.
Cell culture and transfection.
HEK 293T cells were cultured in Dulbecco modified Eagle medium supplemented with 10% fetal bovine serum. CH12F3 cells (a gift from T. Honjo) were cultured in RPMI, 10% FBS, 2 mM l-glutamine, 1× MEM nonessential amino acids, 1× penicillin-streptomycin, 50 μM 2-mercaptoethanol, and 5% NCTC-109 medium (Invitrogen). HEK 293T cells were transfected with Lipofectamine 2000 (Invitrogen) according to the manufacturer's instructions and cultured for 48 h before harvest. Chloramphenicol acetyltransferase (CAT) assays were performed as previously described (42, 46).
Preparation of nuclear, cytoplasmic, and whole-cell extracts.
Total protein was extracted with RIPA buffer (50 mM Tris, pH 7.5, 150 mM NaCl, 0.1% sodium dodecyl sulfate, 1% Nonidet P-40, 0.5% sodium deoxycholate). Nuclear and cytoplasmic protein extractions from HEK 293T cells were performed as described previously (55). Splenic B cells were harvested after a 3-day culture with LPS and IL-4. Live cells were purified from dead and apoptotic cells with the Dead Cell Removal Kit (Miltenyi Biotec), washed twice with cold PBS, and then incubated in ice-cold cell lysis buffer (50 mM Tris, pH 8.0, 0.3 M sucrose, 4 mM magnesium acetate, 12.5 mM KCl, 1 mM dithiothreitol [DTT], 10 mM 2-mercaptoethanol, and protease inhibitors) for 5 min. Nonidet P-40 was added to a final concentration of 0.3%, and samples were incubated for 5 min on ice and centrifuged at 500 × g for 10 min. The nuclear pellets were resuspended in 1× buffer B (50 mM Tris, pH 7.5, 25 mM KCl, 5 mM MgCl2) supplemented with 0.3% Nonidet P-40 and, after a 5-min incubation on ice, overlaid on a sucrose cushion (50 mM Tris, pH 7.5, 25 mM KCl, 5 mM MgCl2, 0.2 M sucrose) and centrifuged at 2,000 × g for 5 min. Nuclear proteins were extracted on ice with buffer C (20 mM HEPES, pH 7.9, 0.4 M NaCl, 1 mM EDTA, 1 mM EGTA, 2 mM DTT) for 30 min. All buffers were supplemented with fresh 1 mM phenylmethylsulfonyl fluoride and 1× mammalian protease inhibitor cocktail (Sigma).
Immunoprecipitation and Western blots.
For HEK 293T cells, 500 μg to 1 mg of protein lysate diluted in RIPA buffer with 1% bovine serum albumin was precleared with CL-4B Sepharose beads (Sigma) or rProtein G agarose (Invitrogen) for 1 to 4 h. EZview red Anti-Flag M2 affinity gel (30 μl; Sigma) or GAL4DBD agarose (Santa Cruz Biotechnology) was used for immunoprecipitation overnight at 4°C. The beads were washed 5 times with RIPA buffer containing 0.5% Nonidet P-40. Samples were eluted from the beads with 2× Laemmli buffer supplemented with 200 mM 2-mercaptoethanol. For CH12F3 coimmunoprecipitation experiments, cells were stimulated to switch from IgM to IgA with 0.5 μg/ml anti-CD40 (eBioscience), 20 ng/ml IL-4 (Peprotech), and TGF-β (R&D systems) for 4 days. Total cell extracts from stimulated or unstimulated cells were prepared in cell lysis buffer (20 mM Tris, pH 7.5, 150 mM NaCl, 1 mM EDTA, 1 mM EGTA, 1% Triton X-100) supplemented with 1 mM phenylmethylsulfonyl fluoride (PMSF) and 1× protease and phosphatase inhibitor cocktail (Thermo Scientific) for 30 min on ice. Total cell lysates (1.5 mg) precleared with protein A-agarose (Invitrogen) were used for immunoprecipitation with anti-YY1 antibody (H414; Santa Cruz), anti-Bcl6 antibody (N-3; Santa Cruz), or nonimmune IgG (Jackson ImmunoResearch). The immunoprecipitated complexes were analyzed by SDS-PAGE and Western blotting with anti-AID antibody (L7E7; Cell Signaling) and heavy-chain-specific anti-mouse secondary antibody (Jackson ImmunoResearch). Immunoreactive proteins were visualized using the ECL kit (Amersham Biosciences) or Supersignal West Pico chemiluminescent substrate (Thermo Scientific) and subjected to autoradiography. The films were scanned on an Epson Perfection 4490 photo scanner in positive film mode.
Antibodies.
The antibodies used included anti-PU.1 (T-21; sc-352; Santa Cruz), anti-YY1 (H414; sc-1703; Santa Cruz), mouse monoclonal anti-AID (L7E7; Cell Signaling), anti-TFIIB (C-18; sc-225; Santa Cruz), mouse monoclonal anti-α-tubulin (T5168; Sigma), mouse monoclonal anti-β-actin (A1978; Sigma), mouse monoclonal anti-Flag M2 (F3165; Sigma), anti-Flag–horseradish peroxidase (HRP) (Sigma), anti-rabbit and anti-mouse HRP-conjugated IgG (GE Healthcare UK Limited), anti-mouse HRP-conjugated IgG, and Fcγ fragment-specific antibody (Jackson ImmunoResearch). PAX5 1166 sera were prepared by Cocalico Biologicals.
siRNA knockdown.
YY1 mRNA was knocked down in CH12F3 cells by electroporation of small interfering RNA (siRNA) 709 oligonucleotide (Sigma); 24 h after the knockdown, the cells were stimulated to switch isotype to IgA. Cells were harvested 48 h after the knockdown. Nuclear and cytoplasmic extracts were analyzed for AID and YY1 expression by SDS-PAGE and Western blotting (using AID L7E7 antibody and YY1 H414 antibody). Signals were normalized to nuclear actin with β-actin antibody (Sigma).
Cycloheximide stability experiments.
HEK 293T cells were transfected with 6 μg Flag-AID and 18 μg CMV-YY1 using Lipofectamine 2000 reagent. The cells were trypsinized, mixed, and replated 24 h after transfection in order to normalize transfection efficiency. Cycloheximide (100 μg/ml; Sigma) was added to the cells at 48 h posttransfection to stop protein translation. Nuclear extracts were prepared at 0, 2.5, 5, 7.5, and 10 h after cycloheximide addition, and AID was followed by Western blotting. Nuclear extracts were devoid of cytoplasmic contamination, as evidenced by lack of α-tubulin expression. Nuclear AID levels were normalized to the amounts of nucleolin (C23 antibody) Santa Cruz and Ponceau S staining.
RESULTS
YY1 controls CSR in activated splenic B cells.
YY1 conditional knockout in the B cell lineage results in arrest at the pro-B cell stage and greatly reduced numbers of splenic B cells. We sought to explore whether YY1 played a role in regulating a late B cell function, such as CSR. To determine the effect of YY1 on CSR, we utilized yy1flox/flox mice, which contain the first yy1 exon flanked by loxP sites (Fig. 1A). Splenic mature B cells isolated from these mice can be treated with recombinant TAT-CRE protein ex vivo to result in deletion of the endogenous yy1 gene and subsequent loss of the YY1 protein. Indeed, we found that knockout of the first yy1 exon resulted in a dramatic reduction of YY1 protein 48 h after TAT-CRE treatment and almost complete YY1 protein loss 72 h after TAT-CRE treatment (Fig. 1B, top and bottom). We consistently observed efficient time-dependent loss of YY1 protein in response to TAT-CRE treatment (Fig. 1B, top and bottom). To confirm this was due to yy1 gene deletion, we assayed for disappearance of the yy1 floxed allele (Fig. 1A, primers 1 and 2) and gain of the deletion allele (Fig. 1A, primers 1 and 4) by PCR. As expected, we observed a progressive time-dependent loss of the yy1 floxed alleles and a concomitant gain of the yy1 deletion allele (Fig. 1C). This yy1flox/flox–plus–TAT-CRE system can therefore be used to explore the importance of YY1 function in CSR of IgM to IgG1, which can be induced by treatment with bacterial LPS and IL-4. Indeed, deletion of the yy1 gene resulted in a 2- to 4-fold reduction of surface IgG1 expression in TAT-CRE-treated yy1flox/flox cells compared to control B cells (Fig. 1D). The drop in IgG1 surface expression in 20 independent experiments is shown in Table S1 in the supplemental material.
CSR of IgM to IgG1 leads to generation of postswitch transcripts (PSTs) containing juxtaposed μ and γ1 switch sequences. Consistent with our fluorescence-activated cell sorter (FACS) data, TAT-CRE treatment led to a 5-fold reduction in the level of postswitch transcripts, indicative of the IgM-to-IgG1 switch (Fig. 2A, IgG1). To ensure that this represented a rearrangement event at the DNA level, we performed DC-PCR with DNA isolated from TAT-CRE- or vehicle-treated yy1flox/flox splenic B cells. As expected, DC-PCR revealed a 5-fold decrease in PCR product in TAT-CRE-treated cells, indicative of the DNA deletion generated in IgG1 CSR (Fig. 2C).
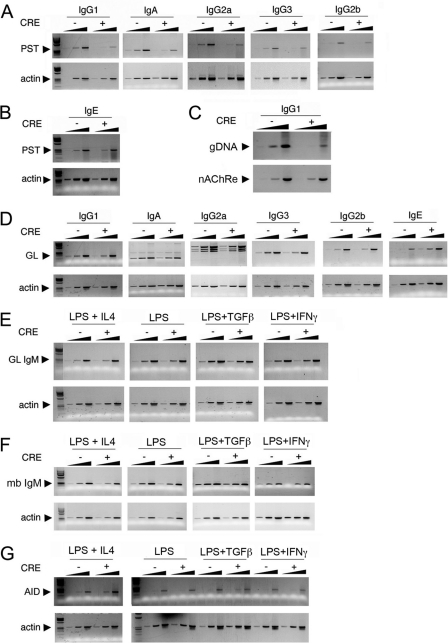
Fig 2.
YY1 regulates CSR, but not germ line, IgM or Aicda transcripts. (A to G) Splenic B cells purified from yy1flox/flox mice were treated with bacterial TAT-CRE, resulting in YY1 knockout (CRE +), or were mock treated (CRE −). The cells were treated with LPS plus IL-4 to induce CSR to IgG1 and IgE, with LPS to induce CSR to IgG3 and IgG2b, with LPS plus TGF-β to induce CSR to IgA, and with LPS plus IFN-γ to induce CSR to IgG2a. RNA was isolated and assayed for PSTs (A and B) or germ line switch region transcripts (GL) (D) by reverse transcription (RT)-PCR. Fivefold-increasing sample amounts are shown for each isotype or loading control. The data are representative of three independent experiments. (A) Loss of YY1 reduces CSR to IgG1, IgA, IgG2a, IgG3, and IgG2b as assayed by RT-PCR of PSTs. (B) Loss of YY1 does not affect CSR to IgE. (C) Digestion circularization PCR of DNA from splenic B cells activated with LPS and IL-4 for 4 days. Primers that detect switching to IgG1 (genomic DNA [gDNA]) were used in the top gel, and primers for the loading control nicotinic acetylcholine receptor (nAChRe) β-subunit gene were used in the bottom gel. The amount of gDNA was increased by 5-fold in each lane. (D) Loss of YY1 has no impact on switch region germ line transcripts (GL), except for a 5-fold increase in IgE transcripts. (E and F) IgM germ line transcripts (GL IgM) and membrane-bound transcripts (mb IgM) are not impacted by loss of YY1. (G) AID mRNA expression is not impacted by loss of YY1.
To determine if YY1 affected CSR to other immunoglobulin isotypes, we assayed for PSTs that quantify switching to various isotypes induced by treatment of cells with various cytokines. Primary yy1flox/flox splenic B cells treated with TAT-CRE were incubated with IL-4 and LPS to induce switching to IgG1 and IgE, with LPS to induce switching to IgG3 and IgG2b, with TGF-β plus LPS to induce switching to IgA, and with IFN-γ plus LPS to induce switching to IgG2a. Interestingly, YY1 knockout resulted in an approximately 5-fold reduction of PST, indicative of CSR to IgG1, IgA, IgG2a, and IgG3, demonstrating that YY1 has a general role in Ig CSR (Fig. 2A). Switching to IgG2B was reduced approximately 2-fold (Fig. 2A). On the other hand, switching to IgE was unaffected by loss of YY1 (Fig. 2B). The reason for this differential effect is discussed below. We confirmed our PST data by assaying CSR to two other isotypes by FACS. Consistent with our PST data, after conditional YY1 knockout, we observed a dramatic loss of surface IgG3 and IgA as assayed by FACS (see Fig. S1 in the supplemental material). In addition, we also observed a reduction in plasma cell differentiation after treatment with IL-4 plus LPS and TAT-CRE as measured by surface CD138 (syndecan) expression (see Fig. S2 in the supplemental material). This reduction was relatively small (40%) but was statistically significant (P < 0.001). In total, our results indicate that loss of YY1 significantly reduces CSR to multiple Ig isotypes.
YY1 does not impact levels of switch region germ line transcripts, IgM transcripts, or Aicda gene expression.
Germ line transcription through switch region DNA sequences is required for CSR, and it is believed that this transcription increases switch region DNA accessibility to AID and DNA repair factors. Loss of YY1 might reduce the levels of these switch region germ line transcripts, leading to reduced CSR. To gain more insight into the molecular mechanisms underlying the impact of YY1 on CSR, we analyzed the effect of YY1 conditional knockout on expression of germ line transcripts that originate from each switch region promoter in response to various cytokines. Splenic B cells from yy1flox/flox mice were treated with TAT-CRE or mock treated, and RNA was collected from 4-day cultures induced to switch to IgG1, IgA, IgG2a, IgG3, IgE, and IgG2b. Interestingly, YY1 knockout did not reduce expression of any germ line switch region transcripts, indicating that YY1 does not impact CSR by regulating germ line transcription (Fig. 2D). On the contrary, YY1 knockout actually elevated IgE switch region transcripts approximately 5-fold (Fig. 2D). As discussed below, elevated IgE germ line transcription may explain the lack of reduced IgE switching after YY1 knockout. YY1 knockout also had no impact on the levels of IgM germ line or IgM coding transcripts under various cytokine induction treatments, indicating that YY1 does not control the levels of these transcripts for regulating CSR (Fig. 2E and F).
Levels of AID can impact the frequency of CSR (13, 15, 54, 56, 63, 64). We hypothesized that YY1 might regulate transcription of the Aicda gene, leading to reduced CSR. However, loss of YY1 had no impact on the expression of AID transcripts in any of the cytokine mixtures used to induce CSR (Fig. 2G). The above results demonstrate that although YY1 generally functions as a regulator of transcription, it does not affect expression of Ig germ line transcripts needed for CSR or expression of the Aicda or IgM gene.
Loss of YY1 does not impact proliferation of splenic B cells.
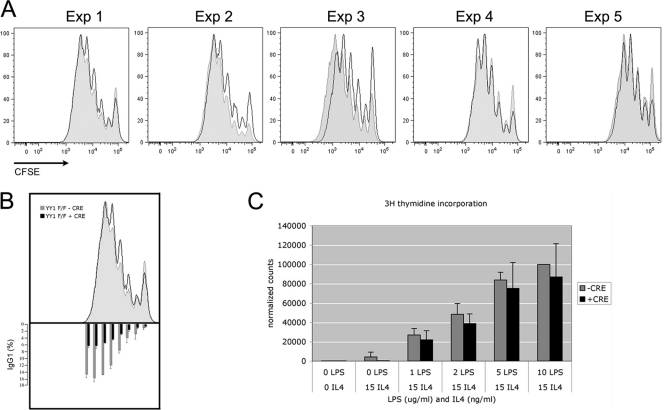
Cell division is necessary for CSR, and Aicda expression ex vivo increases in each division cycle of induced splenic B cells, correlating well with the frequency of CSR (54). Therefore, we tested if loss of YY1 affected the proliferation of activated splenic B cells. We loaded splenic B cells with a cell division tracking dye, CFSE; added TAT-CRE to knock out YY1; and evaluated proliferation after 3 days in culture with LPS and IL-4. No significant differences in proliferation between YY1 conditional knockout cells and mock-treated control cells were observed in five independent experiments (Fig. 3A), even though CSR was still impaired (Fig. 3B, bottom). Similarly, incorporation of tritiated thymidine showed no impact of YY1 deletion on cell growth (Fig. 3C). We conclude that YY1 does not regulate cell division of splenic B cells activated ex vivo.
Fig 3.
Loss of YY1 does not affect proliferation of activated B cells. (A) Splenic B cells from yy1flox/flox mice were stained with CFSE and either untreated (shaded profiles) or treated with TAT-CRE (open profiles). After a 3-day stimulation with LPS and IL-4, CFSE levels in each cell were determined by FACS. The results of 5 independent experiments are shown. (B) Surface IgG1 expression in cells stained with CFSE in panel A was determined by FACS (bottom). Averages from 5 experiments are presented, normalized to surface IgG1 expression in division cycle number 5. The error bars indicate standard deviations. (C) [3H]thymidine incorporation of splenic B cells from yy1flox/flox mice cultured for 3 days in LPS and IL-4 media and treated with TAT-CRE (+CRE) or mock treated (−CRE). The averages of 3 experiments reflect counts normalized to mock-treated cells cultured in 10 μg/ml LPS and 15 ng/ml IL-4. The error bars indicate standard deviations.
YY1 physically interacts with AID.
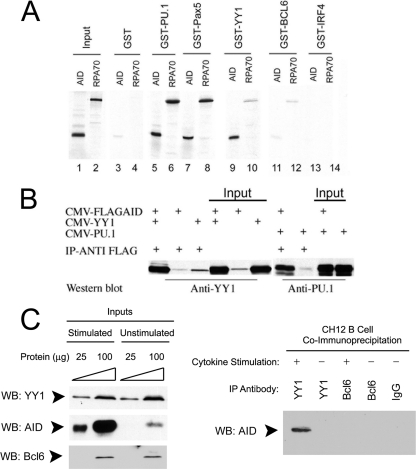
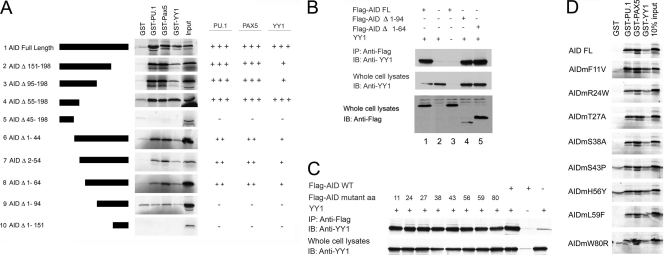
We reasoned that YY1 might control CSR by physically interacting with AID to regulate its function. We assayed for physical interactions by GST pulldown assays between AID, or AID-associated RPA70 protein, and YY1, as well as with a panel of other transcription factors that bind Ig(κ) light chain 3′ or intron enhancers (PU.1, PAX5, BCL6, and IRF4) (32, 47–49, 67). We observed strong interactions between AID and YY1, PU.1, and PAX5, while RPA70 physically interacted with PU.1 and PAX5 (Fig. 4A, lanes 1 to 10). On the other hand, BCL6 and IRF4 failed to interact with either AID or RPA70 (lanes 11 to 14).
Fig 4.
AID interacts with PU.1, PAX5, and YY1 in vitro and in vivo. (A) GST pulldown assay. In vitro-translated 35S-labeled AID or RPA70 protein was incubated with GST-tagged PU.1, PAX5, YY1, Bcl6, or IRF4 purified from bacteria in the presence of ethidium bromide to inhibit protein-DNA interactions and evaluated by SDS-PAGE. (B) AID coimmunoprecipitates with YY1 and PU.1 in vivo. 293T cells were transfected with CMV–Flag-AID, CMV-YY1, or CMV-PU.1 plasmid in the combinations indicated. After immunoprecipitation (IP) with anti-Flag antibody, levels of YY1 and PU.1 were detected by Western blotting with anti-YY1 and anti-PU.1 antibodies. Two percent (10 μg) of the sample is shown as input. The data are representative of three independent experiments. (C) AID and YY1 interact in vivo in B cells. Cell lysates from either stimulated or unstimulated CH12 cells were assayed by Western blotting for YY1, AID, and BCL6 expression (left). The same lysates were immunoprecipitated with the antibodies shown (right), and AID was detected by Western blotting.
To corroborate these findings in vivo, we performed coimmunoprecipitation assays with plasmids expressing Flag-AID and either YY1 or PU.1. Indeed, we found that the YY1 and PU.1 proteins coimmunoprecipitated with Flag-AID (Fig. 4B). We also performed coimmunoprecipitation experiments with CH12 B cells, which induce AID expression in response to treatment with anti-CD40, IL-4, and TGF-β. Indeed, we found this treatment stimulated AID expression in CH12 cells but had no impact on the basal levels of YY1 and BCL6 expressed in these cells (Fig. 4C, left). When extracts from stimulated and unstimulated CH12 cells were immunoprecipitated with YY1 antibody, AID was found to coimmunoprecipitate in the stimulated extracts (Fig. 4C, right). No AID coimmunoprecipitate was observed using either anti-BCL6 antibody or nonimmune IgG (Fig. 4C, right). Therefore, we conclude that YY1 and AID interact in vivo in B cells.
To identify the AID domains responsible for binding each transcription factor, we prepared a variety of AID deletion mutants and tested their abilities to interact with YY1, PU.1, and PAX5 by GST pulldown assays. Progressive C-terminal deletions suggested that sequences between residues 45 and 55 are required for interaction with each transcription factor (Fig. 5A, constructs 1 to 5). This was partially confirmed by a series of N-terminal deletions (Fig. 5A, constructs 6 to 10), suggesting that YY1, PU.1, and PAX5 bind to AID within its N-terminal half, and this binding appears to involve AID sequences 45 to 55, as well as residues 64 to 94. We tested whether deletion of these AID sequences would result in lost interaction in vivo as measured by coimmunoprecipitation. As expected, full-length AID interacted with YY1 by coimmunoprecipitation (Fig. 5B, lanes 1 and 2), consistent with our GST data. Flag-AIDΔ1-94, surprisingly, still coimmunoprecipitated with YY1 (Fig. 5B, lane 4), even though the mutant failed to interact with YY1 in vitro by GST pulldown assay. This suggests that AID and YY1 might be part of a larger protein complex in vivo. We also tested a number of AID point mutants corresponding to mutations found in hyper-IgM syndrome type 2 patients (F11V, R24W, S43P, H56Y, L59F, and W80R) or sites where AID can be phosphorylated (T27A and S38A). None of these point mutations affected binding of AID to YY1 in vivo (Fig. 5C) or to YY1, PU.1, or PAX5 in vitro (Fig. 5D). In summary, our results demonstrate that AID interacts with YY1 in vitro and in vivo via residues in the AID N terminus.
Fig 5.
Identification of AID sequences necessary for interaction with YY1, PU.1, and PAX5. (A) GST pulldown assays were performed with GST-tagged YY1, PU.1, or PAX5 and in vitro-translated 35S-labeled AID deletion mutants. Specific AID deletion mutants are shown on the left, and levels of interaction with PU.1, PAX5, and YY1 are summarized on the right (+, about 25%; ++, about 50%; +++, 100%; −, not above background). (B) YY1 coimmunoprecipitates with Flag-AIDΔ1-94. 293T cells were transfected with the expression plasmids shown above each lane. Samples were immunoprecipitated with anti-Flag antibody and subjected to the Western blot (IB) procedure with anti-YY1 antibody. At the bottom are shown various Flag-AID and mutant inputs. FL, full length. (C) Various hyper-IgM AID point mutants coimmunoprecipitate with YY1. The Flag-AID wild-type and point mutants shown above the lanes were cotransfected into 293T cells with or without YY1 expression plasmid. Samples were immunoprecipitated with anti-Flag antibody and subjected to the Western blot procedure with anti-YY1 antibody. (D) Various hyper-IgM syndrome AID point mutants interact with YY1, PU.1, and Pax5. GST pulldown assays were performed with the 35S-labeled proteins shown on the left and GST fusion proteins shown above each lane.
YY1 affects AID nuclear localization.
AID can shuttle between the cytoplasm and the nucleus (25), and an increasing body of evidence suggests that AID levels in the nucleus are limiting for CSR (13, 15, 56, 63, 64). Therefore, factors that regulate AID levels in the nucleus will regulate the efficiency of CSR.
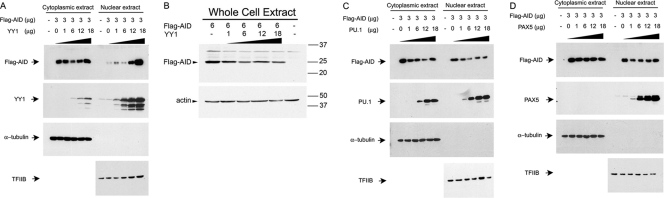
To determine whether YY1 might regulate AID nuclear localization, we cotransfected Flag-tagged AID with increasing amounts of CMV-YY1. Interestingly, AID protein levels in the nucleus increased with increasing nuclear YY1 (Fig. 6A). AID levels in whole-cell lysates did not change, indicating that YY1 expression specifically affected nuclear levels of AID and not total amounts of AID protein (Fig. 6B). Since the fraction of AID typically observed in the nucleus is very small compared to the total cellular AID, lack of change on Western blots of total cell or cytoplasmic extracts is not surprising. In order to detect strong signals of AID in our nuclear fractions, we have used 10 to 20 times more cell equivalents of nuclear extract than of cytoplasmic or whole-cell extracts. The effect of increasing nuclear AID levels was specific to YY1, as transfection with plasmids expressing PU.1 or PAX5 had no impact on AID nuclear or cytoplasmic levels (Fig. 6C and D).
Fig 6.
YY1 affects AID nuclear localization in transfected 293T cells. (A to D) Flag-AID was transfected to 293T cells with increasing amounts of YY1 (A and B), PU.1 (C), or PAX5 (D) expression plasmids. Two days after transfection, nuclear, cytoplasmic, or whole-cell extracts were prepared and assayed by Western blotting for expression of AID with anti-Flag antibody, as well as for expression of YY1, PU.1, and PAX5 by Western blotting with appropriate antibodies. Sixty micrograms of protein assayed in each sample represents about 0.3 million cells of cytoplasmic extract and 3 million cells of nuclear extracts. The purity of nuclear extracts is shown by absence of cytoplasmic α-tubulin, and sample loading is controlled by levels of TFIIB. The data are representative of three independent experiments.
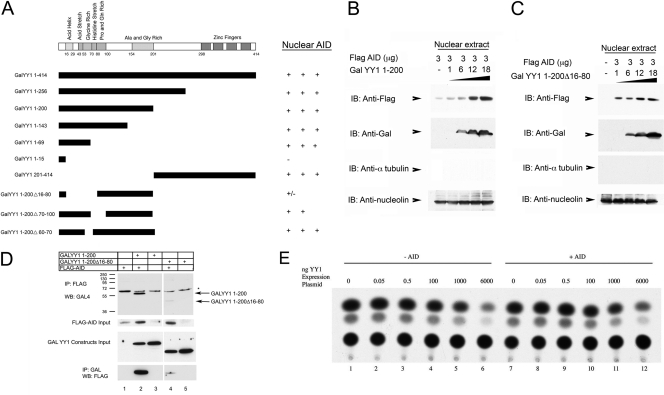
To map the YY1 domain responsible for AID nuclear accumulation, cells were cotransfected with Flag-AID and increasing levels of GAL-tagged YY1 deletion mutants. These studies indicated that both N- and C-terminal regions of YY1 were sufficient for AID nuclear translocation (GALYY1 1 to 200 and GALYY1 201 to 414) (Fig. 7A). Progressive carboxy-terminal YY1 deletions, 1 to 256, 1 to 200, 1 to 143, and 1 to 69, were all capable of increasing AID levels in the nucleus. Deletion to YY1 residue 15, however, ablated this effect. In the context of YY1 sequences 1 to 200, deletion of residues 16 to 80 greatly reduced AID nuclear accumulation, whereas mutants Δ70-100 and Δ60-70 continued to yield elevated nuclear AID (Fig. 7A). Representative Western blot data for the summary presented in Fig. 7A are shown in Fig. S3 in the supplemental material.
Fig 7.
Mapping of YY1 domains needed for increased AID nuclear accumulation. (A) Various GALYY1 mutants (diagrammed on the left) were cotransfected into 293T cells with Flag-AID. Levels of nuclear AID accumulation are represented on the right. (B and C) GALYY1 1-200 or GALYY1 1-200Δ16-80 was cotransfected with Flag-AID into 293T cells. While GALYY1 1-200 expression led to increased AID nuclear accumulation (B), GALYY1 1-200Δ16-80 did not (C). (D) AID nuclear accumulation correlates with YY1 physical interaction. GALYY1 1-200 or GALYY1 1-200Δ16-80 was cotransfected with Flag-AID into 293T cells. Whole-cell extracts of the transfected cells immunoprecipitated with anti-Flag antibody and then blotted with anti-GAL antibody showed coprecipitation of GALYY1 1-200 with Flag-AID but not GALYY1 1-200Δ16-80 (top). Three percent input blots are shown in the middle gels. The lower gel shows the reciprocal experiment, in which immunoprecipitation was performed with anti-GAL antibody followed by blotting with anti-Flag antibody. Again, only GALYY1 1-200 interacted strongly with Flag-AID. (E) AID does not impact YY1 transcription functions. Increasing amounts of CMV-YY1 were transfected in the presence and absence of CMV–Flag-AID and assayed for transcriptional activity of a YY1-responsive promoter containing a multimerized YY1 binding site adjacent to the TK promoter, (YY1)4TKCAT. This promoter is activated by low levels of YY1 but repressed by high levels (6, 42). 293T cells were transfected with (YY1)4TKCAT reporter plasmid and increasing amounts of YY1 expression plasmid either with (+) or without (−) AID expression plasmid. The levels of YY1 transcriptional activation and repression were analyzed by chloramphenicol acetyltransferase assay.
The nuclear accumulation of AID is likely regulated by direct physical interaction with YY1, because a YY1 construct that drives nuclear AID accumulation (GALYY1 1 to 200) (Fig. 7A and B) coimmunoprecipitated with AID (Fig. 7D), whereas a YY1 construct that fails to support nuclear AID accumulation (GALYY1 1-200Δ16-80) (Fig. 7A and C) only weakly interacted with AID (Fig. 7D). Our results indicate that increased nuclear YY1 levels result in increased AID nuclear levels and that YY1 sequences 16 to 80 represent a YY1 domain involved in this phenotype. This region of YY1 includes the transcriptional activation domain (6).
Since AID physically interacts with the YY1 transcriptional activation domain, we set out to determine if AID could control YY1 transcriptional functions. We transfected increasing amounts of CMV-YY1 in the presence and absence of CMV–Flag-AID and assayed for transcriptional activity of a YY1-responsive promoter containing a multimerized YY1 binding site adjacent to the thymidine kinase (TK) promoter. This promoter is activated by low levels of YY1 but repressed by high levels (6, 42). We observed no impact on YY1 transactivation function by AID expression (Fig. 7E). Thus, while YY1 expression apparently can regulate AID-mediated functions (CSR and nuclear accumulation), AID had no impact on YY1 transcriptional functions (activation and repression).
Loss of YY1 in B cells reduces nuclear AID levels.
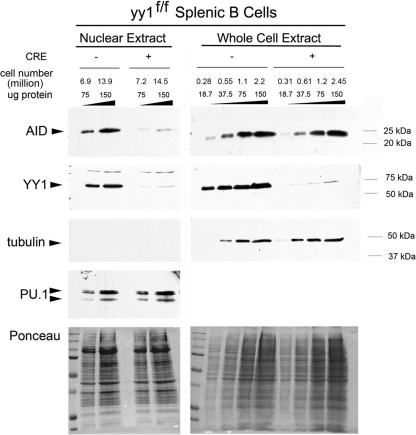
Our data mentioned above showed that elevated YY1 levels result in increased abundance of AID in the nucleus. To determine if reduced YY1 levels would lower the level of nuclear AID, we conditionally knocked out YY1 in mature yy1flox/flox splenic B cells with recombinant TAT-CRE protein and measured AID levels in nuclear extracts and whole-cell lysates after stimulation for 3 days with LPS plus IL-4. YY1 conditional knockout in splenic B cells resulted in a dramatic 91 to 94% reduction in the level of AID in nuclear extracts, whereas total AID levels were the same in control and conditional knockout cells (Fig. 8). Nuclear extracts were assayed for the cytoplasmic marker protein tubulin and were found to be devoid of cytoplasmic contamination (Fig. 8). Protein loading was equal, as evidenced by Ponceau staining, PU.1 in the nuclear extracts, and tubulin in the whole-cell extracts. We conclude that YY1 can modulate soluble AID protein levels in splenic B cell nuclei to control CSR.
Fig 8.
YY1 regulates AID levels in nuclear extracts from activated mouse splenic B cells. Splenic B cells from yy1flox/flox mice were treated with bacterial TAT-CRE (+CRE) or mock-treated (−CRE) as a control and activated ex vivo with LPS plus IL-4. Nuclear and whole-cell extracts were made after 3 days in culture. Endogenous AID protein levels were determined by Western blotting with an anti-AID antibody (Cell Signaling). AID signals were normalized to tubulin and PU.1 levels, as well as the signal from reversible staining of Western blot membranes with Ponceau S solution. A representative of 3 independent experiments is shown. The representative number of cell equivalents for each lane is shown to indicate the much larger cell numbers needed for the nuclear extract lanes compared to whole-cell extracts.
We also performed YY1 siRNA knockdown studies in CH12 B cells. Knockdown of YY1 was much less efficient in these cells (about 50% knockdown compared to 95% in our splenic B cells), but consistent with our splenic B cell conditional knockout experiments, YY1 knockdown in CH12 cells resulted in reduced levels of nuclear AID (see Fig. S4 in the supplemental material).
YY1 controls AID nuclear stability.
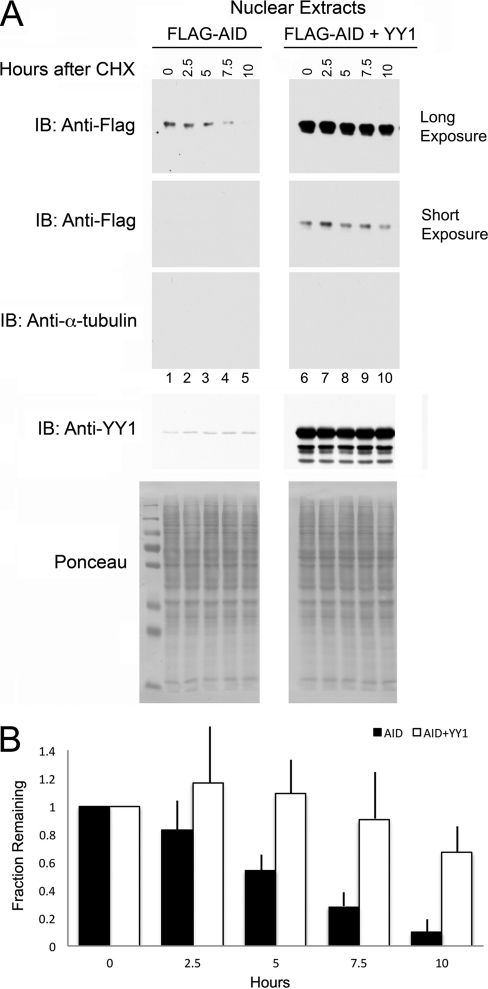
As YY1 is able to regulate the amount of AID in the nucleus and the shorter half-life of AID in the nucleus is important for regulating nuclear AID levels, we set out to determine if YY1 could regulate nuclear AID stability. We performed cycloheximide half-life studies with cells transfected with Flag-AID alone or Flag-AID cotransfected with CMV-YY1. Two days after transfection, cycloheximide was added to inhibit protein synthesis, and nuclear extracts were prepared at various times. Interestingly, we found that cotransfection of AID with YY1 caused a significant increase in nuclear AID stability (Fig. 9A, compare top gel, lanes 1 to 5, to second gel from top, lanes 6 to 10). Samples were devoid of cytoplasmic contamination as evidenced by a lack of α-tubulin (third gel from top), and protein loading was comparable in each lane (bottom two gels). Quantitative data from 8 independent experiments (Fig. 9B) indicated that the half-life of AID can be increased by YY1 to approximately 12 to 13 h in the nucleus compared to reports of about 2.5 h in the literature (1) and about 5 h here. Therefore, YY1 can increase the level of nuclear AID, at least in part, by stabilizing AID in the nucleus.
Fig 9.
YY1 controls AID stability in the nucleus. (A) Flag-AID or Flag-AID plus YY1 was cotransfected into 293T cells, and 2 days later, the cells were treated with 100 μg/ml cycloheximide (CHX) to inhibit protein synthesis. Cells were harvested at the times indicated above the lanes, and nuclear extracts were immunoblotted with antibodies to Flag (top two gels; long and shorter exposures, respectively), α-tubulin to detect potential cytoplasmic contamination (third gel from top), and YY1 (fourth gel from top). Comparable loading of each lane is shown by Ponceau staining in the bottom gel. The exposures of the top two gels were adjusted to enable comparison of the stability of AID alone (top gel, lanes 1 to 5) with that of AID plus YY1 (second gel from top, lanes 6 to 10). (B) The fractions of AID remaining normalized to nucleolin and Ponceau S staining are shown for AID versus AID plus YY1 for 8 independent experiments. The error bars show the standard deviations of the mean.
DISCUSSION
We found that depletion of YY1 from activated splenic B cells compromised CSR. We hypothesized that YY1 could modulate CSR by controlling (i) germ line switch region transcription, (ii) transcription of the Aicda or IgM gene, (iii) proliferation of activated B cells, (iv) AID protein levels, or (v) levels of nuclear AID. We found that YY1 does not regulate expression of switch region germ line transcripts, transcription of the Aicda or IgM gene, splenic B cell proliferation, or total AID protein levels. On the other hand, we found that YY1 modified the levels of AID protein in nuclear extracts, at least in part, by increasing nuclear AID stability.
During B cell development, YY1 expression levels remain relatively constant, as defined by transcript levels (http://www.immgen.org). However, YY1 protein levels are regulated in some systems yielding biological responses. This is best studied in skeletal muscle differentiation systems, where YY1 expression levels drop as a result of proteolysis (66), and in cardiac disease conditions (59, 60). Here, we found that changes in YY1 levels can regulate the nuclear levels of AID.
Knockout of YY1 affected CSR to numerous isotypes, consistent with YY1 regulating nuclear AID levels. However, switching to IgE was unaffected (Fig. 2B). Potentially, this switch region may be less sensitive to low levels of nuclear AID. We observed elevated IgE switch region transcription after deletion of YY1, and this might make the switch region more accessible to the low levels of nuclear AID. IgE CSR could therefore occur regardless of the relatively low level of nuclear AID.
The mechanism of increasing nuclear AID need not be B cell specific, since we found YY1 can control nuclear AID levels in B cells, as well as in HEK 293T cells. A number of non-mutually exclusive mechanisms could explain the function of YY1 in controlling AID levels in the nucleus. First, YY1 might form a complex with AID to control translocation of AID through the nuclear pore. Several studies demonstrated that YY1 subcellular localization is regulated during cell cycle progression and development (17, 18, 41, 53, 72). In addition, apoptotic stimuli promote rapid translocation of YY1 from the cytoplasm to the nucleus in asynchronous HeLa cells (29). Thus, YY1 might function to increase transport of AID from the cytoplasm to the nucleus via the nuclear pore.
Second, YY1 might sequester AID in the nucleus, preventing AID nuclear export. AID C-terminal amino acids 188 to 198 contain a well-characterized nuclear export sequence recognized by the CRM-1 receptor that actively exports AID from the nucleus (34). YY1 is a component of multiple protein complexes, and our data suggest that AID and YY1 are part of a protein complex (Fig. 5B). Thus, YY1 could sequester AID in a complex in the nucleus, reducing AID nuclear export and leading to increased nuclear accumulation, whereas loss of YY1 would enable AID nuclear export.
Finally, AID levels in the nucleus are also regulated by compartment-specific polyubiquitination and degradation (1). Notably, there is an 8-fold difference in nuclear and cytoplasmic AID stability, with the AID half-life in the nucleus being about 2.5 h compared to 20 h in the cytoplasm. Recently, it was found that hsp90 can control the cytoplasmic stability of AID, thus regulating the amount of AID available for nuclear import (40). In the same way, we found that YY1 stabilizes AID in the nucleus. This represents an effective way of increasing AID nuclear accumulation, as AID is much less stable in the nucleus than in the cytoplasm (1). The mechanism of this stabilization is not yet clear, but YY1 might interact with AID and inhibit nuclear AID polyubiquitination, leading to AID stabilization. Loss of YY1 would allow increased AID polyubiquitination and degradation. We favor this hypothesis, because our preliminary results suggest that YY1 reduces AID polyubiquitination. Thus, we favor the hypothesis that YY1 increases nuclear AID levels by inhibiting AID polyubiquitination and subsequent degradation. Further experiments are needed to better define this mechanism. Control of nuclear levels of AID is crucial, not only for regulating antibody maturation processes (CSR and SHM), but also for maintaining the integrity of the mammalian genome. Elevated levels of YY1 could cause aberrant accumulation of AID in germinal center B cells, leading to increased mutagenesis and lymphomagenesis. Indeed, YY1 levels are elevated in germinal center-derived human diffuse large B cell lymphomas (8), suggesting that YY1 contributes to disease progression.
It is also tempting to hypothesize that YY1 plays a role in targeting AID to the Ig loci. YY1 binding to the Ig heavy chain 3′ hypersensitive site 3 (hs3), as well as to the Eμ enhancer, is inducible by LPS (21). In this case, YY1 appears to be sequestered from DNA in resting B lymphocytes through interaction with hypophosphorylated retinoblastoma protein (Rb). However, after LPS induction, Rb becomes hyperphosphorylated and releases YY1, enabling it to bind to the hs3 and Eμ enhancers. Interestingly, hs3b and -4 hypersensitive sites are crucial for formation of Eμ: 3′Eα complexes with germ line switch region promoters after cytokine treatment (70). In AID-deficient mice, this S-S synaptosome complex (Eμ: 3′-Eα complex with the associated switch region promoter) is not formed, suggesting that AID acts as a scaffold to hold the DNA sequences together. It would be very interesting to test whether YY1 is part of this synaptosome complex. We hypothesize that LPS induction of CSR might result from AID recruitment to the synaptosome complex through induction of YY1 binding to the hs3 and Eμ enhancers. This interaction with YY1 could lead to AID stabilization at the Ig loci to create a locally increased concentration of AID molecules, leading to efficient CSR. Further experiments will be needed to test this hypothesis.
In summary, for the first time, we have identified YY1 as a novel regulator of Ig CSR. The mechanism of this regulation involves the ability of YY1 to control AID stability and nuclear AID levels. As the levels of nuclear AID correlate with the levels of DNA mutagenesis and lymphomagenesis, YY1 may provide a new target for therapies against lymphomas of germinal center B cell origin, such as diffuse large B cell lymphoma.
Supplementary Material
ACKNOWLEDGMENTS
We thank Hua Ding from the Protein Core at Children's Hospital of Philadelphia for purification of TAT-CRE. We thank Fred Alt for the AID cDNA, T. Honjo for CH12F3 cells, Matthew Thomas and Dave Allman for advice on CSR assays, and Bruce Freedman and Craig Bassing for helpful discussions and advice.
This work was supported by NIH grants RO1 AI079002 and GM082841.
Footnotes
Published ahead of print 30 January 2012
Supplemental material for this article may be found at http://mcb.asm.org.
REFERENCES
- 1. Aoufouchi S, et al. 2008. Proteasomal degradation restricts the nuclear lifespan of AID. J. Exp. Med. 205:1357–1368 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Atchison L, Ghias A, Wilkinson F, Bonini N, Atchison ML. 2003. The YY1 transcription factor functions as a PcG protein in vivo. EMBO J. 22:1347–1358 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Ballantyne J, Henry DL, Marcu KB. 1997. Antibody class switch recombinase activity is B cell stage specific and functions stochastically in the absence of ‘targeted accessibility’ control. Int. Immunol. 9:963–974 [DOI] [PubMed] [Google Scholar]
- 4. Basu U, et al. 2011. The RNA exosome targets the AID cytidine deaminase to both strands of transcribed duplex DNA substrates. Cell 144:353–363 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Brar SS, Watson M, Diaz M. 2004. Activation-induced cytosine deaminase (AID) is actively exported out of the nucleus but retained by the induction of DNA breaks. J. Biol. Chem. 279:26395–26401 [DOI] [PubMed] [Google Scholar]
- 6. Bushmeyer S, Park K, Atchison ML. 1995. Characterization of functional domains within the multifunctional transcription factor, YY1. J. Biol. Chem. 270:30213–30220 [DOI] [PubMed] [Google Scholar]
- 7. Bushmeyer SM, Atchison ML. 1998. Identification of YY1 sequences necessary for association with the nuclear matrix and for transcriptional repression functions. J. Cell Biochem. 68:484–499 [PubMed] [Google Scholar]
- 8. Castellano G, et al. 2010. Yin Yang 1 overexpression in diffuse large B-cell lymphoma is associated with B-cell transformation and tumor progression. Cell Cycle 9:557–563 [DOI] [PubMed] [Google Scholar]
- 9. Chu CC, Paul WE, Max EE. 1992. Quantitation of immunoglobulin mu to gamma 1 heavy chain switch region recombination by a digestion-circularization polymerase chain reaction method. Proc. Natl. Acad. Sci. U. S. A. 89:6978–6982 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Cobb RM, Oestreich KJ, Osipovich OA, Oltz EM. 2006. Accessibility control of V(D)J. recombination. Adv. Immunol. 91:45–109 [DOI] [PubMed] [Google Scholar]
- 11. Conticello SG, et al. 2008. Interaction between antibody-diversification enzyme AID and spliceosome-associated factor CTNNBL1. Mol. Cell 31:474–484 [DOI] [PubMed] [Google Scholar]
- 12. Demorest Z, et al. 2010. The interaction between AID and CIB1 is nonessential for antibody gene diversification by gene conversion or class switch recombination. PLoS One 5:e11660. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. de Yebenes VG, et al. 2008. miR-181b negatively regulates activation-induced cytidine deaminase in B cells. J. Exp. Med. 205:2199–2206 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Donohoe ME, Zhang L-F, Xu N, Shi Y, Lee JT. 2007. Identification of a Ctcf cofactor, YY1, for the X chromosome binary switch. Mol. Cell 25:43–56 [DOI] [PubMed] [Google Scholar]
- 15. Dorsett Y, et al. 2008. MicroRNA-155 suppresses activation-induced cytidine deaminase-mediated Myc-Igh translocation. Immunity 28:630–638 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Ellyard JI, Benk AS, Taylor B, Rada C, Neuberger MS. 2011. The dependence of Ig class-switching on the nuclear export sequence of AID likely reflects interaction with factors additional to Crm1 exportin. Eur. J. Immunol. 41:485–490 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Ficzycz A, et al. 2001. Expression, activity, and subcellular localization of the Yin Yang1 transcription factor in Xenopus oocytes and embryos. J. Biol. Chem. 276:22819–22825 [DOI] [PubMed] [Google Scholar]
- 18. Ficzycz A, Ovsenek N. 2002. The Yin Yang 1 transcription factor associates with ribonucleoprotein (mRNP) complexes in the cytoplasm of Xenopus oocytes. J. Biol. Chem. 277:8382–8387 [DOI] [PubMed] [Google Scholar]
- 19. Fugmann SD, Lee AI, Shockett PE, Villey IJ, Schatz DG. 2000. The RAG proteins and V(D)J. recombination: complexes, ends, and transposition. Annu. Rev. Immunol. 18:495–527 [DOI] [PubMed] [Google Scholar]
- 20. Gordon S, Akopyan G, Garban H, Bonavida B. 2006. Transcription factor YY1: structure, function, and therapeutic implications in cancer biology. Oncogene 25:1125–1142 [DOI] [PubMed] [Google Scholar]
- 21. Gordon SJ, Saleque S, Birshtein BK. 2003. Yin Yang 1 is a lipopolysaccharide-inducible activator of the murine 3′ Igh enhancer, hs3. J. Immunol. 170:5549–5557 [DOI] [PubMed] [Google Scholar]
- 22. Green MR, et al. 2011. Signatures of murine B-cell development implicate Yy1 as a regulator of the germinal center-specific program. Proc. Natl. Acad. Sci. U. S. A. 108:2873–2878 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Han L, Masani S, Yu K. 2010. CTNNBL1 is dispensible for Ig class switch recombination. J. Immunol. 185:1379–1381 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Hasler J, Rada C, Neuberger MS. 2011. Cytoplasmic activation-induced cytidine deaminase (AID) exists in stoichiometric complex with translation elongation factor 1a (eEF1A). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Ito S, et al. 2004. Activation-induced cytidine deaminase shuttles between nucleus and cytoplasm like apolipoprotein B mRNA editing catalytic polypeptide 1. Proc. Natl. Acad. Sci. U. S. A. 101:1975–1980 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Jeevan-Raj BP, et al. 2011. Epigenetic tethering of AID to the donor switch region during immunoglobulin class switch recombination. J. Exp. Med. 208:1649–1660 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Jung D, Giallourakis C, Mostoslavsky R, Alt FW. 2006. Mechanism and control of V(D)J recombination at the immunoglobulin heavy chain locus. Annu. Rev. Immunol. 24:541–570 [DOI] [PubMed] [Google Scholar]
- 28. Kim JD, Hinz AK, Choo JH, Stubbs L, Kim J. 2007. YY1 as a controlling factor for the Peg3 and Gnas imprinted domains. Genomics 89:262–269 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Krippner-Heidenreich A, et al. 2005. Caspase-dependent regulation and subcellular redistribution of the transcriptional modulator YY1 during apoptosis. Mol. Cell. Biol. 25:3704–3714 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Liu H, et al. 2007. Yin Yang 1 is a critical regulator of B-cell development. Genes Dev. 21:1179–1189 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Liu M, Schatz DG. 2009. Balancing AID and DNA repair during somatic hypermutation. Trends Immunol. 30:173–181 [DOI] [PubMed] [Google Scholar]
- 32. Maitra S, Atchison M. 2000. BSAP can repress enhancer activity by targeting PU.1 function. Mol. Cell Biol. 20:1911–1922 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Martin A, Scharff MD. 2002. AID and mismatch repair in antibody diversification. Nat. Rev. Immunol. 2:605–614 [DOI] [PubMed] [Google Scholar]
- 34. McBride KM, Barreto V, Ramiro AR, Stavropoulos P, Nussenzweig MC. 2004. Somatic hypermutation is limited by CRM1-dependent nuclear export of activation-induced deaminase. J. Exp. Med. 199:1235–1244 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Muramatsu M, et al. 2000. Class switch recombination and hypermutation require activation-induced cytidine deaminase (AID), a potential RNA editing enzyme. Cell 102:553–563 [DOI] [PubMed] [Google Scholar]
- 36. Muramatsu M, et al. 1999. Specific expression of activation-induced cytidine deaminase (AID), a novel member of the RNA-editing deaminase family in germinal center B cells. J. Biol. Chem. 274:18470–18476 [DOI] [PubMed] [Google Scholar]
- 37. Nagulapalli S, Pongubala JMR, Atchison ML. 1995. Multiple proteins physically interact with PU.1. J. Immunol. 155:4330–4338 [PubMed] [Google Scholar]
- 38. Nowak U, Matthews AJ, Zheng S, Chaudhuri J. 2011. The splicing regulator PTBP2 interacts with the cytidine deaminase AID and promotes binding of AID to switch-region DNA. Nat. Immunol. 12:160–166 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Okazaki I-M, Kotani A, Honjo T. 2007. Role of AID in tumorigensis. Adv. Immunol. 94:245–273 [DOI] [PubMed] [Google Scholar]
- 40. Orthwein A, et al. 2010. Regulation of activation-induced deaminase stability and antibody gene diversification by Hsp90. J. Exp. Med. 207:2751–2765 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Palko L, Bass HW, Beyrouthy MJ, Hurt MM. 2004. The Yin Yang-1 (YY1) protein undergoes a DNA-replication-associated switch in localization from the cytoplasm to the nucleus at the onset of S phase. J. Cell Sci. 117:465–476 [DOI] [PubMed] [Google Scholar]
- 42. Park K, Atchison ML. 1991. Isolation of a candidate repressor/activator, NF-E1 (YY-1, δ), that binds to the immunoglobulin κ 3′ enhancer and the immunoglobulin heavy-chain μE1 site. Proc. Natl. Acad. Sci. U. S. A. 88:9804–9808 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Pasqualucci L, et al. 2008. AID is required for germinal center-derived lymphomagenesis. Nat. Gen. 40:108–112 [DOI] [PubMed] [Google Scholar]
- 44. Pavri R, et al. 2010. Activation-induced cytidine deaminase targets DNA at sites of RNA polymerase II stalling by interaction with Spt5. Cell 143:122–133 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Perkel JM, Atchison ML. 1998. A two-step mechanism for recruitment of Pip by PU.1. J. Immunol. 160:241–252 [PubMed] [Google Scholar]
- 46. Pongubala JMR, Atchison ML. 1991. Functional characterization of the developmentally controlled immunoglobulin kappa 3′ enhancer: regulation by Id, a repressor of helix-loop-helix transcription factors. Mol. Cell. Biol. 11:1040–1047 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Pongubala JMR, Atchison ML. 1997. PU.1 can participate in an active enhancer complex without its transcriptional activation domain. Proc. Natl. Acad. Sci. U. S. A. 94:127–132 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Pongubala JMR, et al. 1992. PU.1 recruits a second nuclear factor to a site important for immunoglobulin k 3′ enhancer activity. Mol. Cell. Biol. 12:368–378 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Pongubala JMR, et al. 1993. Effect of PU.1 phosphorylation on interaction with NF-EM5 and transcriptional activation. Science 259:1622–1625 [DOI] [PubMed] [Google Scholar]
- 50. Rada C, Jarvis JM, Milstein C. 2002. AID-GFP chimeric protein increases hypermutation of Ig genes with no evidence of nuclear localization. Proc. Natl. Acad. Sci. U. S. A. 99:7003–7008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Ranjit S, et al. 2011. AID binds cooperatively with UNG and Msh2-Msh6 to Ig switch regions dependent upon the AID C terminus. J. Immunol. 187:2464–2475 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Revy P, et al. 2000. Activation-induced cytidine deaminase (AID) deficiency causes the autosomal recessive form of the hyper-IgM syndrome (HIGM2). Cell 102:565–575 [DOI] [PubMed] [Google Scholar]
- 53. Rizkallah R, Hurt MM. 2009. Regulation of the transcription factor YY1 in mitosis through phosphorylation of its DNA-binding domain. Mol. Cell. Biol. 20:4766–4776 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Rush JS, Liu M, Odegard VH, Unniraman S, Schatz DG. 2005. Expression of activation-induced cytidine deaminase is regulated by cell division, providing a mechanistic basis for division-linked class switch recombination. Proc. Natl. Acad. Sci. U. S. A. 102:13242–13247 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Schreiber E, Matthias P, Muller MM, Schaffner W. 1989. Rapid detection of octamer binding proteins with ‘mini-extracts’, prepared from a small number of cells. Nucleic Acids Res. 17:6419. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Sernandez IV, de Yebenes VG, Dorsett Y, Ramiro AR. 2008. Haploinsufficiency of activation-induced deaminase for antibody diversification and chromosome translocations both in vitro and in vivo. PLoS One 3:e3927. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57. Stavnezer J. 2011. Complex regulation and function of activation-induced cytidine deaminase. Trends Immunol. 32:194–201 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58. Storck S, Aoufouchi S, Weill J-C, Reynaud C-A. 2011. AID and partners: for better and (not) for worse. Curr. Opin. Immunol. 23:337–344 [DOI] [PubMed] [Google Scholar]
- 59. Sucharov CC, et al. 2004. The Ku protein complex interacts with YY1, is up-regulated in human heart failure, and represses alpha myosin heavy-chain gene expression. Mol. Cell. Biol. 24:8705–8715 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60. Sucharov CC, Mariner P, Long C, Bristow M, Leinwand L. 2003. Yin Yang 1 is increased in human heart failure and represses the activity of the human alpha-myosin heavy chain promoter. J. Biol. Chem. 278:31233–31239 [DOI] [PubMed] [Google Scholar]
- 61. Sui G, et al. 2004. Yin Yang 1 is a negative regulator of p53. Cell 117:859–872 [DOI] [PubMed] [Google Scholar]
- 62. Ta V-T, et al. 2003. AID mutant analyses indicate requirement for class-switch-specific cofactors. Nat. Immunol. 4:843–848 [DOI] [PubMed] [Google Scholar]
- 63. Takizawa M, et al. 2008. AID expression levels determine the extent of cMyc oncogenic tranlocations and the incidence of B cell tumor development. J. Exp. Med. 205:1949–1957 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64. Teng G, et al. 2008. MicroRNA-155 is a negative regulator of activation-induced cytidine deaminase. Immunity 28:621–629 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65. Thomas MJ, Seto E. 1999. Unlocking the mechanisms of transcription factor YY1: are chromatin modifying enzymes the key? Gene 236:197–208 [DOI] [PubMed] [Google Scholar]
- 66. Walowitz JL, Bradley ME, Chen S, Lee T. 1998. Proteolytic regulation of the zinc finger transcription factor YY1, a repressor of muscle-restricted gene expression. J. Biol. Chem. 273:6656–6661 [DOI] [PubMed] [Google Scholar]
- 67. Wei F, Zaprazna K, Wang J, Atchison ML. 2009. PU.1 can recruit BCL6 to DNA to repress gene expression in germinal center B cells. Mol. Cell. Biol. 29:4612–4622 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68. Wilkinson FH, Park K, Atchison ML. 2006. Polycomb recruitment to DNA in vivo by the YY1 REPO domain. Proc. Natl. Acad. Sci. U. S. A. 103:19296–19301 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69. Wu S, et al. 2007. A YY1-INO80 complex regulates genomic stability through homologous recombination-based repair. Nat. Struct. Biol. 14:1665–1672 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70. Wuerffel R, et al. 2007. S-S synapsis during class switch recombination is promoted by distantly located transcriptional elements and activation-induced deaminase. Immunity 27:711–722 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71. Xu Z, et al. 2010. 14-3-3 adaptor proteins recruit AID to 5′-AGCT-3′-rich switch regions for class switch recombination. Nat. Struct. Mol. Biol. 17:1124–1135 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72. Yue R, et al. 2009. Beta-arrestin1 regulates zebrafish hematopoiesis through binding to YY1 and relieving polycomb group repression. Cell 139:535–546 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.