Background: The Get4/Get5 protein complex is a homodimer mediated by the Get5 carboxyl domain.
Results: The Get5 homodimerization motif forms a structurally conserved helical domain allowing Get4/Get5 to adopt an extended solution conformation.
Conclusion: Get5 homodimerization is mediated by a 35-residue sequence stabilized by a few conserved hydrophobic interactions.
Significance: The Get5 carboxyl domain contains a novel example of a stable dimerization motif.
Keywords: Membrane Proteins, Nuclear Magnetic Resonance, Protein Structure, Protein Targeting, X-ray Crystallography, Get Pathway, Tail Anchor, Ubl4a
Abstract
Tail-anchored trans-membrane proteins are targeted to membranes post-translationally. The proteins Get4 and Get5 form an obligate complex that catalyzes the transfer of tail-anchored proteins destined to the endoplasmic reticulum from Sgt2 to the cytosolic targeting factor Get3. Get5 forms a homodimer mediated by its carboxyl domain. We show here that a conserved motif exists within the carboxyl domain. A high resolution crystal structure and solution NMR structures of this motif reveal a novel and stable helical dimerization domain. We additionally determined a solution NMR structure of a divergent fungal homolog, and comparison of these structures allows annotation of specific stabilizing interactions. Using solution x-ray scattering and the structures of all folded domains, we present a model of the full-length Get4/Get5 complex.
Introduction
The targeted delivery of trans-membrane proteins to the proper membrane is a critical cellular process. The signal recognition particle pathway delivers the majority of trans-membrane proteins co-translationally in all organisms. Tail-anchored (TA)2 proteins are important exceptions to this pathway (1). TA proteins contain a single trans-membrane helix within 30 residues of the carboxyl terminus, and this feature necessitates post-translational targeting. After insertion, the N terminus remains in the cytoplasm. TA proteins are found in all membranes exposed to the cytoplasm and have a wide variety of roles, such as vesicle fusion, regulating apoptosis, and protein translocation (2, 3).
Eukaryotic pathways for TA protein delivery to the ER have been elucidated and are best described for yeast (for review, see Refs. 4, 5). The majority of TA proteins are targeted via the GET (guided entry of TA proteins) pathway. Targeting progresses from the Get4/Get5/Sgt2 sorting complex that loads ER destined TA protein onto the ATPase Get3 (6–8). Mitochondrial TA proteins appear to be initially retained on Sgt2-bound heat-shock cognate protein chaperones. Get3 then targets the TA protein to the ER membrane via Get1/Get2 (9).
Get4 and Get5 form an obligate heterodimer mediated by the amino domain of Get5 (Get5-N) and the carboxyl domain of Get4 (7, 10). Get4 is an α-helical repeat protein that binds Get3 through a conserved basic face. Following the Get5 amino domain in sequence is a ubiquitin-like domain (Get5-Ubl) that mediates interaction with Sgt2 (10, 11). The carboxyl domain of Get5 (Get5-C) is a homodimerization domain, resulting in a heterotetrameric Get4/Get5 complex (7).
A similar pathway for TA targeting exists in mammals. TRC35 and Ubl4A, homologs of Get4 and Get5, respectively, form a stable complex with the protein Bag-6/Bat-3/Scythe (12). This Bag-6 complex is required for efficient TA protein targeting by transferring them to TRC40, the Get3 homolog, after synthesis is complete (12, 13). It is also involved in the degradation of defective nascent polypeptides and the stabilization of hydrophobic segments of proteins retrotranslocated from the ER prior to degradation by the proteasome (14, 15). The human homolog of Sgt2, SGTA, may also interact with this complex, suggesting a shared mechanism of TA sorting with yeast (16, 17).
Structural studies of GET pathway members continue to provide details on the molecular series of events that occur in TA targeting. To understand the basis for specificity of homodimerization by Get5, we determined both a crystal and solution structure of the carboxyl domain of Get5 along with the solution structure of a fungal homolog. These structures reveal the nature of the conserved dimerization motif. We characterize the oligomeric state of human Ubl4A, which is amendable to the alternate architecture of the mammalian complex. Moreover, we use solution small angle x-ray scattering (SAXS) to define the overall structure of the full Get4/Get5 heterotetramer, providing the first molecular framework of this complex.
EXPERIMENTAL PROCEDURES
Cloning, Expression, and Purification
Get5-C residues 152–212 or 175–212 from Saccharomyces cerevisiae S288C or 159–230 from Aspergillus fumigatus 118 were amplified from vectors described previously (7). The coding sequences were then inserted into a pET33b-derived plasmid. Unlabeled proteins were expressed in Escherichia coli BL21(DE3)Star (Invitrogen) for 3 h at 37 °C after induction with 250 μm isopropyl-β-d-thiogalactopyranoside (Affymetrix). Uniformly 15N-labeled proteins were produced using N-5052 autoinduction media (18), and uniformly 13C/15N-labeled proteins were produced using the method of Marley et al. (19). Cells were lysed using an S-4000 sonicator (Misonix) and purified by immobilized metal affinity chromatography (Qiagen). The full-length Get4/Get5 complex, Get5-Ubl-C (residues 74–212), and AfGet5-Ubl-C (residues 66–230) were prepared as described previously (7). (For clarity, residues that are italicized will refer specifically to the A. fumigatus homolog.)
Human Ubl4A was amplified from a cDNA-containing plasmid (ATCC) and inserted into a pET33b-derived plasmid. Expression and purification were as for Get5-C. Domain swap experiments and pulldown assays were performed as described previously (7). Briefly, Get5-Ubl-C or AfGet5-Ubl-C was incubated in 2-fold stoichiometric excess with polyhistidine-tagged Ubl4A in 50 mm HEPES, 100 mm NaCl and 30 mm imidazole, pH 7.3. Ubl4A was precipitated with nickel-nitrilotriacetic acid-agarose beads (Qiagen). The beads were washed twice with incubation buffer and proteins eluted with 20 mm EDTA.
Crystallization, Data Collection, and Structure Determination
Get5-C residues 175–212 were concentrated to 30 mg/ml in 20 mm sodium phosphate, pH 6.1. Crystallization screening was performed using the sitting-drop vapor diffusion method with commercially available screens (Qiagen) and a Mosquito robot (TTP Labtech). Clusters of plate-like crystals grew after 1 week in a 1:1 ratio drop of protein solution to a reservoir of 3.4 m ammonium sulfate and 0.1 m sodium citrate, pH 5.0, at 22 °C. The clusters were broken apart, and individual crystals were transferred to reservoir solution supplemented with 10% glycerol for 5 min and then cryopreserved in liquid nitrogen. Iodide derivatives were generated by soaking crystals in freshly prepared 2.9 m ammonium sulfate, 0.5 m ammonium iodide, 0.1 m sodium citrate, and 10% glycerol, pH 5.0, for 5 min prior to cryopreservation.
X-ray diffraction data from a single native crystal were collected on beam line 12-2 at the Stanford Synchrotron Radiation Lightsource (SSRL) at 100 K using a Pilatus 6M detector and a microbeam. Positions on the crystal were screened for diffraction using an automated raster scanning protocol. The best position allowed for collection of a near complete dataset to maximum 1.23 Å resolution. Diffraction data from an iodide derivative were collected using a Micromax-007 HF rotating copper-anode generator and an R-axis IV++ image plate detector (Rigaku) to a maximum 1.6 Å resolution and ∼12-fold redundancy. Data were integrated, scaled, and merged using XDS (20), iodide substructure determination, phasing, and initial model building and refinement were performed in PHENIX (21), and manual building and model refinement were performed using COOT (22). Structure figures were prepared using PyMOL (Schrödinger).
NMR Spectroscopy
All NMR measurements were collected using a Varian INOVA 600 MHz spectrometer at 25 °C with a triple resonance probe. Uniformly 13C/15N-labeled Get5-C residues 152–212 or AfGet5-C residues 159–230 were concentrated to a monomer concentration of 3.5 mm in 20 mm sodium phosphate, pH 6.1. Chemical shift assignments were determined using standard triple-resonance experiments (23). Data were processed using either TopSpin (Bruker) or NMRPipe (24) and analyzed using CCPN (25). The PINE web server aided initial assignments (26). Distance restraints were derived from 13C- and 15N-edited NOESY spectra. An asymmetrically labeled dimer of AfGet5-C was prepared by mixing 4 mm unlabeled protein with 2 mm uniformly 13C/15N-labeled protein and incubating at room temperature for 3 days. Intermolecular distance restraints were then determined using a 13C/15N-filtered 13C-edited NOESY spectrum.
HN-N residual dipolar couplings were measured using the IPAP-HSQC experiment. A 2.5 mm solution of 15N-labeled Get5-C was aligned in a 4% strained polyacrylamide gel, and a 2 mm solution of 15N-labeled AfGet5-C was aligned in a 5% strained polyacrylamide gel (27).
Solution Structure Determination
ARIA2.3 was used for automated NOE cross-peak assignment and structure calculation with hydrogen bond, dihedral and residual dipolar coupling-derived restraints (28). Every NOE cross-peak was treated ambiguously as an inter- or intramolecular contact, with initial sets of unambiguous intermolecular contacts determined by constraints imposed by secondary structure (29). For AfGet5-C, experimentally determined intermolecular restraints were also utilized. Restraints for ϕ/ψ dihedral angles were predicted from chemical shifts using TALOS+ (30). An energy term is included to maintain symmetry within each model. Hydrogen bond restraints were initially assigned to the amide protons most resistant to deuterium exchange. After structure calculation, additional hydrogen bonds that were supported by the ensemble of models were added within the helical segments and the calculations repeated. The initial structures were also used to determine the axial and rhombic components of the alignment tensors with the program REDCAT (31). Residual dipolar couplings were added as restraints during subsequent calculations. A log-harmonic energy potential was used during the second Cartesian cooling phase of the simulated anneal protocol with automatic determination of weights for NOE-derived and hydrogen bond distance restraints (32). 100 models were generated in the final iteration of ARIA, and the 10 lowest energy models were selected for refinement in explicit water.
SAXS
Multiple concentrations of S. cerevisiae Get4/Get5 were prepared as described previously (11), with dialysis against 50 mm Tris, 300 mm NaCl, and 5 mm 2-mercaptoethanol, pH 8.0. Data were collected at SSRL beam line 4-2 using a Rayonix MX225-HE detector, 1.13-Å wavelength x-rays, and a detector distance of 2.5 m for a momentum transfer range of 0.0055–0.3709 Å−1. Data were processed using MARPARSE (33), and Guiner analysis was performed with PRIMUS (34). The distance distribution function was determined with GNOM (35). Ten independent ab initio models were generated using DAMMIF (36) and real space data. These were superposed, averaged, and filtered using DAMAVER (37). High resolution structures of Get4 and Get5 components were used as input into the program CORAL (38) for rigid body fitting with reciprocal space data. All calculations used imposed 2-fold symmetry.
Circular Dichroism Spectroscopy
Thermal denaturation measurements were collected using an Aviv 62A DS circular dichroism spectrometer. Get5-C or AfGet5-C at 10 μm in 20 mm sodium phosphate, pH 7.5, was heated from 25 °C to 99 °C in 1 °C increments, and ellipticity was measured at 227 nm or 221 nm, respectively. Fractions of unfolded and folded protein were approximated using plateau values at low and high temperature.
RESULTS
Conservation of the Get5 Carboxyl Domain
Get5 contains an amino domain, a ubiquitin-like domain, and a carboxyl domain that mediates homodimerization (Fig. 1A) (7). Get5-C, 152–212, contains the entire sequence from the end of the Get5-Ubl domain to the carboxyl terminus. Residues 152–176 have poor overall conservation in sequence identity or length. Residues 177–212 form an ∼35-residue conserved motif that is found in Get5 homologs from two of the three subphyla of Ascomycota, the largest described fungal phylum (supplemental Fig. S1) (39). The motif has greater variability within Saccharomycotina than in Pezizomycotina. Taphrinomycotina, the third subphylum that includes Schizosaccharomyces pombe, does not appear to have homologs of Get5 that contain a carboxyl-terminal domain.
FIGURE 1.
Crystal structure of Get5-C. A, schematic of the domain organization of Get5 and sequence alignment of the carboxyl domain in fungi. Lines indicate flexible regions. Sequences are: S. cere, S. cerevisiae; A. goss, Ashbya gossypii; S. scle, Sclerotina sclerotiorum; A. fumi, A. fumigatus. Residues that mediate intermolecular contacts in both Get5 and AfGet5 dimers are highlighted in red, and residues specific to Get5 or AfGet5 are highlighted in blue or green, respectively. B, dimerization interface of a monomer of Get5-C with σa-weighted 2|Fo| −|Fc| electron density contoured at 1.5σ. C, asymmetric unit of Get5-C. One monomer is color ramped from amino (blue) to carboxyl (red) terminus. The 2-fold axis is indicated with a dotted line. Side chains that make intermolecular contacts are shown (left).
Ubl4A lacks the amino Get4 binding domain of Get5, with the Ubl domain as the amino terminus (7). Ubl4A is well conserved in vertebrates. The carboxyl domain of Ubl4A contains the 35-residue dimerization motif seen in Get5 (supplemental Fig. S1). The linker between the dimerization motif and the Ubl domain is generally shorter than in fungal homologs, and Ubl4A has an additional 30 conserved residues following the motif.
Structure of Get5 Dimerization Motif
We cloned, expressed, and purified Get5-C for NMR investigation. The main chain chemical shifts (1HN, 15NN, 13CCO, 13Cα, 13Cβ, and 1Hα) of residues 152–175 have random coil character, whereas those of 179–190 and 194–210 are characteristic of helices (supplemental Fig. S2A). These helices are designated H1 and H2, respectively. Relaxation rates and heteronuclear NOE values also indicate rapid motions for residues 152–175. Consistent with this secondary structure, 1HN of residues 182–187 and 200–208 were the most protected from solvent deuterium exchange whereas residues 152–177 readily exchanged. Therefore, we concluded that residues 152–175 were unstructured and not significantly contributing to the stability of the folded dimerization domain.
Based on these results a truncated version of the carboxyl domain consisting of residues 175–212 was generated for crystallographic studies. This variant crystallized in space group P21 and diffracted to 1.23 Å resolution. Experimental phases were determined by the single-wavelength anomalous diffraction technique using an iodide-soaked crystal (40). Two copies of Get5-C are present in the asymmetric unit of the native crystal. Unambiguous electron density allowed modeling of residues 175–212 of one copy and 175–211 of the second (Fig. 1B). The structure was refined to an Rfree of 0.204. Crystallographic statistics are presented in supplemental Table 1.
The two copies of Get5 in the asymmetric unit interact extensively, burying 870 Å2 of solvent-accessible surface area (∼25% of total) per monomer and sequester the majority of hydrophobic residues (Fig. 1C). In contrast, the most extensive crystallographic contact buries 410 Å2; therefore, the asymmetric unit contains the physiologically relevant dimer.
A 2-fold axis relates one monomer onto the other (Fig. 1C). The H1 helices pack antiparallel to one another. A four-residue linker connects to H2, and the H2 helices cross at conserved Gly-207 forming an ∼95° angle. The association between the two subunits in the dimer is almost entirely by hydrophobic interactions. The dimer axis is lined by Ile-182, Leu-185, Leu-204, and Gly-207. Leu-185 and Pro-178 extend from H1 of one copy into hydrophobic pockets formed by additional H1 residues the second copy. Asp-181 and Asn-189 form the only intermolecular hydrogen bond across the dimer.
There are several interactions that restrict the conformation of H1 relative to H2 within each monomer. Phe-190, which begins the connecting loop, fits in a pocket formed by Leu-186, Ala-196, and Val-200 of the same copy (Fig. 1C). The indole ring of Trp-179 is normal to the ring of Trp-208 of the same copy, and the side chain of Val-177 fits into the resulting pocket. Trp-208 points toward the dimer interface, where the side chain of Arg-203 extends from the opposite copy to form a cation-pi interaction. Arg-203 is conserved as an arginine or lysine in Saccharomycotina, and Trp-208 is conserved across all eukaryotes (supplemental Fig. S1). The side chain of Trp-208 also contacts Val-200 and Leu-204 of the opposite copy.
Solution NMR Structure of Get5-C
We determined the solution structure of Get5-C using NOE-derived and hydrogen bond distance restraints, ϕ and ψ dihedral angle restraints, and residual dipolar couplings restraints. Residues 152–172 did not converge to a consistent structure due to a lack of interresidue NOE-derived distance restraints; therefore, they were omitted in the final model calculations. Statistics for the structure and restraints are summarized in supplemental Table 2, and the ensembles of the 10 lowest energy structures are shown in Fig. 2A.
FIGURE 2.
Solution structures of Get5-C and AfGet5-C. A and B, ensembles of the 10 lowest energy solution structures of the ordered regions of Get5-C (A) and AfGet5-C (B). C, ribbon diagram of AfGet5-C with side chains making intermolecular contacts. AfGet5-C is rotated from the orientation in the top of B as indicated by the sphere in the corner. Inset, view of the environment around Val-192.
The solution structure is very similar to the crystal structure, with an average backbone root mean square deviation of 0.94 ± 0.06 Å across residues 177–212 (supplemental Fig. 3A). The interaction between Arg-203 and Trp-208 and the interactions between the methyl groups of Val-177 with Trp-179 and Trp-208 are maintained in solution (supplemental Fig. 3B). The unique environment created by the two aromatic residues is reflected in unusual upfield chemical shifts of interacting residues, with Val-177 methyl protons at −0.994 ppm and −0.613 ppm, Arg-205 1Hδ at 1.573 ppm and Gln-205 1Hγ, and a side chain amide proton at −0.056 ppm and 4.656 ppm, respectively.
Two significant differences exist between the solution and crystal structures, and both can be explained by the crystal lattice. First, the loop connecting H1 and H2 is rearranged (supplemental Fig. S3C). For the x-ray structure, crystals grew in pH 5.0, and this pH allows Glu-191 and Asp-193 to form hydrogen bonds with Asp-181 and Glu-202, respectively, from two different crystallographic copies. This results in an alternate conformation of the loop, predominantly by a rotation of the peptide bond plane between Glu-191 and Gln-192. The second difference is at the N terminus of the crystal structure, which has a three-residue cloning artifact prior to residue 175 of the crystallization construct. Leu-175 and nonnative Val-173 form hydrophobic contacts with two different crystallographic copies (supplemental Fig. S3D), resulting in the terminus turning outward from the helices. In solution, residues 173–176 are extended (supplemental Fig. S3D). This full domain results in 1090 Å2 of solvent-accessible surface area buried per monomer (∼30% of total).
Structure of Get5-C Domain Homolog
We additionally investigated the carboxyl domain of the Get5 homolog from the filamentous fungi A. fumigatus (AfGet5-C). Several structures of other GET pathway members have been determined for this organism (11, 41). A. fumigatus is in the subphylum Pezizomycotina, and Get5 homologs from this group have conserved features distinct from Saccharomycotina (supplemental Fig. 1). There is more conservation in the residues immediately preceding the tryptophan at the amino terminus of H1, and there are three additional conserved phenylalanines. We cloned, expressed, and purified AfGet5-C residues 159–230, comprising the full sequence of the carboxyl domain for NMR investigation.
Main chain chemical shifts indicated that residues 159–186 are likely random coil whereas 212–228 and 195–208 formed helices corresponding to H1 and H2 (supplemental Fig. 2B). An additional short coil is predicted at residues Ser-190 and Val-191, designated H0. We determined the solution structure of AfGet5-C. Residues 159–185 did not converge to a consistent structure due to a lack of interresidue NOE-derived distance restraints and were omitted from the final structure calculation. Structure and restraint statistics are provided in supplemental Table 2, and the ensembles of the 10 lowest energy structures are shown in Fig. 2B.
The overall structure of AfGet5-C is similar to the Get5-C (Fig. 2, B and C). However, AfGet5-C has an additional turn at the amino terminus of H1, comprising residues Glu-195, Ala-196, and Pezizomycotina-specific Phe-197 (supplemental Fig. 1). Coil H0 caps the helical bundle on either end, and Val-191 and Val-192 extend into the core. The two other conserved phenylalanines, Phe-204, and Phe-222, line the symmetry axis within the dimer along with Leu-201 and Ala-225. The dimer interface is homologous to Get5-C, with Phe-204 forming interactions that are equivalent to those of Leu-185. Val-191 and Phe-197 together occupy a position similar to Pro-178. There is a single intermolecular electrostatic interaction made by Arg-208 and Asp-200, similar to the hydrogen bond between Asp-181 and Asn-189. The H2 helices cross at an ∼110° angle and cross between Phe-222 and Ala-225. This reconfiguration accommodates the aromatic rings of Phe-204 and Phe-222 and the side chains extending from H0 in the core.
AfGet5-C has an arrangement of tryptophans that is distinct from the Val-177/Trp-179/Trp-208 interaction of Get5 (Fig. 2C, inset). The extra turn in H1 positions the aromatic ring of Phe-197 normal to the ring of Trp-198 and results in upfield shifts of 5.799 ppm and 5.919 ppm for 1Hδ and 1Hϵ, respectively, of Phe-197. The side chain of Val-192 occupies a pocket formed by Phe-197, Trp-198, and Trp-226. AfGet5-C does not have an intermolecular cation-pi interaction, and the side chain of Trp-226 is flipped relative to the equivalently positioned Trp-208; the increased distance of the main chains between the two copies prevents any extended side chain interaction to this position. Arg-203 is not conserved in Pezizomycotina.
Get5 Carboxyl Domain Is Highly Stable Dimer
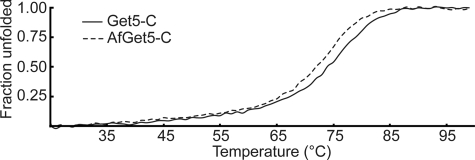
The thermal stabilities of Get5-C and AfGet5-C were determined by circular dichroism (CD) spectroscopy (Fig. 3). Get5-C has a thermal melting curve with a single transition from folded to unfolded states. The midpoint of this transition, Tm, is ∼74 °C. AfGet5-C has a surprisingly similar thermal melting curve and a Tm of 72 °C. After heating to 99 °C, both proteins can refold completely (supplemental Fig. S4).
FIGURE 3.
Get5-C is a stable dimer. Thermal melting curves of Get5-C (solid line) and AfGet5-C (dashed line) measured by CD spectroscopy.
Mammalian Get5 Homolog Ubl4A Does Not Homodimerize
We expressed and purified recombinant human Ubl4A from E. coli. Ubl4A, with a molecular mass of 20.0 kDa, elutes later from a size exclusion column than a dimeric Get5-Ubl-C construct that is lacking the amino-terminal Get4 binding domain (15.9 kDa), and slightly earlier than the monomeric Get5-Ubl domain alone (10.0 kDa) (supplemental Fig. S5B). Surprisingly, these data are most consistent with Ubl4A existing as a monomer in solution. Alone, Ubl4A is unstable and precipitates over several days when stored at 4 °C.
It has been demonstrated previously that Get5 dimers exchange monomers within hours at room temperature (7). In that experiment, we were unable to demonstrate the formation of a heterodimer between Get5 and AfGet5. We tested whether Ubl4A can form heterodimeric complexes with either homolog (supplemental Fig. S5B). A heterodimer could be detected with Get5 but not AfGet5, suggesting that the Ubl4A dimerization domain is most similar to Get5-C.
Get4/Get5 Complex Is Extended in Solution
SAXS can be used to generate low resolution models of protein complexes in solution when high resolution structures of individual domains are available. We collected SAXS data on the full-length Get4/Get5 heterotetramer, which has a molecular mass of 120 kDa. The pair-distance distribution function obtained by an indirect Fourier transform, P(r), is characteristic of an elongated particle (42) as there is an asymmetric peak that decreases to a large distance in a linear fashion (Fig. 4A). Compact particles have a more symmetric, parabolic peak. The radius of gyration and maximum particle diameter obtained from this analysis are 68.1 ± 0.1 Å and 240 Å, respectively. These values are consistent with the observation that Get4/Get5 appears atypically large by size exclusion chromatography (7). An ab initio reconstruction using the indirect Fourier transform generates the surface shown in Fig. 4A. This surface is the average of 10 individual models and represents the most probable volume.
FIGURE 4.
SAXS of Get4/Get5. A, pair-distance distribution function of the Get4/Get5 heterotetramer derived from SAXS. B, averaged ab initio reconstruction of Get4/Get5 shown as a gray surface in two orientations. C, rigid body models generated by independent simulated annealing calculations using known structures against SAXS data. In each model, the two copies of Get5 are colored magenta and orange, and the two copies of Get4 are colored cyan and blue. The three top models are shown ranked according to χ2 fit to the experimental SAXS curve.
With high resolution structures now available for each ordered domain of the Get4/Get5 complex, rigid body fitting against the SAXS data can generate unbiased models of the heterotetramer in solution. The crystal structure of Get4 and Get5-N (Protein Data Base ID 3LKU), a homology model of Get5-Ubl generated from the Ubl domain of Ubl4A (Protein Data Base ID 2DZI) (7), and the solution structure of Get5-C were used for fitting by the program CORAL. This program models flexible termini and interdomain linkers as chains of dummy residues (38), restraining the possible orientations of the rigid domains fit using simulated annealing. We performed the calculation eight times. The model with the best fit to the data (Fig. 4C, top) is in agreement with the dimensions of the averaged ab initio model (Fig. 4B). In all trials there is a similar extended spatial arrangement of the ordered domains, although there is variation in the overall rotations of the Get5-Ubl domain and Get4/Get5-N relative to Get5-C, as well as in the angle that Get4 makes with the long axis of the particle (Fig. 4C). The ab initio model likely reflects an average of these orientations rather than a single species.
DISCUSSION
Get5 homodimerization is mediated by an ∼35-residue motif within its carboxyl domain. Two copies of this motif form a domain with a novel arrangement of four α-helices. The sequence of the motif has some similarity to coiled-coils (7); however, rather than arranged in a zipper-like fashion, hydrophobic residues are buried within a small core. Elements of the motif are conserved across the eukaryotic kingdom, such as the positions of hydrophobic residues that mediate the dimer interface and the aromatic residues toward the beginning and end of the motif (supplemental Fig. S1). Of 13 positions that are at the dimer interface, 6 are conserved across eukaryotes (Fig. 1A and supplemental Fig. S1). There is extensive divergence of the remaining sequence, especially at surface positions, yet an overall conservation of structure as well as thermal stability.
The overall solution data of the Get4/Get5 complex points to an extended conformation in solution. With this architecture, the two copies of the amino-terminal face of Get4, which binds Get3, point outward at opposite ends of the particle. Get5-C presumably plays a structural role in this by orienting the Ubl domain that in turn orients Get4.
Ubl4A is a component of the Bag-6 complex along with Bag-6/Bat-3/Scythe and TRC35. This complex is involved in directing TA protein biogenesis (12, 13) and has more recently been implicated in the stabilization of proteins retrotranslocated from the ER prior to degradation (15). TRC35 is believed to mask a nuclear localization signal within Bag-6 and modulates the population of cytoplasmic and nuclear resident protein (15); however, nothing is known regarding the physical interactions of components of the complex. Purified Ubl4A is an unstable monomer but can form an artificial heterodimer with Get5-Ubl-C. This suggests that the carboxyl domain of Ubl4A has the conserved, exposed protein-protein interaction face and may mediate its interaction with the Bag-6 complex. We were unable to identify a feature in Bag-6 that contained the Get5-C motif; therefore, one might expect a unique heterodimeric interaction. The architecture of the Bag-6 complex is distinct from the Get4/Get5/Sgt2 complex. Certain steps, such as the recognition of TA proteins by SGTA, the binding of SGTA to the Ubl domain of Ubl4A, and the subsequent loading of TRC40 are likely mechanistically analogous with yeast. The rearrangement to include Bag-6/Bat-3/Scythe could allow interplay between TA targeting pathways, protein degradation and apoptosis.
Oligomerization in biology plays an important role, and there are many examples of dimerization domains; however, there are few examples of small (less than 70 residues) globular dimerization motifs. We searched for other examples of helix-turn-helix homodimers similar to Get5-C in that they are neither intercalated nor have extensive coiled-coils. There are four proteins in this category with high resolution structures available. They are the Qua1 domain from STAR proteins (43, 44), the protein kinase A (PKA) type Iα and type IIα regulatory subunits (45, 46), and the Siah-interacting protein (SIP) (47). Of these structures, Get5-C has the shortest sequence and the highest ratio of buried to exposed surface area (30% versus 28% for the next most, the crystal structure of the PKA type IIα regulatory subunit) (48). It is significantly more thermostable than the only other dimer where this was measured, the Qua1 domain, with a Tm of 74 °C compared with 63 °C (43). Therefore, this domain is currently unique in biology.
Engineering dimerization into proteins is an important goal (49, 50). Much of this type of work focuses on the use of coiled-coils in the form of leucine zippers (51). The motif we describe here may provide a useful alternative. Recombinant Get5-C expresses very well and is easily purified. The 35-residue sequence is also amenable to chemical synthesis. The Get4/Get5 complex proves that Get5-C is sufficient in maintaining dimers of significantly larger protein assemblies. The requirement of only a few key residues suggests that there is ample opportunity for protein design to alter stability, specificity, and surface properties.
It has recently been proposed that TA protein binding to Get3 causes two dimers of Get3 to assemble into a tetrameric complex (52). This is supported by crystal structures of a tetrameric archael Get3 homolog and by SAXS analysis of TA protein-bound Get3 complexes that show similar overall size and shape. This tetramer has a longest dimension of ∼150 Å, which makes it possible for the two copies of Get4 within the Get4/Get5 complex to interact with the two different Get3 dimers prior to loading of TA proteins. Sgt2, bound to at least one Get5 ubiquitin-like domain (11), would then be positioned near the hydrophobic cavity created by Get3 (Fig. 5). Alternatively, the two copies of Get4 may interact with the two Get3 subunits within a single lower order dimer. This necessitates a dramatic conformational change within Get4/Get5 to bring the Get4 N terminus into proximity. Flexibility within Sgt2 may be amendable with this model. The exact mechanism of TA protein transfer from Sgt2 to Get3 awaits further characterization.
FIGURE 5.
Model of the Get4/Get5/Sgt2 sorting complex and its interaction with Get3. Schemes are drawn to scale, with the exception of chaperones bound to Sgt2. The arrow indicates the path of the TA protein from chaperones to Sgt2 to Get3, which is held by Get4.
Get5 plays a central role in localizing the various components involved in TA targeting forming the nexus of the so-called sorting complex. The unusually stable dimerization domain of Get5 is likely critical to the sorting function. It remains unclear whether this domain plays a broader role beyond simple dimerization. The novelty of the architecture provides ample opportunity for future studies.
Supplementary Material
Acknowledgments
We thank S. O. Shan, M. E. Rome, C. J. M. Suloway, and H. B. Gristick for critical reading of the manuscript; members of the laboratory for support and useful discussions; Julie Hoy for help in data collection on our home source; Graeme Card, Ana Gonzalez, and Michael Soltice for help with data collection at SSRL BL12-2; and Tsutomu Matsui and Hiro Tsuruta (1962–August 2011) for help with bioSAXS data collection and processing at SSRL BL4-2.
The Molecular Observatory at Caltech is supported by the Gordon and Betty Moore Foundation, the Beckman Institute, and the Sanofi-Aventis Bioengineering Research Program. Operations at SSRL are supported by the United States Department of Energy and the National Institutes of Health.

This article contains supplemental Tables 1 and 2 and Figs. S1–S5.
- TA
- tail-anchored
- ER
- endoplasmic reticulum
- GET
- guided entry of TA proteins
- SAXS
- small angle x-ray scattering
- Ubl
- ubiquitin-like.
REFERENCES
- 1. Shan S. O., Walter P. (2005) Co-translational protein targeting by the signal recognition particle. FEBS Lett. 579, 921–926 [DOI] [PubMed] [Google Scholar]
- 2. Kutay U., Hartmann E., Rapoport T. A. (1993) A class of membrane proteins with a C-terminal anchor. Trends Cell Biol. 3, 72–75 [DOI] [PubMed] [Google Scholar]
- 3. Borgese N., Brambillasca S., Colombo S. (2007) How tails guide tail-anchored proteins to their destinations. Curr. Opin. Cell Biol. 19, 368–375 [DOI] [PubMed] [Google Scholar]
- 4. Rabu C., Schmid V., Schwappach B., High S. (2009) Biogenesis of tail-anchored proteins: the beginning for the end? J. Cell Sci. 122, 3605–3612 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Simpson P. J., Schwappach B., Dohlman H. G., Isaacson R. L. (2010) Structures of Get3, Get4, and Get5 provide new models for TA membrane protein targeting. Structure 18, 897–902 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Battle A., Jonikas M. C., Walter P., Weissman J. S., Koller D. (2010) Automated identification of pathways from quantitative genetic interaction data. Mol. Syst. Biol. 6, 379. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Chartron J. W., Suloway C. J., Zaslaver M., Clemons W. M., Jr. (2010) Structural characterization of the Get4/Get5 complex and its interaction with Get3. Proc. Natl. Acad. Sci. U.S.A. 107, 12127–12132 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Wang F., Brown E. C., Mak G., Zhuang J., Denic V. (2010) A chaperone cascade sorts proteins for posttranslational membrane insertion into the endoplasmic reticulum. Mol. Cell 40, 159–171 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Schuldiner M., Metz J., Schmid V., Denic V., Rakwalska M., Schmitt H. D., Schwappach B., Weissman J. S. (2008) The GET complex mediates insertion of tail-anchored proteins into the ER membrane. Cell 134, 634–645 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Chang Y. W., Chuang Y. C., Ho Y. C., Cheng M. Y., Sun Y. J., Hsiao C. D., Wang C. (2010) Crystal structure of Get4-Get5 complex and its interactions with Sgt2, Get3, and Ydj1. J. Biol. Chem. 285, 9962–9970 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Chartron J. W., Gonzalez G. M., Clemons W. M. (2011) A structural model of the Sgt2 protein and its interactions with chaperones and the Get4/Get5 complex. J. Biol. Chem. 286, 34325–34334 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Mariappan M., Li X., Stefanovic S., Sharma A., Mateja A., Keenan R. J., Hegde R. S. (2010) A ribosome-associating factor chaperones tail-anchored membrane proteins. Nature 466, 1120–1124 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Leznicki P., Clancy A., Schwappach B., High S. (2010) Bat3 promotes the membrane integration of tail-anchored proteins. J. Cell Sci. 123, 2170–2178 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Minami R., Hayakawa A., Kagawa H., Yanagi Y., Yokosawa H., Kawahara H. (2010) BAG-6 is essential for selective elimination of defective proteasomal substrates. J. Cell Biol. 190, 637–650 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Wang Q., Liu Y., Soetandyo N., Baek K., Hegde R., Ye Y. (2011) A ubiquitin ligase-associated chaperone holdase maintains polypeptides in soluble states for proteasome degradation. Mol. Cell 42, 758–770 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Winnefeld M., Grewenig A., Schnölzer M., Spring H., Knoch T. A., Gan E. C., Rommelaere J., Cziepluch C. (2006) Human SGT interacts with Bag-6/Bat-3/Scythe and cells with reduced levels of either protein display persistence of few misaligned chromosomes and mitotic arrest. Exp. Cell Res. 312, 2500–2514 [DOI] [PubMed] [Google Scholar]
- 17. Hegde R. S., Keenan R. J. (2011) Tail-anchored membrane protein insertion into the endoplasmic reticulum. Nat. Rev. Mol. Cell Biol. 12, 787–798 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Studier F. W. (2005) Protein production by auto-induction in high density shaking cultures. Protein Expr. Purif. 41, 207–234 [DOI] [PubMed] [Google Scholar]
- 19. Marley J., Lu M., Bracken C. (2001) A method for efficient isotopic labeling of recombinant proteins. J. Biomol. NMR 20, 71–75 [DOI] [PubMed] [Google Scholar]
- 20. Kabsch W. (2010) XDS. Acta Crystallogr. D Biol. Crystallogr. 66, 125–132 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Adams P. D., Afonine P. V., Bunkóczi G., Chen V. B., Davis I. W., Echols N., Headd J. J., Hung L. W., Kapral G. J., Grosse-Kunstleve R. W., McCoy A. J., Moriarty N. W., Oeffner R., Read R. J., Richardson D. C., Richardson J. S., Terwilliger T. C., Zwart P. H. (2010) PHENIX: a comprehensive Python-based system for macromolecular structure solution. Acta Crystallogr. D Biol. Crystallogr. 66, 213–221 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Emsley P., Lohkamp B., Scott W. G., Cowtan K. (2010) Features and development of Coot. Acta Crystallogr. D Biol. Crystallogr. 66, 486–501 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Sattler M., Schleucher J., Griesinger C. (1999) Prog. Nucl. Magn. Reson. Spectrosc. 34, 93–158 [Google Scholar]
- 24. Delaglio F., Grzesiek S., Vuister G. W., Zhu G., Pfeifer J., Bax A. (1995) NMRPipe: a multidimensional spectral processing system based on UNIX pipes. J. Biomol. NMR 6, 277–293 [DOI] [PubMed] [Google Scholar]
- 25. Vranken W. F., Boucher W., Stevens T. J., Fogh R. H., Pajon A., Llinas M., Ulrich E. L., Markley J. L., Ionides J., Laue E. D. (2005) The CCPN data model for NMR spectroscopy: development of a software pipeline. Proteins 59, 687–696 [DOI] [PubMed] [Google Scholar]
- 26. Bahrami A., Assadi A. H., Markley J. L., Eghbalnia H. R. (2009) Probabilistic interaction network of evidence algorithm and its application to complete labeling of peak lists from protein NMR spectroscopy. PLoS Comput. Biol. 5, e1000307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Chou J. J., Gaemers S., Howder B., Louis J. M., Bax A. (2001) A simple apparatus for generating stretched polyacrylamide gels, yielding uniform alignment of proteins and detergent micelles. J. Biomol. NMR 21, 377–382 [DOI] [PubMed] [Google Scholar]
- 28. Rieping W., Habeck M., Bardiaux B., Bernard A., Malliavin T. E., Nilges M. (2007) ARIA2: automated NOE assignment and data integration in NMR structure calculation. Bioinformatics 23, 381–382 [DOI] [PubMed] [Google Scholar]
- 29. Bardiaux B., Bernard A., Rieping W., Habeck M., Malliavin T. E., Nilges M. (2009) Influence of different assignment conditions on the determination of symmetric homodimeric structures with ARIA. Proteins 75, 569–585 [DOI] [PubMed] [Google Scholar]
- 30. Shen Y., Delaglio F., Cornilescu G., Bax A. (2009) TALOS+: a hybrid method for predicting protein backbone torsion angles from NMR chemical shifts. J. Biomol. NMR 44, 213–223 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Valafar H., Prestegard J. H. (2004) REDCAT: a residual dipolar coupling analysis tool. J. Magn. Reson. 167, 228–241 [DOI] [PubMed] [Google Scholar]
- 32. Nilges M., Bernard A., Bardiaux B., Malliavin T., Habeck M., Rieping W. (2008) Accurate NMR structures through minimization of an extended hybrid energy. Structure 16, 1305–1312 [DOI] [PubMed] [Google Scholar]
- 33. Smolsky I. L., Liu P., Niebuhr M., Ito K., Weiss T. M., Tsuruta H. (2007) J. Appl. Crystallogr. 40, S453–458 [Google Scholar]
- 34. Konarev P. V., Volkov V. V., Sokolova A. V., Koch M. H., Svergun D. I. (2003) J. Appl. Crystallogr. 36, 1277–1282 [Google Scholar]
- 35. Svergun D. I. (1992) J. Appl. Crystallogr. 25, 495–503 [Google Scholar]
- 36. Franke D., Svergun D. I. (2009) J. Appl. Crystallogr. 42, 342–346 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Volkov V. V., Svergun D. I. (2003) J. Appl. Crystallogr. 36, 860–864 [Google Scholar]
- 38. Petoukhov M. V., Svergun D. I. (2005) Global rigid body modeling of macromolecular complexes against small-angle scattering data. Biophys. J. 89, 1237–1250 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. James T. Y., Kauff F., Schoch C. L., Matheny P. B., Hofstetter V., Cox C. J., Celio G., Gueidan C., Fraker E., Miadlikowska J., Lumbsch H. T., Rauhut A., Reeb V., Arnold A. E., Amtoft A., Stajich J. E., Hosaka K., Sung G. H., Johnson D., O'Rourke B., Crockett M., Binder M., Curtis J. M., Slot J. C., Wang Z., Wilson A. W., Schüssler A., Longcore J. E., O'Donnell K., Mozley-Standridge S., Porter D., Letcher P. M., Powell M. J., Taylor J. W., White M. M., Griffith G. W., Davies D. R., Humber R. A., Morton J. B., Sugiyama J., Rossman A. Y., Rogers J. D., Pfister D. H., Hewitt D., Hansen K., Hambleton S., Shoemaker R. A., Kohlmeyer J., Volkmann-Kohlmeyer B., Spotts R. A., Serdani M., Crous P. W., Hughes K. W., Matsuura K., Langer E., Langer G., Untereiner W. A., Lücking R., Büdel B., Geiser D. M., Aptroot A., Diederich P., Schmitt I., Schultz M., Yahr R., Hibbett D. S., Lutzoni F., McLaughlin D. J., Spatafora J. W., Vilgalys R. (2006) Reconstructing the early evolution of fungi using a six-gene phylogeny. Nature 443, 818–822 [DOI] [PubMed] [Google Scholar]
- 40. Dauter Z., Dauter M., Rajashankar K. R. (2000) Novel approach to phasing proteins: derivatization by short cryo-soaking with halides. Acta Crystallogr. D Biol. Crystallogr. 56, 232–237 [DOI] [PubMed] [Google Scholar]
- 41. Suloway C. J., Chartron J. W., Zaslaver M., Clemons W. M., Jr. (2009) Model for eukaryotic tail-anchored protein binding based on the structure of Get3. Proc. Natl. Acad. Sci. U.S.A. 106, 14849–14854 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Mertens H. D., Svergun D. I. (2010) Structural characterization of proteins and complexes using small-angle x-ray solution scattering. J. Struct. Biol. 172, 128–141 [DOI] [PubMed] [Google Scholar]
- 43. Beuck C., Szymczyna B. R., Kerkow D. E., Carmel A. B., Columbus L., Stanfield R. L., Williamson J. R. (2010) Structure of the GLD-1 homodimerization domain: insights into STAR protein-mediated translational regulation. Structure 18, 377–389 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Meyer N. H., Tripsianes K., Vincendeau M., Madl T., Kateb F., Brack-Werner R., Sattler M. (2010) Structural basis for homodimerization of the Src-associated during mitosis, 68-kDa protein (Sam68) Qua1 domain. J. Biol. Chem. 285, 28893–28901 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Newlon M. G., Roy M., Morikis D., Hausken Z. E., Coghlan V., Scott J. D., Jennings P. A. (1999) The molecular basis for protein kinase A anchoring revealed by solution NMR. Nat. Struct. Biol. 6, 222–227 [DOI] [PubMed] [Google Scholar]
- 46. Banky P., Roy M., Newlon M. G., Morikis D., Haste N. M., Taylor S. S., Jennings P. A. (2003) Related protein-protein interaction modules present drastically different surface topographies despite a conserved helical platform. J. Mol. Biol. 330, 1117–1129 [DOI] [PubMed] [Google Scholar]
- 47. Santelli E., Leone M., Li C., Fukushima T., Preece N. E., Olson A. J., Ely K. R., Reed J. C., Pellecchia M., Liddington R. C., Matsuzawa S. (2005) Structural analysis of Siah1-Siah-interacting protein interactions and insights into the assembly of an E3 ligase multiprotein complex. J. Biol. Chem. 280, 34278–34287 [DOI] [PubMed] [Google Scholar]
- 48. Gold M. G., Lygren B., Dokurno P., Hoshi N., McConnachie G., Taskén K., Carlson C. R., Scott J. D., Barford D. (2006) Molecular basis of AKAP specificity for PKA regulatory subunits. Mol. Cell 24, 383–395 [DOI] [PubMed] [Google Scholar]
- 49. Kuhlman B., O'Neill J. W., Kim D. E., Zhang K. Y., Baker D. (2001) Conversion of monomeric protein L to an obligate dimer by computational protein design. Proc. Natl. Acad. Sci. U.S.A. 98, 10687–10691 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Bolon D. N., Grant R. A., Baker T. A., Sauer R. T. (2005) Specificity versus stability in computational protein design. Proc. Natl. Acad. Sci. U.S.A. 102, 12724–12729 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Apostolovic B., Danial M., Klok H. A. (2010) Coiled coils: attractive protein folding motifs for the fabrication of self-assembled, responsive and bioactive materials. Chem. Soc. Rev. 39, 3541–3575 [DOI] [PubMed] [Google Scholar]
- 52. Suloway C. J., Rome M. E., Clemons W. M. (2012) Tail-anchor targeting by a Get3 tetramer. The structure of an archaeal homologue. EMBO J. 31, 707–719 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.