Abstract
Using a novel experimental system that allows control of the matric potential of an agar slab, we explored the hydration conditions under which swarming motility is possible. If there is recognition that this physical parameter is a key determinant of swarming, it is usually neither controlled nor measured rigorously but only manipulated through proxies, namely, the agar concentration and the drying time of “soft” agar plates (swarming plates). We contend that this not only obscures the biophysical mechanisms underlying swarming but also impedes a full assessment of its clinical and environmental significances. Our results indicate that swarming motility is restricted to a narrow range of high matric water potentials in the three pseudomonads tested (Pseudomonas sp. DSS73, Pseudomonas syringae B728a, and Pseudomonas aeruginosa PA14). The threshold below which no swarming was observed was about −0.45 kPa for the first and about −0.1 kPa for the latter two. Above the threshold, the expansion rate of DSS73 swarms increased exponentially with the matric potential. Mutants deficient in surfactant production were totally or partially unable to expand rapidly on the surface of the agar slab. Our results thus suggest that swarming motility in pseudomonads is restricted to (micro)sites where ambient humidity is very high (relative humidity of >99.99%). The spatiotemporal occurrence of such sites is limited in many types of terrestrial environments.
INTRODUCTION
Swarming, a type of motility by which cells rapidly and collectively colonize a wet solid surface, has received considerable attention owing to the impressive patterns it generates and to its link to virulence in many pathogens (14). The last few decades have seen great progress in the description of the genetic bases of this mechanism (29, 32, 35). The underlying biophysics has, however, remained relatively obscure until very recently, when some clever experimental systems (34, 37) and microscopic techniques (5, 31, 36) provided important insights into the nature of swarming.
It is well established that swarming requires the presence of a thin liquid film to allow flagellum rotation (5, 31) and/or the overcoming of drag and viscous forces (8). This film has been observed microscopically, and its thickness has been recently estimated (37). The origin of this liquid film is debated, however, and it is thought to originate from the excretion of biosurfactant(s) and/or osmolyte(s) (4, 37).
In any case, the ability for bacteria to swarm and thus, presumably, generate fluid films at the surface of solid culture medium has been shown to be strongly dependent on the agar concentration. Most Gram-negative organisms optimally swarm at agar concentrations of 4 to 7 g liter−1, although swarming has been reported to occur at concentrations as high as 15 g liter−1 (28). Swarming is notoriously affected by small variations in the drying time of the so-called swarming plates, to the point that standardization efforts have been attempted (30). Presumably, agar concentration and plate drying time directly affect the energy state of water at the surface of the solid medium. In the field of physics, the rigorous expression of this energy state is as water potential, an equivalent to the water activity used in classical microbiology (24). At the surface of the hydrogel that constitutes a solid culture medium, the main (additive) components of the water potential are the osmotic and matric potentials. The agar, which is not a solute but the gelling agent, is expected to contribute mostly to the matric potential, albeit in an ill-defined manner. A recent paper describes how the relative humidity (RH; directly convertible into water potential) over an agar gel decreases with increasing agar concentrations (12). Unfortunately, no data are available for concentrations lower than 20 g liter−1, which was found to generate a water potential of about −126 kPa. To our knowledge, this value would be the best available estimate of the lower bound of the water potential range supporting swarming. This bound is, however, underestimated, as swarming in many bacteria typically requires agar concentrations lower than 10 g liter−1 (14). We argue that to understand the biophysical basis of swarming motility and predict its occurrence in a host or in the environment, a rigorous measure of the matric potential range enabling this type of motility is needed. We attempted this determination for three strains belonging to the genus Pseudomonas, using agar slabs placed in a system allowing full control of the matric potential down to values close to water saturation (i.e., 0 kPa or 100% RH).
MATERIALS AND METHODS
Strains and media.
Three pseudomonad strains and their isogenic mutants, deficient in biosurfactant synthesis, were studied. The wild-type strains were Pseudomonas syringae B728a, Pseudomonas aeruginosa PA14, and Pseudomonas sp. DSS73. This last strain can be tentatively affiliated with the Pseudomonas fluorescens species based on the 16S sequence (GenBank accession number GQ334363) and phenotypic observations (20). These strains thus encompass three species and have been isolated from different environments (B728a from the bean phyllosphere [18], PA14 from human patient skin [26], and DSS73 from the sugar beet rhizosphere [20]). These strains were compared with the following mutants deficient in biosurfactant production: B728a ΔsyfA (3), PA14 ΔrhlA (17), and DSS73 ΔamsY (16). The B728a and DSS73 strains were constitutively expressing gfp (2, 19).
All strains were maintained on FAB solid mineral medium (11) supplemented with 5 mM citrate. For the agar slab porous surface model (PSM) experiments (see below), SWM medium was used for B728a (15), modified M9 medium was used for PA14 (29), and FAB with 5 mM citrate was used for DSS73, as these media successfully induced swarming of their respective strains in standard swarming assays.
Agar slab PSM.
We have previously described the porous surface model (PSM), which allows growth of cells on the surface of a porous ceramic plate under a controlled matric potential (6). In this system, the matric potential is directly imposed on the bacteria by setting the height of the hanging column of liquid medium that connects the ceramic plate to a bottle serving as the medium reservoir. Here, we have added an agar slab on the top of the ceramic plate so that its inoculated surface can be subjected to prescribed matric potentials. The agar slabs were obtained by pouring 5.5 ml of freshly autoclaved agar medium, as described above, into the lid of a small plastic petri dish (diameter, 40 mm; height, 12.5 mm; Phoenix Biomedical Products, Mississauga, Canada). The gel was left to dry for 5 min under a laminar flow bench before being transferred upside down onto the ceramic plate of a preautoclaved PSM, its reservoir filled with 200 ml of the appropriate liquid medium. While swarming medium typically contains less than 7 g agar liter−1, we used a concentration of 15 g agar (Fluka; Sigma-Aldrich) liter−1 in the slab, unless noted otherwise. This provides sufficient strength to the gel for it to be easily transferred onto the PSM.
Inoculation.
A suction of 0.7 kPa was applied to the agar slab at least 20 min before inoculation. The inoculum was prepared by scraping cells from a 24-h-old plate before suspending them in sterile saline solution and adjusting the suspension optical density at 600 nm (OD600) to about 1. Seven microliters of this suspension was then carefully pipetted into the depression in the center of the slab (about 4 mm in diameter) which results from the presence of a protuberance in the wall of the lid of the petri dish used to cast the slab.
After inoculation, the agar slab PSM was capped with the lid of a 55-mm-diameter petri dish to maintain the system as aseptic while allowing visual inspection of the agar surface. A picture of the full system is presented as supporting information (see Fig. S1 in the supplemental material).
Incubation and swarming observations.
Incubation was carried out at room temperature. During the first 15 h, the agar slabs were always incubated at −0.7 kPa, and then the matric potential was modified and the slabs were checked for the presence of swarms. The occurrence of swarming was either assessed visually and documented with a digital camera or, for DSS73 and its derivative, followed using a Leica MZ16 FA epifluorescent stereomicroscope equipped for green fluorescent protein (GFP) detection and fitted with a charge-coupled device (CCD) camera.
Experimental design and data analysis.
For DSS73 strains, successive images of a colony (or swarm) were measured using Image Pro Plus (version 5.1) to estimate its radial expansion rate. From each image, the average colony radius was calculated by taking the square root of the value of the swarm surface area divided by π, even though swarms were not always strictly circular and often presented tendrils. The expansion rate of a colony at a given matric potential was estimated by linear regression on at least 4 successive data points. Each rate determination was performed on separate colonies on separate PSMs. No systematic replication was performed; instead, we covered a range of matric potentials to be able to perform accurate regression analysis of the expansion rate against the matric potential. The regression analysis was carried out in Sigma Plot (version 11).
For B728a and PA14, no measurements were performed and our qualitative description of the swarm dynamics relied on a minimum of three replicate agar slab PSMs.
RESULTS
Physical validation of the agar slab PSM system.
We first explored the response of the agar slab to variations in matric potential in the absence of inoculated organisms by imposing cycles of matric potentials from −0.25 to −1.5 kPa. We observed changes in volume (not measured) and in weight (by about 9% between the two extremes) in the slab as water was drawn in or out when the matric potential was varied. Note that after a change of matric potential, the weight took several hours to stabilize, as expected for hydrogels submitted to drying or wetting (21). We thus concluded that the PSM was successful at imposing a prescribed matric potential to the agar slab.
Swarming of DSS73.
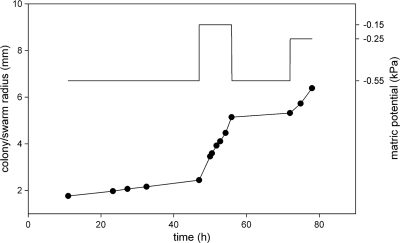
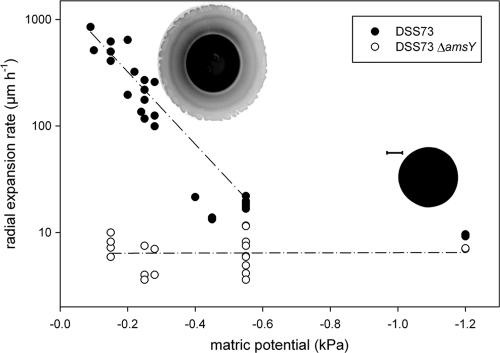
For a given matric potential, the radii of DSS73 colonies increased quasilinearly with time (Fig. 1), a finding which allowed us to derive expansion rates by linear regression. The expansion rate of DSS73 colonies was affected by the matric potential (Fig. 1 and 2), while such an influence was absent for the DSS73 amsY mutant (Fig. 2, slope of linear regression not statistically different from 0 [P = 0.954]).
Fig 1.
Spatial dynamic of a DSS73 colony (bottom curve, left axis) as affected by the matric potential imposed by the PSM (top curve, right axis). Note that the actual changes in matric potential experienced by the colony are more gradual than shown on the top graph because the equilibration between the agar slab and the PSM is not immediate.
Fig 2.
Radial expansion rates of DSS73 and DSS73 ΔamsY, an isogenic mutant that is unable to produce surfactant, as affected by the matric potential imposed via the PSM. The dashed lines illustrate a fitting of the data by regression (linear regression for the mutant; exponential regression for the wild type, after excluding data points outside the swarming range). Inset are representative inverted images of a typical colony (right) and swarm (top left), as observed with epifluorescence microscopy. Bar, 1 mm. The contrast of the images has been digitally altered to make the tendrils visible.
While DSS73 and the DSS73 amsY mutant presented similar expansion rates (in the 4- to 20-μm h−1 range) for matric potentials more negative than −0.5 kPa, under moister conditions, the wild type displayed faster expansion than the mutant and developed tendrils (Fig. 2). This indicated that the water potential threshold for the initiation of swarming for this strain was about −0.4 kPa. Above this threshold, the radial expansion rate of DSS73 swarms increased exponentially with the matric potential, as illustrated by the nonlinear regression presented in Fig. 2. The fitted model (radial expansion = 1,543 × exp[7.8 × matric potential], where “exp” indicates e raised to the power of the value in brackets and both parameters are significantly different from 0 with a P value of <0.0001) explained 85% of the variance of the data. The transition from slow colony expansion to swarming upon modification of the matric potential was relatively fast (typically less than 20 min) and fully reversible (Fig. 1).
To verify that our results were independent of the initial agar concentration of the agar slab, we measured the expansion rate of DSS73 at several matric potentials on slabs containing 9 or 20 g agar liter−1 instead of 15. As the matric potential thresholds for swarming initiation were similar irrespective of the initial agar concentration (see Fig. S2 in the supplemental material), we conclude that the imposed matric potential is the main controlling factor in our experimental system. We note, however, that the slabs with 9 g agar liter−1 presented lower expansion rates (parameter a of the exponential regression y = a × e−b×x was estimated at 659 [standard deviation {SD}, 88], which is significantly different from that obtained for 15 g agar liter−1, 1,543 [SD, 244]; P < 0.05). We hypothesize that this is due to the minor surface irregularities often visible on the fragile 9-g agar liter−1 slabs which can originate from their transfer onto the PSM system and/or from dehydration when subjected to the initial −0.7-kPa incubation.
Swarming of PA14 and B728a.
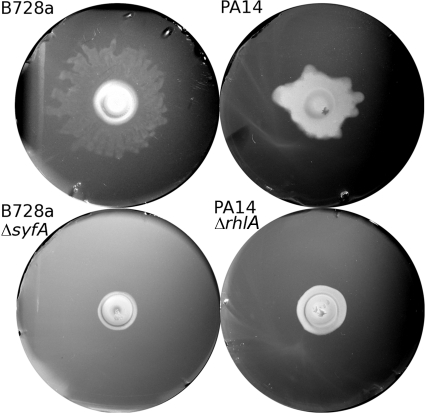
PA14 and B728a swarmed only under extremely wet conditions, i.e., for matric potentials higher than −0.1 kPa (RH > 99.99992%). We determined the threshold for tendril formation for both strains at about −0.07 kPa, but it was with some uncertainty because our experimental system has a precision of ±0.02 kPa. The mutant strains did not present tendrils (Fig. 3). Recognizing the challenge associated with accurately prescribing matric potentials in this very high range, we did not attempt measurements of expansion rates for these strains. We did, however, confirm qualitatively that, as for DSS73, the expansion speed of the swarms increased with the matric potential.
Fig 3.
Representative photographs of colonies or swarms on agar slabs maintained at about −0.02 kPa after the initial 15 h of incubation at −0.7 kPa. The pictures on the left were acquired after 9 h of incubation, and those on the right were acquired after 23 h. The agar slabs are 40 mm in diameter. The contrast of the images has been improved digitally.
DISCUSSION
Our agar slab PSM system allowed us to measure for the first time the wetness range that supports swarming on an agar surface. These ranges were not identical for the 3 strains, with DSS73 being able to swarm under slightly drier conditions than the two other strains. This difference is consistent with the behavior of the strains on standard swarming plates. Indeed, although PA14 and B728a typically favor “soft” swarming plates (4 to 6 g agar liter−1 [15, 29]), DSS73 is capable of rapid swarming on plates solidified with as much as 10 to 12 g agar liter−1, as illustrated in Fig. S3 in the supplemental material. The superior swarming ability of DSS73 can probably be explained by the very strong surface tension reduction activity of amphisin, the cyclic lipopeptide it produces (20). Despite these interstrain differences, swarming was shown to require very moist conditions (i.e., very close to 100% RH) in all organisms considered here. Such extremely humid conditions have a limited occurrence, both spatially and temporally, in most nonaquatic bacterial habitats, such as the phyllosphere or the rhizosphere. At these high matric potentials, swimming motility is typically possible—and efficient—in soils (9, 33) and sand (7) matrixes, on the ceramic surface of the PSM (8), and presumably in grooves present on most plant leaves (1). This points to the fact that the agar surface presents physical characteristics that hinder microbial motion more than many environmental surfaces. We speculate that such characteristics may include viscosity or the absence of marked microtopography, which would limit the thickness of surficial liquid film at a given matric potential (22). It is possible that swarming has evolved to permit bacterial motion on surfaces that share these unfavorable characteristics. Such surfaces might be found in animal or vegetal tissues since swarming has been shown to be important for seed and straw colonization by DSS73 (19). Alternatively, one can hypothesize that swarming has mainly evolved to permit the spatial structuring of biofilms fully immersed in liquid. Swarming has indeed been demonstrated to be integral to the development of mature P. aeruginosa biofilms in flow cells (23, 27). Another open question is that of the mechanisms underlying the sensitivity of swarming to modest changes of humidity that were quantified here and highlighted previously (e.g., references 13 and 30). The rapidity and reversibility of the transition from swarming to nonswarming states upon variations in matric potential, as illustrated in Fig. 1, are reminiscent of those observed for the Pseudomonas putida KT2440 transition from swimming to nonswimming states (8). It is thus tempting to suggest that they similarly are merely manifestations of the physical pinning (or release) of the cells on (or from) the gel surface as a result of variations in liquid film thickness dictated by the prescribed matric potentials (8). However, we cannot rule out that these transitions result from some active behavior and that the cells are able to sense minute variations in hydration conditions and adapt their transcriptional activity accordingly. Indeed, it has recently been reported that the swarm edge in P. aeruginosa presents much higher rhlA expression on soft agar (0.4%) than on slightly harder agar (0.6%) (13). This raises the question of the sensing mechanism that triggers this transition, especially considering that these variations in matric potential (less than 1 kPa) are minor compared to the total water potential experienced by the colony in our system, which is dominated by the osmotic potential of the culture medium (typically more negative than −300 kPa [24] and not affected by operating the PSM).
From a methodological point of view, the agar slab PSM system presents several advantages over the standard swarming plates. First, it allows the creation of constant, reproducible, and rigorously characterized moisture conditions which swarming plates fail to yield, jeopardizing the interlaboratory reproducibility of this standard assay (see, for example, the controversies in references 25 and 10). Particularly problematic is the drying of the swarming plates, either performed on purpose after plate pouring (30) or happening uncontrollably during subsequent incubation. Even the use of humidity-controlled chambers for incubation does not guarantee that the organisms are truly exposed to a known matric potential because the humidity of the air phase of the swarming plates will not equilibrate rapidly with that of the chamber. Such an absence of equilibrium must have happened in the work of Hamze and collaborators (10), who incubated their swarming plates (7 g agar liter−1) in a chamber set at either 40 or 70% RH (−115 and −45 MPa, respectively), values unlikely to sustain swarming or even continued growth. The second benefit of the agar slab PSM is that it permits subjecting swarms to variations of ambient moisture conditions while maintaining online microscopic observability. This holds promise for identifying the genetic determinants and, possibly, the sensing machinery contributing to swarming by exploring the transcriptional dynamic as it is affected by hydration conditions by using gfp bioreporters, as in reference 13.
Supplementary Material
ACKNOWLEDGMENTS
We express our gratitude to Ole Nybroe (Copenhagen University), Adrien Y. Burch (University of California, Berkeley), and Eric Déziel (INRS-Institut Armand-Frappier, Québec) for kindly providing us with the strains used here.
This work was supported by the Villum Kann Rasmussen Foundation Center of Excellence, the Center for Environmental and Agricultural Microbiology (CREAM), and a grant from the Danish Council for Strategic Research (2104-08-0012, MIRESOWA).
Footnotes
Published ahead of print 10 February 2012
Supplemental material for this article may be found at http://aem.asm.org/.
REFERENCES
- 1. Beattie GA. 2011. Water relations in the interaction of foliar bacterial pathogens with plants. Annu. Rev. Phytopathol. 49:533–555 [DOI] [PubMed] [Google Scholar]
- 2. Burch AY, Browne PJ, Dunlap CA, Price NP, Lindow SE. 2011. Comparison of biosurfactant detection methods reveals hydrophobic surfactants and contact-regulated production. Environ. Microbiol. 13:2681–2691 [DOI] [PubMed] [Google Scholar]
- 3. Burch AY, Shimada BK, Browne PJ, Lindow SE. 2010. Novel high-throughput detection method to assess bacterial surfactant production. Appl. Environ. Microbiol. 76:5363–5372 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Chen BG, Turner L, Berg HC. 2007. The wetting agent required for swarming in Salmonella enterica serovar Typhimurium is not a surfactant. J. Bacteriol. 189:8750–8753 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Darnton NC, Turner L, Rojevsky S, Berg HC. 2010. Dynamics of bacterial swarming. Biophys. J. 98:2082–2090 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Dechesne A, Or D, Gülez G, Smets BF. 2008. The porous surface model: a novel experimental system for online quantitative observation of microbial processes under unsaturated conditions. Appl. Environ. Microbiol. 74:5195–5200 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Dechesne A, et al. 2010. Biodegradation in a partially saturated sand matrix: compounding effects of water content, bacterial spatial distribution, and motility. Environ. Sci. Technol. 44:2386–2392 [DOI] [PubMed] [Google Scholar]
- 8. Dechesne A, Wang G, Gülez G, Or D, Smets BF. 2010. Hydration-controlled bacterial motility and dispersal on surfaces. Proc. Natl. Acad. Sci. U. S. A. 107:14369–14372 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Griffin DM, Quail G. 1968. Movement of bacteria in moist, particulate systems. Aust. J. Biol. Sci. 21:579–582 [DOI] [PubMed] [Google Scholar]
- 10. Hamze K, et al. 2011. Single cell analysis in situ in a B. subtilis swarming community identifies distinct spatially separated subpopulations differentially expressing hag (flagellin), including specialized swarmers. Microbiology 157:2456–2469 [DOI] [PubMed] [Google Scholar]
- 11. Hansen SK, et al. 2007. Characterization of a Pseudomonas putida rough variant evolved in a mixed-species biofilm with Acinetobacter sp. strain C6. J. Bacteriol. 189:4932–4943 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Jost D, Winter J, Gallert C. 2011. Water and oxygen dependence of Pseudomonas putida growing in silica sand capillary fringes. Vadose Zone J. 10:532–540 [Google Scholar]
- 13. Kamatkar NG, Shrout JD. 2011. Surface hardness impairment of quorum sensing and swarming for Pseudomonas aeruginosa. PLoS One 6:e20888. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Kearns DB. 2010. A field guide to bacterial swarming motility. Nat. Rev. Microbiol. 8:634–644 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Kinscherf TG, Willis DK. 1999. Swarming by Pseudomonas syringae B728a requires gacS (lemA) and gacA but not the acyl-homoserine lactone biosynthetic gene ahlI. J. Bacteriol. 181:4133–4136 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Koch B, et al. 2002. Lipopeptide production in Pseudomonas sp. strain DSS73 is regulated by components of sugar beet seed exudate via the Gac two-component regulatory system. Appl. Environ. Microbiol. 68:4509–4516 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Liberati NT, et al. 2006. An ordered, nonredundant library of Pseudomonas aeruginosa strain PA14 transposon insertion mutants. Proc. Natl. Acad. Sci. U. S. A. 103:2833–2838 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Loper JE, Lindow SE. 1987. Lack of evidence for in situ fluorescent pigment production by Pseudomonas syringae pv syringae on bean leaf surfaces. Phytopathology 77:1449–1454 [Google Scholar]
- 19. Nielsen TH, Nybroe O, Koch B, Hansen M, Sorensen J. 2005. Genes involved in cyclic lipopeptide production are important for seed and straw colonization by Pseudomonas sp. strain DSS73. Appl. Environ. Microbiol. 71:4112–4116 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Nielsen TH, et al. 2002. Antibiotic and biosurfactant properties of cyclic lipopeptides produced by fluorescent Pseudomonas spp. from the sugar beet rhizosphere. Appl. Environ. Microbiol. 68:3416–3423 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Or D, Phutane S, Dechesne A. 2007. Extracellular polymeric substances affecting pore-scale hydrologic conditions for bacterial activity in unsaturated soils. Vadose Zone J. 6:298–305 [Google Scholar]
- 22. Or D, Smets BF, Wraith JM, Dechesne A, Friedman SP. 2007. Physical constraints affecting bacterial habitats and activity in unsaturated porous media—a review. Adv. Water Resour. 30:1505–1527 [Google Scholar]
- 23. Pamp SJ, Tolker-Nielsen T. 2007. Multiple roles of biosurfactants in structural biofilm development by Pseudomonas aeruginosa. J. Bacteriol. 189:2531–2539 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Papendick RI, Campbell GS. 1981. Theory and measurement of water potential, p 1–23 In Parr JF, Gardner WR, Elliot LF. (ed), Water potential relations in soil microbiology. Soil Science Society of America, Madison, WI [Google Scholar]
- 25. Patrick JE, Kearns DB. 2009. Laboratory strains of Bacillus subtilis do not exhibit swarming motility. J. Bacteriol. 191:7129–7133 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Rahme LG, et al. 1995. Common virulence factors for bacterial pathogenicity in plants and animals. Science 268:1899–1902 [DOI] [PubMed] [Google Scholar]
- 27. Shrout JD, et al. 2006. The impact of quorum sensing and swarming motility on Pseudomonas aeruginosa biofilm formation is nutritionally conditional. Mol. Microbiol. 62:1264–1277 [DOI] [PubMed] [Google Scholar]
- 28. Takahashi C, et al. 2008. Swarming of Pseudomonas aeruginosa PAO1 without differentiation into elongated hyperflagellates on hard agar minimal medium. FEMS Microbiol. Lett. 280:169–175 [DOI] [PubMed] [Google Scholar]
- 29. Tremblay J, Deziel E. 2010. Gene expression in Pseudomonas aeruginosa swarming motility. BMC Genomics 11:587. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Tremblay J, Deziel E. 2008. Improving the reproducibility of Pseudomonas aeruginosa swarming motility assays. J. Basic Microbiol. 48:509–515 [DOI] [PubMed] [Google Scholar]
- 31. Turner L, Zhang RJ, Darnton NC, Berg HC. 2010. Visualization of flagella during bacterial swarming. J. Bacteriol. 192:3259–3267 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Verstraeten N, et al. 2008. Living on a surface: swarming and biofilm formation. Trends Microbiol. 16:496–506 [DOI] [PubMed] [Google Scholar]
- 33. Wong PTW, Griffin DM. 1976. Bacterial movement at high matric potentials. I. In artificial and natural soils. Soil Biol. Biochem. 8:215–218 [Google Scholar]
- 34. Wu Y, Hosu BG, Berg HC. 2011. Microbubbles reveal chiral fluid flows in bacterial swarms. Proc. Natl. Acad. Sci. U. S. A. 108:4147–4151 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Xavier JB, Kim W, Foster KR. 2011. A molecular mechanism that stabilizes cooperative secretions in Pseudomonas aeruginosa. Mol. Microbiol. 79:166–179 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Zhang HP, Be'er A, Smith RS, Florin EL, Swinney HL. 2009. Swarming dynamics in bacterial colonies. Europhys. Lett. 87:48011 [Google Scholar]
- 37. Zhang R, Turner L, Berg HC. 2010. The upper surface of an Escherichia coli swarm is stationary. Proc. Natl. Acad. Sci. U. S. A. 107:288–290 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.