Abstract
The food-borne pathogen Listeria monocytogenes experiences osmotic stress in many habitats, including foods and the gastrointestinal tract of the host. During transmission, L. monocytogenes is likely to experience osmotic stress at different temperatures and may adapt to osmotic stress in a temperature-dependent manner. To understand the impact of temperature on the responses this pathogen uses to adapt to osmotic stress, we assessed genome-wide changes in the L. monocytogenes H7858 transcriptome during short-term and long-term adaptation to salt stress at 7°C and 37°C. At both temperatures, the short-term response to salt stress included increased transcript levels of sigB and SigB-regulated genes, as well as mrpABCDEFG, encoding a sodium/proton antiporter. This antiporter was found to play a role in adaptation to salt stress at both temperatures; ΔmrpABCDEFG had a significantly longer lag phase than the parent strain in BHI plus 6% NaCl at 7°C and 37°C. The short-term adaptation to salt stress at 7°C included increased transcript levels of two genes encoding carboxypeptidases that modify peptidoglycan. These carboxypeptidases play a role in the short-term adaptation to salt stress only at 7°C, where the deletion mutants had significantly different lag phases than the parent strain. Changes in the transcriptome at both temperatures suggested that exposure to salt stress could provide cross-protection to other stresses, including peroxide stress. Short-term exposure to salt stress significantly increased H2O2 resistance at both temperatures. These results provide information for the development of knowledge-based intervention methods against this pathogen, as well as provide insight into potential mechanisms of cross-protection.
INTRODUCTION
The food-borne pathogen Listeria monocytogenes is a food safety and public health concern, as it causes life-threatening infections in at-risk human populations, including pregnant women, neonates, and the elderly. L. monocytogenes is commonly present in farm and food-processing environments and is resistant to diverse environmental conditions, including low pH, high salt, and low temperatures (49). The ability to survive and/or proliferate under these stresses contributes to the persistence of L. monocytogenes both in foods and food-processing environments, elevating the risk of transmission of this pathogen through foods to humans.
Osmotic stress is among the adverse conditions that L. monocytogenes may experience in foods and in the host gastrointestinal (GI) tract. L. monocytogenes is capable of growing at salt concentrations up to 10% (49), yet the entire repertoire of mechanisms this pathogen uses to adapt to osmotic stress is unknown (44). As L. monocytogenes may experience osmotic stress at different temperatures, such as refrigeration temperature when present on foods and host body temperature when in the GI tract, there is the potential for this pathogen to utilize different mechanisms to adapt to osmotic stress in the various environments encountered during transmission. In addition, exposure to osmotic stress can lead to cross-protection against other stresses; for example, some mechanisms known to contribute to osmotic stress resistance, such as the compatible solute transporters and cold shock proteins, also contribute to L. monocytogenes ' ability to grow at low temperature (41, 44). This overlap in osmotic stress and cold stress resistance mechanisms is supported by phenotypic data showing that growth at low temperature provides cross-protection to subsequent salt stress (8). At 37°C, exposure of L. monocytogenes to osmotic stress increases its resistance to subsequent exposure to bile salts (5), highlighting that the activation of different stress response mechanisms during adaptation to osmotic stress may lead to increased resistance to subsequent stresses in a temperature-dependent manner. While it is clear that osmotic stress-induced cross-protection occurs, the mechanisms responsible for cross-protection are largely unknown. A functional genomics approach has the potential to identify mechanisms involved in cross-protection and can lead to knowledge-based intervention strategies (6).
While osmotic stress-induced changes in expression have been investigated for specific genes, identifying and understanding temperature-dependent, genome-wide changes in the transcriptome due to salt stress will provide insights into how this pathogen adapts to and survives this stress, leading to the development of new intervention strategies. We assessed transcriptional changes in L. monocytogenes H7858 during adaptation to salt stress at 7°C and 37°C to identify (i) the short-term response to salt stress, (ii) the long-term response to salt stress, and (iii) temperature-dependent differences in adaptation to salt stress. Our data suggest new targets for growth inhibition strategies against L. monocytogenes on foods and highlight the potential mechanisms for cross-protective effects of exposure to salt against subsequent stresses.
MATERIALS AND METHODS
Growth conditions.
L. monocytogenes strain H7858, a serotype 4b strain from the 1998 frankfurter outbreak (33), was stored at −80°C in brain heart infusion (BHI) broth with 15% glycerol, and this stock was used for all experiments. H7858 from frozen stock was streaked onto BHI agar and incubated at 37°C overnight. A single colony was inoculated into 5 ml BHI broth, which was incubated at 37°C, 230 rpm for 20 h. For growth at 37°C, this culture was transferred 1:100 into a final volume of 100 ml BHI and incubated for 3 h at 37°C without aeration in 300-ml flasks. This culture was then transferred 1:10 into a final volume of 100 ml BHI plus 6% NaCl at 37°C and incubated at 37°C without aeration in 300-ml flasks. Samples of this culture were taken every 1 h for 10 h and plated onto BHI agar with an Autoplate 4000 (Spiral Biotech, Norwood, MA). For growth at 7°C, the 20-h BHI culture was transferred at 1:100 into a final volume of 100 ml BHI 7°C and incubated at 7°C for 50 h without aeration in 300-ml flasks. This culture was then transferred 1:10 into a final volume of 100 ml BHI plus 6% NaCl and incubated at 7°C without aeration in 300-ml flasks. Samples of this culture were taken every 24 h for 192 h and plated onto BHI agar. Plates were incubated at 37°C overnight, and colonies enumerated with a Q-Count (Spiral Biotech). This experiment was conducted for two independent cultures at each growth temperature, and data were used to calculate growth parameters using a modified logistic model (3). Deletion mutant strains were grown under the same conditions except that the inoculation level into BHI plus 6% NaCl was 1:1,000, as previously described (8).
RNA extraction.
RNA was extracted from cultures growing in BHI, as well as cultures growing in BHI plus 6% NaCl, as described above. RNA synthesis and degradation was stopped by adding 10 ml phenol-ethanol (1:10, vol/vol) to 100 ml culture, mixing, and then centrifuging at 10,000 rpm for 20 min at 4°C. Cell pellets were suspended in TriReagent (Ambion, Austin, TX) and homogenized for 4 min in a BeadBeater (BioSpec Products, Bartlesville, OK) with 0.1 mm acid-washed zirconium beads. After homogenization, the manufacturer's protocol was used to extract RNA. After extraction, RNA was treated with RQ1 DNase (Promega, Madison, WI), followed by purification with RNeasy minikit columns (Qiagen, Valencia, CA). RNA quality was assessed on a Bioanalyzer (Agilent, Santa Clara, CA), and only samples with a RNA integrity number (RIN) of >8.0 were used for subsequent analyses.
Determination of lag-phase sampling points.
RNA was extracted from cultures exposed to 6% NaCl in BHI at 7°C and 37°C after 7.5, 15, 30, and 60 min (absolute time scale) and also at 2.5%, 5%, 10%, and 20% of lag-phase duration for each culture. This relative time scale resulted in sampling at 7.5, 15, 30, and 60 min for 37°C cultures and at 50, 100, 200, and 400 min for 7°C cultures. RNA was extracted from three independent cultures for each sampling scheme. cDNA was synthesized from 500 ng total RNA using the TaqMan reverse transcription kit (Applied Biosystems, Foster City, CA) as described previously (8). Ten-fold serial dilutions of cDNA were used as the input for quantitative PCR (qPCR) assays for opuCA and rpoB. qPCR mixtures contained 1× Universal TaqMan mastermix (Applied Biosystems), 900 nmol each primer, and 250 nmol TaqMan probe and were run on the ABI Prism 7000 (Applied Biosystems) under the following conditions: 40 cycles at 95°C for 15 s and 60°C for 1 min. Cycle threshold (CT) values and reaction efficiencies were determined using ABI SDS software, version 1.0. qPCRs were performed in duplicate for each cDNA sample tested.
Target gene copy numbers were quantified using genomic DNA standard curves and normalized to the copy numbers of rpoB (12). The results from all samples were then compared to the transcript levels of the H7858 log-phase culture prior to salt exposure at 37°C. The average normalized log copy number and standard deviation from three independent RNA samples are reported for each time point tested. Based on the results, we chose to use the relative time scale to collect samples for the microarray experiments. RNA was extracted from exponential-phase cultures at each growth temperature prior to exposure to salt stress and then at 2.5%, 5%, 10%, and 20% of lag-phase duration at each temperature during salt stress, as well as during exponential growth under salt stress (defined as the time it took for the number of cells to increase by 2 doublings in BHI plus 6% NaCl, as calculated from the growth parameters). The resulting RNA samples were used for cDNA synthesis and subsequent microarray hybridization.
cDNA synthesis and microarray hybridization.
cDNA labeling and array hybridization were performed essentially as previously described (35). Briefly, the J. Craig Venter Institute Pathogen Functional Genomics Resource Center (JCVI PFGRC) Microbial RNA Aminoallyl Labeling for Microarrays (standard operating procedure [SOP] no. M007; http://pfgrc.jcvi.org/index.php/microarray/protocols), was used to label 4 μg total RNA with Cy3/Cy5 fluorescent dyes. JCVI PFGRC L. monocytogenes full-genome microarrays (version 4) were hybridized using the JCVI PFGRC Hybridization of Labeled DNA and cDNA Probes (SOP no. M008), and slides were scanned with a GenePix 4000B (Molecular Devices, Sunnyvale, CA). Lag-phase and exponential-phase cDNA from cells grown in BHI plus 6% NaCl at each temperature were directly hybridized with the control cDNA (exponential-phase cells grown in BHI) for each temperature, and samples from the same time point and from each temperature were directly hybridized (e.g., a sample from the 2.5% lag phase at 37°C was directly hybridized with a sample from the 2.5% lag phase at 7°C). cDNAs generated from 4 independent RNA isolations from each sample type were used, for a total of 60 hybridizations.
Microarray data analysis.
Raw intensity values for each array were normalized using pin-tip LOWESS in R, version 2.2.1, with the MAANOVA package, version 0.98. Signals from the two replicate probes on each array were averaged, and log2 transformation applied. Differences in transcript levels were determined using a mixed-model analysis of variance (ANOVA) that tested for significant differences due to growth temperature (7°C and 37°C), time after salt stress (0%, 2.5%, 5%, 10%, and 20% of lag phase and during exponential growth), and the interaction of these two factors using the following linear model: array + dye + sample (biological replicate) + temperature + time + temperature × time. The random effects were biological replicate and array, whereas the fixed effects were dye, temperature, and time. Significant changes in expression were determined by calculating the P values for the Fs statistic for each gene using 1,000 random permutations, and the P values were adjusted for multiple comparisons using the Benjamini-Hochberg correction. Only probes that matched the H7858 genome were analyzed further; for genes with more than one probe, only data from the probe with the best match were considered. Due to inconsistencies in the print batches of microarrays, 384 probes were removed from the analysis, as these probes were not present on all of the arrays. To facilitate comparison between the results for H7858 and those from other published work, the open reading frames (ORFs) probed by the oligonucleotides on the array were annotated according to orthologous genes in the strain EGD-e reference genome wherever possible, as determined by orthologous protein sequences using OrthoMCL (31). When an oligonucleotide probed an ORF with an ortholog in EGD-e, the probe was annotated with the EGD-e locus identifiers, as described previously (47). The expression profiles of significant genes were analyzed using quality threshold (QT) clustering, which groups genes with similar expression profiles based on jackknife correlations (24); QT clustering was implemented in MeV, version 4.01 (JCVI), with a diameter of 0.5 and a minimum cluster size of 5.
qRT-PCR confirmation of microarray results.
Quantitative reverse transcriptase (qRT) PCR was used to confirm microarray results for selected genes. TaqMan primers and probes were designed in Primer Express, version 1.0 (Applied Biosystems) (see Table S1 in the supplemental material). cDNA was synthesized, qPCRs were conducted, and data were analyzed as described above.
Mutant construction.
In-frame deletion mutations were constructed in LMOh7858_3019 (lmo2574), LMOh7858_1689 (lmo1580), LMOh7858_1017 (liaH), LMOh7858_1089 (liaR), LMOh7858_2527-2533 (mrpABCDEFG), and LMOh7858_3076 (lmo2812) using the splicing by overlap extension (SOE) method as previously described (Table 1; for primer sequences, see Table S2 in the supplemental material) (11). All mutations were confirmed by PCR and by sequencing the chromosomal copy of the deletion allele.
Table 1.
Strains and plasmids used in this study
| Strain | Relevant genotype | Reference, source, or construction |
|---|---|---|
| pTMB1 | ΔLMOh7858_1689 | This work |
| pTMB2 | ΔLMOh7858_3019 | This work |
| pTMB3 | ΔLMOh7858_1017 | This work |
| pBMB78 | ΔLMOh7858_1089 | This work |
| pBMB79 | ΔLMOh7858_3076 | This work |
| pBMB82 | ΔLMOh7858_mrp | This work |
| H7858 | Parent strain | 33 |
| FSL B2-289 | ΔLMOh7858_1689 | pTMB1 → H7858 |
| FSL B2-291 | ΔLMOh7858_3019 | pTMB2 → H7858 |
| FSL B2-323 | ΔLMOh7858_1017 | pTMB3 → H7858 |
| FSL B2-315 | ΔLMOh7858_1089 | pBMB78 → H7858 |
| FSL B2-318 | ΔLMOh7858_3076 | pBMB79 → H7858 |
| FSL B2-319 | ΔLMOh7858_3076 ΔLMOh7858_3019 | pBMB79 → FSL B2-291 |
| FSL B2-320 | ΔLMOh7858_mrp | pBMB82 → H7858 |
Hydrogen peroxide resistance assays.
Resistance to H2O2 was measured for H7858 in BHI and in BHI plus 6% NaCl by adding 30% H2O2 to 10 ml culture for a final concentration of 50 mM H2O2 and measuring the decrease in cell density by plating onto BHI agar after 30 min of incubation at room temperature. The levels of resistance of two independent cultures were measured at the same time points as in the gene expression experiments at each growth temperature.
Microarray data accession number.
Microarray data are available at NCBI GEO, accession number GSE27521.
RESULTS
Determination of sampling points after exposure to BHI plus 6% NaCl.
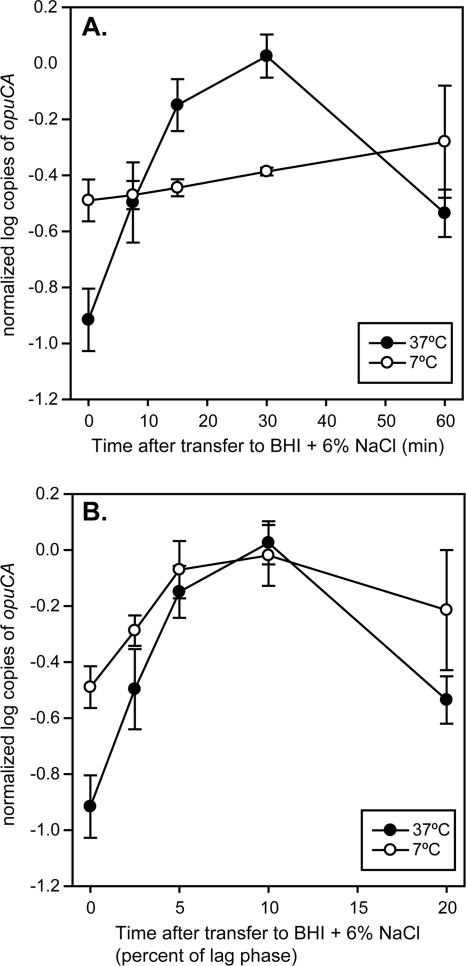
Due to differences in the length of lag phase and growth rates in BHI plus 6% NaCl at the two temperatures (7°C and 37°C), we needed to determine whether samples for transcriptome analysis during lag phase should be taken at the same time point (absolute time scale) or at the same relative points (relative time scale) for both cultures. qRT-PCR was used to quantitatively examine relationships between transcript levels for opuCA, known to be responsive to salt stress, and the absolute and relative time scales for each growth temperature. On the absolute time scale, the opuCA transcript levels increased 10-fold after 30 min of exposure to BHI plus 6% NaCl at 37°C, while the opuCA transcript levels were found to change very little over the first 60 min of exposure to salt stress at 7°C (Fig. 1A). When samples were taken on the relative time scale (i.e., 2.5% of lag phase is 7.5 min and 50 min at 37°C and 7°C, respectively), a similar pattern of opuCA transcript levels was observed for cultures under salt stress at both 7°C and 37°C, indicating that the timing of salt-induced changes in the transcriptome may be temperature dependent (Fig. 1B). Based on these data, the relative time scale was used to assess changes in the transcriptome during salt stress at 7°C and 37°C with microarrays. Exponential-phase cultures in BHI at 7°C and 37°C were used as controls, and cultures at each temperature were sampled at 2.5%, 5%, 10%, and 20% of the length of lag phase after exposure to BHI plus 6% NaCl to determine the short-term response to salt stress. In addition, exponential-phase cultures in BHI plus 6% NaCl were sampled to determine the long-term response to salt stress.
Fig 1.
Comparison of opuCA transcript levels over different time scales during salt stress-induced lag phase at 7°C and 37°C. Transcript levels over absolute time (A) and transcript levels over relative time, represented as percentage of the length of lag phase (B). The mean and standard deviation of 4 independent replicates are reported for each time point.
Adaptation to salt stress leads to numerous, mostly temperature-dependent changes in transcription.
Two-way ANOVA was used to identify genes whose expression was significantly influenced by (i) growth temperature (37°C or 7°C), (ii) time after exposure to salt stress, i.e., where the short-term response consists of significant changes in transcript levels that occur during salt-induced lag phase (compared to the results for the control) and the long-term response consists of significant changes in transcript levels that occur during exponential growth under salt stress (compared to the control), and (iii) temperature-dependent changes during the short- and long-term response to salt stress.
A significant effect of growth temperature on transcript levels was found for 139 genes (see Table S3 in the supplemental material). These genes only had a significant difference in transcript levels due to temperature, with no effect of salt exposure on transcript levels. For example, genes encoding proteins involved in DNA replication, recombination, and repair, as well as genes encoding transposases, had higher transcript levels at 37°C than at 7°C. Genes encoding proteins involved in biosynthesis of cofactors and ethanolamine utilization had higher transcript levels at 7°C than at 37°C. These differences due only to temperature were not considered further, as our main interest was in gene expression changes during adaptation to salt stress, as well as temperature-dependent changes that occurred during adaptation to salt stress.
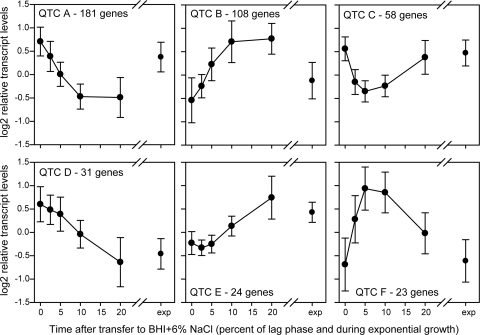
Significant changes in transcript levels during adaptation to salt stress were found for 425 genes (Table 2); transcript levels for these genes were clustered into groups with similar profiles (quality threshold clusters [QTC]) (Fig. 2, QTC A to F). Of these 425 genes, 98 genes also had a significant difference in transcript levels by growth temperature (indicated by “Temperature and time” in Table 2); these genes were further separated by which temperature had higher transcript levels of each gene (Table 2). These genes showed differences in relative transcript levels at 7 and 37°C but showed the same pattern of change during salt exposure. Specifically, 44 and 54 genes showed higher transcript levels at 7 and 37°C, respectively. For example, prfA transcript levels (Table 2, QTC B) increased during exposure to salt stress at both temperatures but were higher overall at 37°C.
Table 2.
Genes with significant changes in transcript levels over time and over time and temperature
| Factora | QTCb | No. of genes | Highlights |
|---|---|---|---|
| Time | QTC A | 144 | DNA replication; energy metabolism; ribosomal proteins; protein degradation; transcriptional regulators |
| B | 89 | Transcriptional regulators (TetR and MarR family); transport and binding proteins, including Na+/H+ antiporter; biosynthesis of cofactors | |
| C | 46 | Cell envelope membrane proteins | |
| D | 16 | Hypothetical proteins and proteins with unknown function | |
| E | 17 | Tetracycline resistance (tetA); membrane proteins; biosynthesis and degradation of polysaccharides | |
| F | 15 | Cell envelope penicillin binding protein lmo0441; hypothetical proteins and proteins with unknown function | |
| Temperature and time | |||
| Higher transcript levels at 37°C | A | 25 | Cell envelope; cold shock family protein; glutamine ABC transporter |
| B | 9 | prfA; protein degradation; transcript similar to B. subtilis YuzB | |
| C | 8 | Energy metabolism (ldh-2, ansA); DegA and TetR family transcriptional regulators; mechanosensitive channel protein (mscL) | |
| D | 3 | Hypothetical proteins; proteins with unknown function | |
| E | 4 | ABC transporter; putative oxidoreductase | |
| F | 5 | Chitin binding protein; alternative sigma factor sigL; acetolactate synthase (alsS) | |
| Higher transcript levels at 7°C | A | 12 | Hypothetical proteins; proteins with unknown function |
| B | 10 | Phosphate ABC transporter; oxidoreductase; hemolysin III (lmo1864) | |
| C | 4 | lspA signal peptidase; hypothetical proteins | |
| D | 12 | Chemotaxis and motility (cheA, cheY); glutamate biosynthesis (gltBD) | |
| E | 3 | Hypothetical proteins; proteins with unknown function | |
| F | 3 | Protein export (prsA-2); hypothetical proteins |
The 425 genes were clustered based on patterns in transcript levels over time (short- and long-term responses). The 327 genes with a significant time effect only (“Time”) were separated from the 98 genes that had both a significant time and a significant temperature effect (“Temperature and time”).
Relative transcript levels for genes in each QTC group are presented in Fig. 2.
Fig 2.
QT clusters for the 425 genes with significant changes in transcript levels over time (both short and long term) after exposure to BHI plus 6% NaCl. Numbers on the x axes represent the time after transfer to BHI plus 6% NaCl as a percentage of the length of lag phase, and “exp” represents exponential growth in BHI plus 6% NaCl. Highlights from the clusters are presented in Table 2.
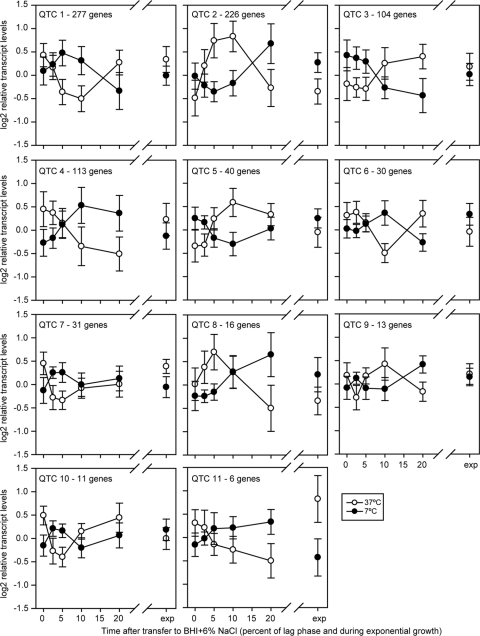
The majority of alterations in transcript levels during adaptation to salt stress were dependent on growth temperature; 888 genes had a significant time-by-temperature interaction effect, indicating that the changes in transcript levels due to exposure to salt stress were different depending on the growth temperature (Table 3). These genes (shown in Table 3) have significant changes in transcript levels over time that are temperature dependent, in contrast to genes with significant yet independent temperature and time effects (shown in Table 2 and described above). Among the 888 genes with a significant time-by-temperature interaction effect, 867 genes were clustered into groups with similar temperature-dependent transcript level profiles (Fig. 3, QTC 1 to 11); 21 genes did not cluster with these groups. These genes show different patterns of transcript levels under salt stress at 7 and 37°C. For example, genes in QTC 3 were induced during salt stress at 37°C, while they were downregulated during salt stress at 7°C. Overall, these data show that the response of L. monocytogenes to salt stress differs considerably by temperature, with 888 genes (i.e., 29% of the 3,015 genes probed by our microarray) showing different patterns in cells exposed to salt at 7 and 37°C.
Table 3.
Genes with temperature-dependent significant changes in transcript levels during adaptation to salt stress
| QTCa | No. of genes with significantly changed transcript levelsb | Highlightsc |
|---|---|---|
| 1 | 277 | Chemotaxis and motility; purine and pyrimidine ribonucleotide biosynthesis; transport and binding proteins (ABC transporter lmo0194, lmo0195, and lmo1636); ribosomal proteins; tRNA aminoacylation; fatty acid and phospholipid biosynthesis (putative lipoprotein lmo0047, acetyl-coenzyme A carboxylase accAD); two-component response system liaSR; toxic ion resistance (lmo1966 and lmo1967) |
| 2 | 226 | Protein degradation (peptidase lmo0265 and lmo1375); protein folding and stabilization (groES, groEL, dnaK, grpE, and clpB); adaptation to atypical conditions (sigB, ctc, opuCABCD, gadD3, bsh, and bilEAB and general stress proteins lmo0515, lmo1580, and lmo2748); pathogenesis (inlA, inlC2, inlD, and inlF); glycerol uptake (glpK and glpF); transport and binding proteins (cation- and iron-carrying compounds lmo2231, lmo2575, and lmo2602) |
| 3 | 104 | Amino acid biosynthesis (hisHBDG, ilvDBNC, and leuABCD); cell envelope membrane proteins |
| 4 | 113 | Transport and binding proteins (cation- and iron-carrying compounds lmo0803, lmo1424, lmo1516, lmo1848, lmo1960, lmo2689); lmaDCBA and A118 bacteriophage-like protein operon lmo0119-lmo0128; agrBCD involved in cell communication and pathogenesis; cell envelope proteins (pencillin binding protein lmo2229, membrane protein lmo2484); LiaSR system proteins (liaH and liaI) |
| 5 | 40 | Transcriptional regulators; electron transport (quinol oxidase, qoxBCD) |
| 6 | 30 | Ribosomal proteins; cell division (ftsX and ftsE) |
| 7 | 31 | Phosphate ABC transporter; cell membrane biosynthesis; PemK family transcriptional regulator |
| 8 | 16 | Catalase (kat); two-component response regulator (lisR); hypothetical proteins; proteins with unknown function |
| 9 | 13 | Glycine betaine transporter betL; proteins with unknown functions |
| 10 | 11 | aminopeptidase; hypothetical proteins |
| 11 | 6 | PrfA-regulated genes (plcA, mpl, actA, and plcB) |
Relative transcript levels for genes in each QTC group are presented in Fig. 3.
Of the 888 significant genes, 867 clustered into 11 QTC groups and 21 genes had transcript patterns that did not fit into these groups.
Fig 3.
QT clusters of 867 genes with a significant temperature-by-time interaction. Numbers on the x axes represent the time after transfer to BHI plus 6% NaCl as a percentage of the length of lag phase, and “exp” represents exponential growth in BHI plus 6% NaCl. Highlights of the clusters are presented in Table 3.
The short-term response to salt stress at 7°C and 37°C included elevated transcription of σB-regulated genes.
Of the 170 genes known to be regulated by σB (35), 55 were significantly upregulated during salt-induced lag phase at both 7°C and 37°C and had similar patterns of induction over time (Table 3 and Fig. 3, QTC 2). While σB-regulated genes were induced during salt exposure at both growth temperatures, the expression profiles for these genes during the short-term response to salt stress were dependent on growth temperature. At 37°C, the transcript levels of sigB- and σB-regulated genes (e.g., ctc, opuCA, bsh, and inlA) were highest during the short term during the lag phase induced by 10% salt, while at 7°C, the transcript levels of these same genes were highest during the lag phase induced by 20% salt (Fig. 3, QTC 2). The significant increases in transcript levels for ctc, opuCA, bsh, inlA, lmo1580, and lmo2748 were confirmed by qRT-PCR (Table 4). Three genes annotated as universal or general stress proteins (lmo0515, lmo1580, and lmo2748) that are known to be regulated by σB were highly expressed during the short-term adaptation to salt stress at both temperatures. A significant role for lmo2748 in the L. monocytogenes salt stress response has already been established (1), so we investigated the role of lmo1580 in salt stress. In BHI plus 6% NaCl, there were no significant effects on either lag phase or growth rate at 7°C or 37°C for the ΔLMOh7858_1689 (Δlmo1580) strain compared to the results for the parent strain (Table 5).
Table 4.
Changes in transcript levels during adaptation to BHI plus 6% NaCl determined by qPCR
| H7858 ORF | EGD-e ORF | Name | Fold change (mean ± SD) in transcript levels during salt stress at indicated temp and % of lag phase durationa |
|||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 37°C |
7°C |
|||||||||||
| 2.5 | 5 | 10 | 20 | Exp | 2.5 | 5 | 10 | 20 | Exp | |||
| LMOh7858_0221 | lmo0201 | plcA | 0.7 ± 0.8 | 0.5 ± 0.4 | 0.7 ± 0.2 | 1.1 ± 0.6 | 20.4 ± 10.5 | 1.1 ± 0.3 | 2.9 ± 1.3 | 5.1 ± 1.9 | 2.7 ± 1.3 | 1.8 ± 0.8 |
| LMOh7858_0231 | lmo0211 | ctc | 9.2 ± 6.5 | 20.4 ± 4.3 | 51.0 ± 19.5 | 10.4 ± 1.9 | 7.1 ± 0.4 | 1.7 ± 0.5 | 3.6 ± 1.5 | 4.2 ± 0.7 | 2.9 ± 0.6 | 1.4 ± 0.6 |
| LMOh7858_0498 | lmo0433 | inlA | 0.8 ± 0.4 | 2.3 ± 0.8 | 10.1 ± 5.5 | 1.4 ± 0.7 | 33.1 ± 14.2 | 1.0 ± 0.2 | 1.6 ± 0.5 | 2.2 ± 0.3 | 2.4 ± 0.3 | 5.0 ± 2.9 |
| LMOh7858_1017 | lmo0955 | liaH | 2.2 ± 1.7 | 4.7 ± 0.2.3 | 12.9 ± 9.3 | 2.1 ± 1.0 | 1.9 ± 0.2 | 3.0 ± 1.4 | 24.8 ± 10.2 | 54.4 ± 18 | 4.6 ± 0.9 | 1.7 ± 0.6 |
| LMOh7858_1081 | lmo1014 | gbuA | 0.8 ± 0.2 | 1.3 ± 0.3 | 5.6 ± 1.3 | 9.8 ± 2.4 | 2.7 ± 0.3 | 1.8 ± 0.4 | 2.4 ± 0.7 | 2.6 ± 0.5 | 2.9 ± 0.4 | 2.4 ± 0.3 |
| LMOh7858_1089 | lmo1022 | liaR | 0.5 ± 0.2 | 0.4 ± 0.2 | 1.6 ± 0.5 | 0.9 ± 0.4 | 1.6 ± 0.3 | 1.0 ± 0.2 | 1.2 ± 0.3 | 2.9 ± 0.6 | 0.4 ± 0.1 | 0.7 ± 0.3 |
| LMOh7858_1523 | lmo1428 | opuCA | 2.8 ± 0.7 | 6.0 ± 2 | 9.4 ± 1.7 | 2.7 ± 1.2 | 3.1 ± 1.6 | 1.7 ± 0.3 | 2.9 ± 0.8 | 3.1 ± 0.7 | 1.9 ± 0.2 | 1.0 ± 0.5 |
| LMOh7858_1570 | lmo1473 | dnaK | 1.1 ± 0.6 | 2.4 ± 0.6 | 5.6 ± 2 | 1.4 ± 0.5 | 0.8 ± 0.1 | 1.0 ± 0.2 | 1.7 ± 0.4 | 4.1 ± 0.5 | 3.8 ± 1.1 | 1.8 ± 0.5 |
| LMOh7858_1689 | lmo1580 | 4.8 ± 3.4 | 14.5 ± 5.5 | 30.0 ± 13.4 | 1.7 ± 0.7 | 5.0 ± 1.1 | 2.4 ± 1 | 5.1 ± 3.0 | 7.2 ± 2.0 | 4.6 ± 1.3 | 1.3 ± 0.8 | |
| LMOh7858_2196 | lmo2067 | bsh | 3.0 ± 1.6 | 5.7 ± 1.1 | 33.1 ± 9.5 | 6.8 ± 2.2 | 10.6 ± 0.9 | 2.4 ± 0.9 | 4.7 ± 2.4 | 7.1 ± 2.1 | 7.2 ± 2 | 4.1 ± 2.8 |
| LMOh7858_2198 | lmo2069 | groES | 1.0 ± 0.4 | 2.3 ± 0.3 | 6.9 ± 2 | 2.6 ± 0.7 | 1.6 ± 0.3 | 1.2 ± 0.5 | 1.9 ± 0.7 | 3.0 ± 0.9 | 4.9 ± 1.7 | 2.6 ± 1.3 |
| LMOh7858_2221 | lmo2092 | betL | 0.6 ± 0.1 | 1.3 ± 0.3 | 5.0 ± 1.5 | 2.2 ± 0.5 | 3.1 ± 0.6 | 1.8 ± 0.3 | 3.0 ± 0.9 | 3.7 ± 0.7 | 3.9 ± 0.4 | 2.5 ± 0.4 |
| LMOh7858_2330 | lmo2196 | oppA | 0.6 ± 0.1 | 0.4 ± 0.1 | 0.2 ± 0.1 | 0.2 ± 0.1 | 0.7 ± 0.2 | 0.9 ± 0.2 | 0.8 ± 0.2 | 0.5 ± 0.1 | 0.1 ± 0.1 | 0.6 ± 0.1 |
| LMOh7858_3012 | lmo2748 | 10.6 ± 4.9 | 47.8 ± 15 | 200 ± 90 | 15.9 ± 8.7 | 8.6 ± 2.5 | 4.4 ± 2.0 | 8.8 ± 3.9 | 12.8 ± 4.7 | 17.5 ± 3.6 | 5.0 ± 2.5 | |
| LMOh7858_3019 | lmo2754 | 0.3 ± 0.2 | 0.3 ± 0.1 | 1.1 ± 0.6 | 1.1 ± 0.5 | 0.9 ± 0.1 | 1.2 ± 0.3 | 1.5 ± 0.7 | 1.7 ± 0.6 | 2.2 ± 0.7 | 2.3 ± 1.1 | |
| LMOh7858_3052 | lmo2785 | kat | 0.8 ± 0.2 | 1.7 ± 0.6 | 4.2 ± 2.3 | 1.7 ± 0.7 | 0.7 ± 0.1 | 1.3 ± 0.2 | 2.3 ± 0.7 | 3.3 ± 0.5 | 3.4 ± 0.8 | 3.3 ± 1 |
| LMOh7858_3076 | lmo2812 | 0.4 ± 0.3 | 0.7 ± 0.4 | 4.5 ± 2.6 | 1.2 ± 0.5 | 1.1 ± 0.3 | 1.2 ± 0.4 | 9.2 ± 4.5 | 95.7 ± 14.7 | 26.9 ± 9.0 | 0.8 ± 0.3 | |
Boldface indicates comparisons where transcript levels were significantly different at that time point under salt stress compared to the transcript levels of the control without salt. Exp, exponential phase.
Table 5.
Growth parameters of mutants in BHI plus 6% NaCl at 7°C and 37°C
| Strain | Genotype | Value (mean ± SD)a for indicated parameter |
|||
|---|---|---|---|---|---|
| 37°C |
7°C |
||||
| Length of lag phase (h) | Growth rate (log10 CFU/ml/h) | Length of lag phase (h) | Growth rate (log10 CFU/ml/h) | ||
| H7858 | Parent strain | 7.1 ± 0.2 | 0.29 ± 0.01 | 30.2 ± 2.0 | 0.027 ± 0.001 |
| FSL B2-289 | ΔLMOh7858_1689 | 7.4 ± 0.4 | 0.28 ± 0.01 | 32.9 ± 1.0 | 0.026 ± 0.001 |
| FSL B2-291 | ΔLMOh7858_3019 | 7.2 ± 0.2 | 0.29 ± 0.01 | 39.5 ± 4.1 | 0.029 ± 0.001 |
| FSL B2-323 | ΔLMOh7858_1017 | 6.7 ± 1.0 | 0.27 ± 0.01 | 27.2 ± 1.7 | 0.028 ± 0.001 |
| FSL B2-315 | ΔLMOh7858_1089 | 6.8 ± 0.9 | 0.26 ± 0.02 | 29.0 ± 1.7 | 0.028 ± 0.001 |
| FSL B2-318 | ΔLMOh7858_3076 | 7.8 ± 0.5 | 0.28 ± 0.01 | 23.8 ± 2.2 | 0.026 ± 0.001 |
| FSL B2-319 | ΔLMOh7858_3076 ΔLMOh7858_3019 | 7.1 ± 0.5 | 0.28 ± 0.01 | 37.3 ± 1.7 | 0.027 ± 0.002 |
| FSL B2-320 | ΔLMOh7858_mrp | 9.1 ± 0.5 | 0.27 ± 0.01 | 55.3 ± 1.1 | 0.009 ± 0.001 |
Values in boldface are significantly different from the growth parameter of the parent strain (H7858) for each growth temperature.
Genes with roles in ion transport have increased transcript levels during the short- and long-term response to salt stress at 7°C and 37°C.
A number of genes annotated as cation- and iron-carrying compounds had increased transcript levels during adaptation to salt stress. At 7°C, 5 genes had elevated transcript levels during the short-term response (lmo0803, lmo1424, lmo1516, lmo1848, and lmo2689), and 3 genes had elevated transcript levels during the long-term response (lmo1424, lmo1848, and lmo1960) (Fig. 3 and Table 3, QTC 4). At 37°C, a different set of cation- and iron-carrying compound-encoding genes were induced (lmo2231, lmo2575, and lmo2231) than at 7°C (Fig. 3, Table 3, QTC 2). The short- and long-term response to salt stress at both temperatures included an ∼2-fold increase in transcript levels of the mrpABCDEFG operon (LMOh7858_2527-2533), which encodes a monovalent cation/proton antiporter (Table 2 and Fig. 2, QTC B). The transcript levels of the mrp operon under salt stress were significantly higher at all the lag-phase sampling points, as well as during exponential growth, than during exponential growth without additional salt. At 37°C in BHI plus 6% NaCl, deletion of mrpABCDEFG resulted in a lag phase that was significantly longer (9.1 ± 0.5 h [mean ± standard deviation], adjusted P value, 0.002) than that of the parent strain (7.1 ± 0.2 h), but the growth rates were similar for the ΔmrpABCDEFG strain and the parent strain (Table 5). At 7°C in BHI plus 6% NaCl, the deletion of mrpABCDEFG resulted in a lag phase that was significantly longer (55.3 ± 1.1 h, adjusted P value, <0.0001) than that of the parent strain (30.2 ± 2.0 h) and growth that was significantly slower (0.009 ± 0.006 log10 CFU/ml/h, adjusted P value, <0.0001) than that of the parent strain (0.027 ± 0.001 log10 CFU/ml/h).
The long-term response to salt stress at 37°C includes upregulation of stress response and virulence genes.
The long-term response at 37°C included increased transcript levels of virulence genes prfA (Fig. 2, QTC B), mpl, actA, plcB (Fig. 3, QTC11), plcA, and inlA (Table 4). At 37°C, the long-term response also included increased transcript levels for all genes of the ilvD-leuD operon (Fig. 3, QTC 3), which are known to be upregulated in L. monocytogenes in the phagosome (13). Genes encoding proteins involved in purine ribonucleotide biosynthesis (lmo1764-lmo1771, purDHNMFLQS) also had ∼2-fold-higher transcript levels during exponential growth in BHI plus 6% NaCl than during exponential growth in BHI. Genes known to be activated by the general stress response (35) also had increased transcript levels during the long-term response to salt stress at 37°C, including ctc, opuCA, lmo1580 (Table 4), gadT2D2, and gadD3 (Fig. 3, QTC 5).
Induction of the cell envelope stress response system LiaFSR during the short-term response to salt stress at 7°C.
LiaFSR is a three-component response system that is known to be induced by cell wall-acting antibiotics, as well as other cell envelope stresses, such as ethanol stress (18, 27). During the short-term adaptation to salt stress at 7°C, the transcript levels of liaR increased (Table 3, QTC 1), and qRT-PCR confirmed a significant, 2.9-fold increase (adjusted P value, <0.0001) in transcript levels during the lag phase induced by 10% salt compared to the transcript levels in the control at 7°C (Table 4). Of the 28 genes known to be regulated by LiaFSR in L. monocytogenes (18), 22 had significantly higher transcript levels during the salt-induced lag phase at 7°C (Table 3; also see Table S5 in the supplemental material), including liaH, which was confirmed by qRT-PCR to have significantly higher transcript levels than the control (adjusted P value, <0.001) at 5% (25-fold), 10% (54-fold), and 20% (4.6-fold) of lag phase (Table 4). To determine whether the LiaFSR system plays a role in adaptation to salt stress, we monitored the growth of the ΔliaR and ΔliaH strains and the parent strain H7858 in BHI plus 6% NaCl at 7°C. Both the ΔliaR and the ΔliaH strain had lag phases and growth rates similar to those of H7858 (Table 5), indicating that, while the LiaFSR system is induced by salt stress, it does not have a direct effect on the salt stress growth phenotype.
The short-term response to salt stress at 7°C induces cell wall modifications.
Genes encoding proteins involved in peptidoglycan biosynthesis were downregulated during the short-term response to salt stress at both 7°C and 37°C (Fig. 3, QTC 1), though most genes encoding proteins involved in peptidoglycan modifications were upregulated during adaptation to salt stress at 7°C. Two genes encoding autolysins had increased transcript levels during the adaptation to salt stress at 7°C; lmo1215 during the short-term response (Fig. 3, QTC 4) and lmo1216 during the short- and long-term responses (Fig. 3, QTC 1). Of the nine penicillin binding proteins (PBP) in L. monocytogenes, 5 were upregulated during the adaptation to salt stress at 7°C, and only one was upregulated at 37°C. Specifically, a class B penicillin binding protein (PBP) encoded by lmo0441 had significantly higher transcript levels during the short-term adaptation to salt stress at both 7°C and 37°C (Fig. 2, QTC F). Another class B PBP, encoded by lm2039, was induced during the short-term adaptation only at 7°C (Fig. 3, QTC 6), and the class A PBP encoded by lmo2229 was also induced only at 7°C (Fig. 3, QTC 4). Two PBPs have known d-alanyl d-alanine (DD) carboxypeptidase activity (lmo2754 and lmo2812) (29); both of these genes were induced during the short-term response to salt stress at 7°C, and lmo2754 also had 2.3-fold-higher transcript levels than the control during exponential growth in BHI plus 6% NaCl (Table 4). To determine the impact of the DD carboxypeptidases on adaptation to salt stress, mutants with single mutations in both genes, as well as a double mutant, were created and their growth parameters compared to the growth of the parent strain. At 37°C, all DD carboxypeptidase mutants had growth parameters similar to those of the parent strain (Table 5). At 7°C, the ΔLMOh7858_3019 (lmo2754) and ΔLMOh7858_3019 ΔLMOh7858_3076 (double mutant) strains had significantly longer lag phases than the parent strain, while the ΔLMOh7858_3076 (lmo2812) strain had a significantly shorter lag phase than the parent strain (Table 5). All mutants had growth rates similar to that of the parent strain, indicating that their main role occurs during the short-term adaptation to salt stress at low temperature and does not contribute significantly to the long-term response.
Increased transcript levels of genes encoding chaperones and proteases during the long-term response to salt stress at 7°C.
The transcription of genes encoding chaperones and proteases was increased during the short-term response to salt stress at both temperatures, including dnaK, groES (Table 4), groEL, grpE, htrA, and clpP (Fig. 3 and Table 3, QTC2). However, the long-term response to salt stress at 7°C included increased transcript levels of dnaK (1.8-fold), groES (2.6-fold), clpE (2.4-fold), lmo2256 (intracellular protease, 1.5-fold), and lmo2391 (similar to Bacillus subtilis yhfK, 1.8-fold) compared to the transcript levels in exponential-phase cultures in BHI at 7°C; this increase in the expression of protease-encoding genes was not observed as part of the long-term response to salt stress at 37°C.
Salt stress induces cross-protection to hydrogen peroxide stress.
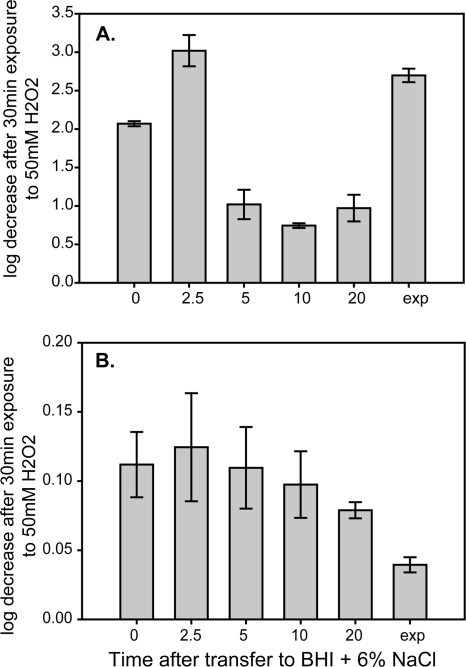
The transcript levels of kat, which encodes catalase, were significantly higher during the short-term response to salt stress at 37°C (4.2-fold, adjusted P value, <0.0001) and at 7°C (3.4-fold, adjusted P value, <0.0001) than the levels during exponential growth at either temperature (Table 4). At 7°C, the kat transcript levels were also significantly higher, by 3.3-fold (adjusted P value, <0.0001), than the levels in the control during the long-term response to salt stress. Resistance to H2O2 was measured during both the short- and long-term adaptation to salt stress at each temperature (Fig. 4). At 37°C, an increase in resistance to 50 mM H2O2 was observed during the short-term adaptation to salt stress, with significantly (adjusted P value, <0.05) higher resistance at 5%, 10%, and 20% of lag phase than in the control. At 7°C, cultures were overall more resistant to 50 mM H2O2 than 37°C cultures. H2O2 resistance at 7°C also increased during the short-term adaptation to salt stress and was significantly (adjusted P value, <0.05) higher during the long-term adaptation to salt stress than it was in the control, coinciding with the significantly higher transcript levels of kat at the same time point.
Fig 4.
Resistance to 50 mM H2O2 before and during salt stress at 37°C (A) and 7°C (B). Mean and standard deviation of four replicates are reported for each time point. Time zero represents resistance of exponential-phase cultures in BHI prior to exposure to NaCl; 2.5, 5, 10, and 20 represent the percentage of lag phase where cultures were assayed for peroxide resistance; and “exp” represents the peroxide resistance of cultures growing exponentially in BHI plus 6% NaCl.
DISCUSSION
Managing osmotic stress is important for L. monocytogenes ' survival and growth in foods, as well as in the host (41, 44). Our data reported here indicate that L. monocytogenes initiates numerous changes in transcription during adaptation to salt stress and that the majority of these adaptations are temperature dependent. Induction of a cell envelope stress response and cell wall modifications occur at 7°C, while virulence gene expression is induced under salt stress at 37°C. At both temperatures, the general stress response is activated during the short-term adaptation to salt stress, and the long-term response includes the upregulation of genes known to be involved in the response to other environmental stresses. The activation of these systems by salt stress leads to cross-protection to subsequent stresses, as demonstrated by the increased H2O2 resistance of cells during the short-term adaptation to salt stress, and has the potential to facilitate the transmission of L. monocytogenes on foods and increase survival in the host.
The general stress response is activated during the short-term response to salt stress.
The general stress response, controlled by the alternative sigma factor SigB, is induced by a number of environmental stresses, including osmotic stress (4). SigB plays a significant role in adaptation to osmotic stress but is not essential for L. monocytogenes to adapt to the stress (4, 17). Here, we observed a transient increase in transcript levels for sigB and SigB-regulated genes as part of the initial response to salt stress at both temperatures. A similar, transitory pattern of induction for sigB and SigB-regulated genes has also been observed in B. subtilis during the short-term adaptation to salt stress at 37°C (23), in L. monocytogenes upon exposure to 0.5 M NaCl (51), and in L. monocytogenes under the combination of low pH and low water activity (55). Critical roles in adaptation to salt stress have been identified for many SigB-regulated genes, including opuCA (17), ctc (20), and gbuA (10), and we observed increased transcript levels of these SigB-regulated genes at both temperatures. In addition, we observed increased transcript levels of SigB-regulated genes encoding general stress proteins (lmo0515, lmo2748, and lmo1580). Lmo2748 plays a role in adaptation to salt stress (1). While we found that transcription of lmo1580 is highly induced by salt stress, the characterization of a null mutant showed that it does not contribute significantly to adaptation to salt stress at either temperature tested, raising the possibility of compensation by the other general stress proteins.
Transcription of virulence and virulence-associated genes is significantly increased during adaptation to salt stress.
Environmental changes are one of the key factors influencing virulence gene expression in bacterial pathogens. Exposure to osmotic stress can induce virulence gene expression in L. monocytogenes, and, along with temperature, is thought to be one of the main signals that indicates presence in the gastrointestinal tract (43). At 7°C, we observed a significant increase in transcript levels of virulence genes, such as plcA and inlA, only during the short-term adaptation to salt stress. While the transcript levels for these genes were significantly higher than in the control culture at 7°C, it is important to note that overall, the transcript levels of virulence and virulence-associated genes were always higher at 37°C than at 7°C. At 37°C, we observed significantly higher transcript levels of virulence and virulence-associated genes during both the short- and long-term response to salt stress. During the short-term response, inlA and inlB transcript levels were significantly increased, along with transcript levels of the virulence-associated genes lisR (Fig. 3, QTC8), inlC2, and inlF (Fig. 3, QTC2). This short-term response at 37°C may represent the initial adaptation to the intestinal environment, where the induction of SigB is thought to play the major role in controlling the expression of stress response genes, as well as virulence genes, including the expression of internalins, thus priming the cell for invasion (21, 36). Even a short-term exposure of L. monocytogenes to salt stress can increase virulence; one study found increased invasion in HT-29 cells of L. monocytogenes after exposure to 0.3 M NaCl for only 10 min (52).
The long-term response to salt stress at 37°C included significantly higher transcript levels of PrfA-regulated virulence genes, including plcA, plcB, hly, and actA; other researchers have also observed increased transcript levels of these genes during growth with 4.5% NaCl at 25°C, which led to significantly higher invasion and intracellular spread in Caco-2 cells (34). We also observed that the long-term response to salt stress included increased transcript levels of genes encoding proteins involved in branched-chain amino acid synthesis (Fig. 3, QTC 3, ilvD operon, lmo1983-lmo1991). The increased expression of these genes is similar to L. monocytogenes gene expression patterns when in the phagosome (13). These findings are particularly relevant in light of the recent discovery that L. monocytogenes is exposed to chloride in the phagosome and the finding that the presence of chloride enhances the ability of L. monocytogenes to escape the phagosome (40). While salt stress poses an osmotic stress that may be relevant to gastrointestinal passage, the increased chloride content as part of salt stress appears to serve as an additional signal during the intracellular life cycle of L. monocytogenes.
The Mrp Na+/H+ antiporter plays a major role in adaptation to salt stress.
Cation transporters play an important role in balancing osmotic stress, and recent transcriptomics experiments have shown that the expression of genes encoding ion transporters, including the ion transporter Mrp, is induced in Bacillus cereus (14) and B. subtilis (23) during adaptation to salt stress. Mrp is a hetero-oligomeric antiporter that is widespread among bacteria and archaea (50) and has known roles in sodium resistance and pH homeostasis in B. subtilis (25), as well as a role in pathogenesis in Pseudomonas aeruginosa (30). The Mrp antiporter is essential for B. subtilis growth at 0.5 M NaCl (28). Our experiments also identified increased transcription of mrpABCDEFG during both the short- and long-term response to salt stress at both 7 and 37°C and showed that the inactivation of this antiporter had severe consequences for the ability of L. monocytogenes to adapt to salt stress, indicated by an increased lag-phase duration at both temperatures and a reduced growth rate at 7°C. A role for Mrp in salt stress has not previously been identified in L. monocytogenes; it appears to play a major role in adaptation to salt stress, as an mrp null mutant had reduced growth even in rich medium, where compatible solutes would be available.
Peptidoglycan modifications are important to the short-term response to salt stress at 7°C.
It is well known that L. monocytogenes modifies the fatty acid content of phospholipids in the membrane and, therefore, the membrane fluidity to adapt to low temperature (2, 56). Here, we monitored gene expression changes during adaptation to salt stress in cultures that were already adapted to low temperature in order to identify temperature-specific adaptation processes to the high-salt environment. We did not observe the induction of genes involved in lipid biosynthesis but did see extensive changes in genes involved in peptidoglycan modifications, which only occurred at low temperature. We observed increased transcript levels of lmo2229, encoding a class A PBP, during the short-term adaptation to salt stress at 7°C; the homologous PBP in B. subtilis contributes to growth under high salinity (37). We observed increased transcript levels of two genes encoding DD carboxypeptidases, lmo2754 and lmo2812, during the short-term response to salt stress at 7°C, and the inactivation of lmo2754 led to a reduced ability to adapt to salt stress, as indicated by a significantly increased lag phase at 7°C. Both of these proteins have been shown to have DD carboxypeptidase activity (29), which functions to cleave peptidoglycan cross-links. In Lactobacillus casei, salt stress leads to decreased peptidoglycan cross-links (38), and DD carboxypeptidase is important for the adaptation of Ochrobactrum bacteria to high salinity (39). The DD carboxypeptidase lmo2754 protein level was significantly higher in L. monocytogenes at 4°C than when grown at 37°C (9), indicating that this may be the allele important for modifications at low temperature. DD carboxypeptidase expression under high-osmolyte and low-temperature conditions has been proposed as a means of maintaining elasticity in the cell wall under these conditions (7). As our data indicate that DD carboxypeptidase is important for L. monocytogenes to adapt to salt stress at low temperature, compounds that interfere with DD carboxypeptidase activity have potential as control agents for L. monocytogenes on foods during refrigerated storage.
Salt stress at low temperature activates the LiaFSR cell envelope stress response.
Many environmental stresses can affect the integrity of the cell envelope and have the potential to induce a cell envelope stress response. A number of cell envelope stress response systems are known in Gram-positive bacteria and are controlled by sigma factors and two-component response systems (26). Salt stress can induce a cell envelope stress response, but the regulators of that response may be different in each organism. For example, in B. subtilis, salt stress activates the cell envelope stress responses controlled by σW, σM, and σX (23). In L. monocytogenes, we observed activation of the LiaFSR system during the short-term adaptation to salt stress at low temperature. In Streptococcus mutans, LiaFSR plays a role in adaptation to salt stress, as well as to other cell envelope stresses; loss of function of LiaR led to reduced growth rates when exposed to ethanol, salt, vancomycin, and bacitracin (48). There is variation in the inducing signals, as well as the regulon genes, in the LiaFSR system among the firmicutes (16, 18, 53). Our data indicate that the LiaFSR system is induced by salt stress but does not contribute directly to the salt stress phenotype, which supports the idea that the LiaFSR system is a cell envelope damage-sensing system rather than a direct stress-sensing mechanism (53). Our data suggest that cell wall-damaging agents, such as bacteriocins, may be less effective in a high-salt environment, as induction of the LiaFSR system by salt stress may provide increased resistance to other cell wall stresses.
Adaptation to salt stress induced the transcription of genes that could lead to cross-protection.
It is well known that cross-protection, the increased resistance to a subsequent stress, can be induced by exposure to a nonlethal stress, and yet the underlying mechanisms that confer this additional stress resistance are just beginning to be revealed (6). Our transcriptome data suggest that opportunities for cross-protection occur both during the short-term response and the long-term response and that these effects may be temperature dependent. Under some stress conditions, cross-protective effects can be attributed to activation of the general stress response. For example, SigB-dependent cross-protection to osmotic stress occurs when L. monocytogenes is first exposed to alkaline stress (22). During the short-term adaptation to salt stress at both temperatures, we observed increased transcript levels of SigB and SigB-regulated genes, which has the potential to provide cross-protection to other stresses that rely on the general stress response for combating the stress.
Our data provide information about the molecular mechanisms that may provide cross-protection under certain stress combinations. In L. monocytogenes, exposure to salt stress has previously been reported to lead to cross-protection against bile stress (5); here, we show that adaptation to salt stress leads to elevated transcript levels of opuCA, betL, bilEAB, and bsh, which are all known to play a role in bile resistance (15, 45, 46). The exposure of L. monocytogenes to salt stress increases subsequent resistance to heat stress (42), and we observed that the short-term response to salt stress included increased transcript levels of dnaK, groES, groEL, htrA, and clpP, which encode proteins involved in the response to heat stress (19, 32, 54). We also showed that adaptation to salt stress led to increased transcript levels of kat, which coincides with increased resistance to hydrogen peroxide, a cross-protective effect that has also been demonstrated in B. cereus (14). The mechanisms involved in providing cross-protection of cells during long-term adaptation may be varied and, in many cases, are unknown. Our data set provides an opportunity to predict and evaluate other cross-protection responses that may occur due to salt stress at different temperatures and can be used to develop knowledge-based intervention strategies to control this pathogen in the food chain.
Supplementary Material
ACKNOWLEDGMENTS
We thank Silin Tang for assistance with growth experiments.
This work was supported by award number R01A1052151 from the National Institute of Allergy and Infectious Diseases.
The content is solely the responsibility of the authors and does not necessarily represent the official views of the National Institute of Allergy and Infectious Diseases or the National Institutes of Health.
Footnotes
Published ahead of print 3 February 2012
Supplemental material for this article may be found at http://aem.asm.org/.
REFERENCES
- 1. Abram F, et al. 2008. Identification of components of the sigma B regulon in Listeria monocytogenes that contribute to acid and salt tolerance. Appl. Environ. Microbiol. 74:6848–6858 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Badaoui Najjar MB, Chikindas M, Montville TJ. 2007. Changes in Listeria monocytogenes membrane fluidity in response to temperature stress. Appl. Environ. Microbiol. 73:6429–6435 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Baranyi J, Roberts TA. 1994. A dynamic approach to predicting bacterial growth in food. Int. J. Food Microbiol. 23:277–294 [DOI] [PubMed] [Google Scholar]
- 4. Becker LA, Cetin MS, Hutkins RW, Benson AK. 1998. Identification of the gene encoding the alternative sigma factor sigmaB from Listeria monocytogenes and its role in osmotolerance. J. Bacteriol. 180:4547–4554 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Begley M, Gahan CG, Hill C. 2002. Bile stress response in Listeria monocytogenes LO28: adaptation, cross-protection, and identification of genetic loci involved in bile resistance. Appl. Environ. Microbiol. 68:6005–6012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Begley M, Hill C. 2010. Food safety: what can we learn from genomics? Annu. Rev. Food Sci. Technol. 1:341–361 [DOI] [PubMed] [Google Scholar]
- 7. Bergholz PW, Bakermans C, Tiedje JM. 2009. Psychrobacter arcticus 273-4 uses resource efficiency and molecular motion adaptations for subzero temperature growth. J. Bacteriol. 191:2340–2352 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Bergholz TM, den Bakker HC, Fortes ED, Boor KJ, Wiedmann M. 2010. Salt stress phenotypes in Listeria monocytogenes vary by genetic lineage and temperature. Foodborne Pathog. Dis. 7:1537–1549 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Cacace G, et al. 2010. Proteomics for the elucidation of cold adaptation mechanisms in Listeria monocytogenes. J. Proteomics 73:2021–2030 [DOI] [PubMed] [Google Scholar]
- 10. Cetin MS, Zhang C, Hutkins RW, Benson AK. 2004. Regulation of transcription of compatible solute transporters by the general stress sigma factor, sigmaB, in Listeria monocytogenes. J. Bacteriol. 186:794–802 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Chan YC, et al. 2008. Contributions of two-component regulatory systems, alternative sigma factors, and negative regulators to Listeria monocytogenes cold adaptation and cold growth. J. Food Prot. 71:420–425 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Chan YC, Raengpradub S, Boor KJ, Wiedmann M. 2007. Microarray-based characterization of the Listeria monocytogenes cold regulon in log- and stationary-phase cells. Appl. Environ. Microbiol. 73:6484–6498 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Chatterjee SS, et al. 2006. Intracellular gene expression profile of Listeria monocytogenes. Infect. Immun. 74:1323–1338 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. den Besten HM, Mols M, Moezelaar R, Zwietering MH, Abee T. 2009. Phenotypic and transcriptomic analyses of mildly and severely salt-stressed Bacillus cereus ATCC 14579 cells. Appl. Environ. Microbiol. 75:4111–4119 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Dussurget O, et al. 2002. Listeria monocytogenes bile salt hydrolase is a PrfA-regulated virulence factor involved in the intestinal and hepatic phases of listeriosis. Mol. Microbiol. 45:1095–1106 [DOI] [PubMed] [Google Scholar]
- 16. Eldholm V, et al. 2010. The pneumococcal cell envelope stress-sensing system LiaFSR is activated by murein hydrolases and lipid II-interacting antibiotics. J. Bacteriol. 192:1761–1773 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Fraser KR, Sue D, Wiedmann M, Boor K, O'Byrne CP. 2003. Role of sigmaB in regulating the compatible solute uptake systems of Listeria monocytogenes: osmotic induction of opuC is sigmaB dependent. Appl. Environ. Microbiol. 69:2015–2022 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Fritsch F, et al. 2011. The cell envelope stress response mediated by the LiaFSR three-component system of Listeria monocytogenes is controlled via the phosphatase activity of the bifunctional histidine kinase LiaS. Microbiology 157:373–386 [DOI] [PubMed] [Google Scholar]
- 19. Gahan CG, O'Mahony J, Hill C. 2001. Characterization of the groESL operon in Listeria monocytogenes: utilization of two reporter systems (gfp and hly) for evaluating in vivo expression. Infect. Immun. 69:3924–3932 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Gardan R, Duche O, Leroy-Setrin S, Labadie J. 2003. Role of ctc from Listeria monocytogenes in osmotolerance. Appl. Environ. Microbiol. 69:154–161 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Garner MR, Njaa BL, Wiedmann M, Boor KJ. 2006. Sigma B contributes to Listeria monocytogenes gastrointestinal infection but not to systemic spread in the guinea pig infection model. Infect. Immun. 74:876–886 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Giotis ES, Julotok M, Wilkinson BJ, Blair IS, McDowell DA. 2008. Role of sigma B factor in the alkaline tolerance response of Listeria monocytogenes 10403S and cross-protection against subsequent ethanol and osmotic stress. J. Food Prot. 71:1481–1485 [DOI] [PubMed] [Google Scholar]
- 23. Hahne H, et al. 2010. A comprehensive proteomics and transcriptomics analysis of Bacillus subtilis salt stress adaptation. J. Bacteriol. 192:870–882 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Heyer LJ, Kruglyak S, Yooseph S. 1999. Exploring expression data: identification and analysis of coexpressed genes. Genome Res. 9:1106–1115 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Ito M, Guffanti AA, Oudega B, Krulwich TA. 1999. mrp, a multigene, multifunctional locus in Bacillus subtilis with roles in resistance to cholate and to Na+ and in pH homeostasis. J. Bacteriol. 181:2394–2402 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Jordan S, Hutchings MI, Mascher T. 2008. Cell envelope stress response in Gram-positive bacteria. FEMS Microbiol. Rev. 32:107–146 [DOI] [PubMed] [Google Scholar]
- 27. Jordan S, Junker A, Helmann JD, Mascher T. 2006. Regulation of LiaRS-dependent gene expression in Bacillus subtilis: identification of inhibitor proteins, regulator binding sites, and target genes of a conserved cell envelope stress-sensing two-component system. J. Bacteriol. 188:5153–5166 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Kajiyama Y, Otagiri M, Sekiguchi J, Kosono S, Kudo T. 2007. Complex formation by the mrpABCDEFG gene products, which constitute a principal Na+/H+ antiporter in Bacillus subtilis. J. Bacteriol. 189:7511–7514 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Korsak D, Markiewicz Z, Gutkind GO, Ayala JA. 2010. Identification of the full set of Listeria monocytogenes penicillin-binding proteins and characterization of PBPD2 (Lmo2812). BMC Microbiol. 10:239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Kosono S, et al. 2005. Characterization of a multigene-encoded sodium/hydrogen antiporter (sha) from Pseudomonas aeruginosa: its involvement in pathogenesis. J. Bacteriol. 187:5242–5248 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Li L, Stoeckert CJ, Jr, Roos DS. 2003. OrthoMCL: identification of ortholog groups for eukaryotic genomes. Genome Res. 13:2178–2189 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Nair S, Derre I, Msadek T, Gaillot O, Berche P. 2000. CtsR controls class III heat shock gene expression in the human pathogen Listeria monocytogenes. Mol. Microbiol. 35:800–811 [DOI] [PubMed] [Google Scholar]
- 33. Nelson KE, et al. 2004. Whole genome comparisons of serotype 4b and 1/2a strains of the food-borne pathogen Listeria monocytogenes reveal new insights into the core genome components of this species. Nucleic Acids Res. 32:2386–2395 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Olesen I, Vogensen FK, Jespersen L. 2009. Gene transcription and virulence potential of Listeria monocytogenes strains after exposure to acidic and NaCl stress. Foodborne Pathog. Dis. 6:669–680 [DOI] [PubMed] [Google Scholar]
- 35. Oliver HF, Orsi RH, Wiedmann M, Boor KJ. 2010. Listeria monocytogenes sigmaB has a small core regulon and a conserved role in virulence but makes differential contributions to stress tolerance across a diverse collection of strains. Appl. Environ. Microbiol. 76:4216–4232 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Ollinger J, Bowen B, Wiedmann M, Boor KJ, Bergholz TM. 2009. Listeria monocytogenes sigmaB modulates PrfA-mediated virulence factor expression. Infect. Immun. 77:2113–2124 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Palomino MM, Sanchez-Rivas C, Ruzal SM. 2009. High salt stress in Bacillus subtilis: involvement of PBP4* as a peptidoglycan hydrolase. Res. Microbiol. 160:117–124 [DOI] [PubMed] [Google Scholar]
- 38. Piuri M, Sanchez-Rivas C, Ruzal SM. 2005. Cell wall modifications during osmotic stress in Lactobacillus casei. J. Appl. Microbiol. 98:84–95 [DOI] [PubMed] [Google Scholar]
- 39. Principe A, Jofre E, Alvarez F, Mori G. 2009. Role of a serine-type D-alanyl-D-alanine carboxypeptidase on the survival of Ochrobactrum sp. 11a under ionic and hyperosmotic stress. FEMS Microbiol. Lett. 295:261–273 [DOI] [PubMed] [Google Scholar]
- 40. Radtke AL, et al. 2011. Listeria monocytogenes exploits cystic fibrosis transmembrane conductance regulator (CFTR) to escape the phagosome. Proc. Natl. Acad. Sci. U. S. A. 108:1633–1638 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Schmid B, et al. 2009. Role of cold shock proteins in growth of Listeria monocytogenes under cold and osmotic stress conditions. Appl. Environ. Microbiol. 75:1621–1627 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Skandamis PN, Yoon Y, Stopforth JD, Kendall PA, Sofos JN. 2008. Heat and acid tolerance of Listeria monocytogenes after exposure to single and multiple sublethal stresses. Food Microbiol. 25:294–303 [DOI] [PubMed] [Google Scholar]
- 43. Sleator RD, Clifford T, Hill C. 2007. Gut osmolarity: a key environmental cue initiating the gastrointestinal phase of Listeria monocytogenes infection? Med. Hypotheses 69:1090–1092 [DOI] [PubMed] [Google Scholar]
- 44. Sleator RD, Gahan CG, Hill C. 2003. A postgenomic appraisal of osmotolerance in Listeria monocytogenes. Appl. Environ. Microbiol. 69:1–9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Sleator RD, Watson D, Hill C, Gahan CG. 2009. The interaction between Listeria monocytogenes and the host gastrointestinal tract. Microbiology 155:2463–2475 [DOI] [PubMed] [Google Scholar]
- 46. Sleator RD, Wemekamp-Kamphuis HH, Gahan CG, Abee T, Hill C. 2005. A PrfA-regulated bile exclusion system (BilE) is a novel virulence factor in Listeria monocytogenes. Mol. Microbiol. 55:1183–1195 [DOI] [PubMed] [Google Scholar]
- 47. Stasiewicz MJ, Wiedmann M, Bergholz TM. 2011. The transcriptional response of Listeria monocytogenes during adaptation to growth on lactate and diacetate includes synergistic changes that increase fermentative acetoin production. Appl. Environ. Microbiol. 77:5294–5306 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Suntharalingam P, Senadheera MD, Mair RW, Levesque CM, Cvitkovitch DG. 2009. The LiaFSR system regulates the cell envelope stress response in Streptococcus mutans. J. Bacteriol. 191:2973–2984 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Swaminathan B, Cabanes D, Zhang W, Cossart P. 2007. Listeria monocytogenes, p 457–492 In Doyle MP, Beuchat LR. (ed), Food microbiology: fundamentals and frontiers, 3rd ed ASM Press, Washington, DC [Google Scholar]
- 50. Swartz TH, Ikewada S, Ishikawa O, Ito M, Krulwich TA. 2005. The Mrp system: a giant among monovalent cation/proton antiporters? Extremophiles 9:345–354 [DOI] [PubMed] [Google Scholar]
- 51. Utratna M, Shaw I, Starr E, O'Byrne CP. 2011. Rapid, transient, and proportional activation of sigma(B) in response to osmotic stress in Listeria monocytogenes. Appl. Environ. Microbiol. 77:7841–7845 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Walecka E, Molenda J, Karpiskova R, Bania J. 2011. Effect of osmotic stress and culture density on invasiveness of Listeria monocytogenes strains. Int. J. Food Microbiol. 144:440–445 [DOI] [PubMed] [Google Scholar]
- 53. Wolf D, et al. 2010. In-depth profiling of the LiaR response of Bacillus subtilis. J. Bacteriol. 192:4680–4693 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Wonderling LD, Wilkinson BJ, Bayles DO. 2004. The htrA (degP) gene of Listeria monocytogenes 10403S is essential for optimal growth under stress conditions. Appl. Environ. Microbiol. 70:1935–1943 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Zhang DL, Ross T, Bowman JP. 2010. Physiological aspects of Listeria monocytogenes during inactivation accelerated by mild temperatures and otherwise non-growth permissive acidic and hyperosmotic conditions. Int. J. Food Microbiol. 141:177–185 [DOI] [PubMed] [Google Scholar]
- 56. Zhu K, Bayles DO, Xiong A, Jayaswal RK, Wilkinson BJ. 2005. Precursor and temperature modulation of fatty acid composition and growth of Listeria monocytogenes cold-sensitive mutants with transposon-interrupted branched-chain alpha-keto acid dehydrogenase. Microbiology 151:615–623 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.