Abstract
To better understand the microbial functional diversity changes with subsurface redox conditions during in situ uranium bioremediation, key functional genes were studied with GeoChip, a comprehensive functional gene microarray, in field experiments at a uranium mill tailings remedial action (UMTRA) site (Rifle, CO). The results indicated that functional microbial communities altered with a shift in the dominant metabolic process, as documented by hierarchical cluster and ordination analyses of all detected functional genes. The abundance of dsrAB genes (dissimilatory sulfite reductase genes) and methane generation-related mcr genes (methyl coenzyme M reductase coding genes) increased when redox conditions shifted from Fe-reducing to sulfate-reducing conditions. The cytochrome genes detected were primarily from Geobacter sp. and decreased with lower subsurface redox conditions. Statistical analysis of environmental parameters and functional genes indicated that acetate, U(VI), and redox potential (Eh) were the most significant geochemical variables linked to microbial functional gene structures, and changes in microbial functional diversity were strongly related to the dominant terminal electron-accepting process following acetate addition. The study indicates that the microbial functional genes clearly reflect the in situ redox conditions and the dominant microbial processes, which in turn influence uranium bioreduction. Microbial functional genes thus could be very useful for tracking microbial community structure and dynamics during bioremediation.
INTRODUCTION
Uranium contamination of groundwater, sediment, and soil, initiated from uranium mining, processing, storage, and nuclear weapon production is a potential threat to human health and the natural environment. Uranium is present in oxic to suboxic waters and soils primarily as soluble uranyl species with high toxicity due to its bioavailability as a heavy metal and radiation source. A proposed method to decrease the risk of uranium contamination is to reduce highly soluble U(VI) to sparingly soluble U(IV) (17). The stimulation of microbial enzymatic reduction of U(VI) has shown a substantial promise for in situ bioremediation of uranium-contaminated groundwater, where organic compounds such as acetate, ethanol, or glucose were injected to the subsurface environment as electron donors (1, 20, 27, 33). Multiple electron acceptors, such as Mn(IV), Fe(III), NO3−, U(VI), and SO42−, in natural subsurface environments are used by microbes typically in sequence of energy yield. For example, Desulfovibrio vulgaris showed utilization of Fe(III) first, followed by U(VI), and finally sulfate in a competition experiment (7). In the field, nitrate has been shown to be reduced prior to the U(VI) and U(VI) reduction that often occurs simultaneously with Fe(III) reduction (1, 13). However, relatively few studies have focused on functional diversity of microbial communities with changes of subsurface redox conditions under in situ field conditions.
The Old Rifle site is located at a former uranium ore processing facility in Rifle, CO, where the subsurface aquifer was contaminated by uranium. The site is part of the uranium mill tailings remedial action (UMTRA) program of the U.S. Department of Energy. Field experiments conducted at the Old Rifle site demonstrate a decrease in soluble U(VI) from groundwater upon the addition of acetate to the subsurface and stimulation of endogenous microorganisms (1). Loss of soluble U(VI) correlated with the stimulation of Fe-reducing conditions in the subsurface and the enrichment of Geobacter spp., microorganisms known to reduce both Fe(III) and soluble U(VI) in the subsurface (1, 11, 15, 21, 28, 30). With continuous injection of acetate, sulfate was then used by microorganisms as the dominant electron acceptor. However, in some cases, an increase in U(VI) concentration was observed to be associated with a shift from Fe-reducing to sulfate-reducing conditions (1, 5). Thus, several questions were raised regarding factors that controlled the bioreduction of U(VI) and the specific microbial populations that were stimulated with a shift of redox conditions in the field experiments.
However, due to temporal and spatial changes in microbial diversity and the heterogeneity of environmental conditions, characterizing the microbial communities in an accurate and comprehensive way remains a challenge. The development and application of genomic tools have greatly advanced characterization and profiling of the microbial communities in complex environments. One such development, GeoChip 2.0 (10), is a comprehensive functional gene array. The GeoChip 2.0 contains 24,243 oligonucleotide probes and covers >10,000 genes in >150 functional groups involved in carbon, nitrogen, phosphorus, and sulfur cycling, metal reduction and resistance, and organic contaminant degradation, and has been demonstrated to be a robust tool for investigating biogeochemical, ecological and environmental processes from different habitats (16, 29, 31, 36).
In this study, GeoChip 2.0 was used to characterize microbial communities under Fe-reducing conditions and the shift from Fe-reducing to sulfate-reducing conditions during in situ uranium bioreduction. Two experimental plots were amended with acetate for stimulating microbial reduction of uranium. One was maintained mainly under Fe-reducing conditions, and the other was intentionally driven to conditions under which sulfate reduction dominated. The objectives of this study were to (i) determine the microbial functional diversity under Fe-reducing conditions and the transition from Fe-reducing to sulfate-reducing conditions and (ii) link geochemical changes to microbial functional diversity. Our results demonstrate a shift in the functional structure of microbial communities from Fe-reducing to sulfate-reducing conditions. The microbial community structure and functional dynamics changed in a manner consistent with geochemical differences associated with different redox conditions.
MATERIALS AND METHODS
Site description and plot design.
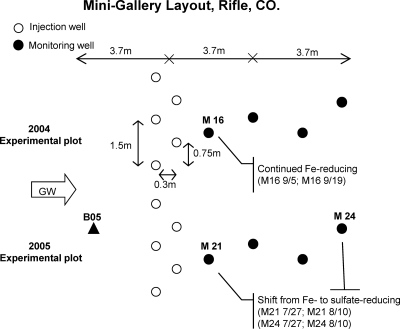
The Old Rifle UMTRA site is a flood plain of the Colorado River consisting of recent alluvium overlying the Eocene Wasatch Formation. The flood plain is approximately 2 km long, and virtually the entire site was contaminated as a result of a long-term vanadium and uranium milling operation. The groundwater flows at 0.46 to >0.61 m/day in a direction that normally parallels the Colorado River. The geology and hydrogeology of the site were described previously (1, 3, 5, 20, 28). In this experiment, two adjacent experimental plots of background, injection and down-gradient wells, one installed in 2004 and the other in 2005, were run simultaneously. The experimental plots had similar layouts to the 2005 experimental plot, 3.8 m to the southeast. Each experimental plot had five injection wells perpendicular to groundwater flow, four monitoring wells down-gradient of acetate injection, and one monitoring well positioned up-gradient of the injection wells (5) (Fig. 1). The 2004 experimental plot was amended with acetate for about 3 weeks to introduce Fe-reducing conditions. Amendment of the 2005 experimental plot was started earlier, and the subsurface was driven to sulfate-reducing conditions. Injections to both experimental plots were stopped simultaneously on 19 September 2006.
Fig 1.
Layout of the experimental plots at the Old Rifle uranium mill tailings site. Each plot had 5 injection wells (open circles) perpendicular to groundwater flow, 4 monitoring wells (filled circles) down-gradient of acetate injection, and 1 monitoring well positioned up-gradient of the injection wells (filled triangle). The 2004 experimental plot was maintained under Fe-reducing conditions, and the 2005 experimental plot was driven to sulfate-reducing conditions by using different durations of biostimulation.
Sampling.
Groundwater was sampled and analyzed from 17 July to 31 October in the two experimental plots (5). Groundwater (2 liters) was collected in sterile glass bottles using a peristaltic pump and kept on ice until it was delivered to the laboratory and then filtered (0.2-μm pore size) to collect biomass. Filters were stored at −80°C until DNA extraction. To better understand the microbial functional structure with subsurface redox changes, 8 groundwater samples, B05 (7/27 and 9/16), M16 (9/5 and 9/19), M21 (7/27 and 8/10), and M24 (7/27 and 8/10) were selected for microbial functional structure analysis with GeoChip. The 9/5 sample and 9/19 sample (M16) corresponded to the early days and the end of acetate injection in the 2004 experimental plot, where there was a continuous decrease of redox potential (Eh) under Fe-reducing conditions. The 7/27 and 8/10 sample set (M21 and M24) corresponded to the shift from iron-reducing to sulfate-reducing conditions in the 2005 experimental plot. The samples of B05 were used as a background control. Groundwater was pumped from the designated depth(s) in the monitoring wells using a portable peristaltic pump (Cole-Palmer Instrument Co.). The geochemical analysis of U(VI), Fe(II), bromide (potassium bromide as tracer), acetate, and sulfate and molecular analysis were described previously (5). The pH, dissolved oxygen (DO), sulfide, conductivity, and redox potential (Eh) of groundwater were determined in the field (5). Since only a trace level of nitrate was detected previously (1, 3), it was not considered in this experiment.
DNA isolation and purification.
Community DNA was extracted from groundwater filters by combining grinding and SDS for cell lysis and was purified as detailed by Zhou et al. (35). Purification was modified by elution of DNA from the resin column two times with 30 μl of hot water (80°C). The purified DNA was quantified with an ND-1000 spectrophotometer (Nanodrop, Inc.) and Quant-It PicoGreen (Invitrogen, Carlsbad, CA).
Sample amplification, labeling, microarray hybridization, and data processing.
An aliquot of DNA (25 ng) of each sample was amplified in triplicate using the TempliPhi kit (Amersham Biosciences, Piscataway, NJ) and labeled as described previously (31, 32). Hybridizations were performed with GeoChip 2.0 (10) on an HS4800 Pro hybridization station (Tecan US, Durham, NC) in triplicate at 45°C for 10 h. Microarrays were scanned on a ScanArray 5000 microarray analysis system (PerkinElmer, Wellesley, MA) at 95% laser power and 68% photomultiplier tube gain (PMT). Signal intensities were measured with ImaGene 6.0 (Biodiscovery, Inc., El Segundo, CA). Background was subtracted from all intensity data used for further analysis. Intensities of three replicates for each set of experiments were normalized with total intensities of all spots with a signal-to-noise ratio (SNR) greater than 1.0, where SNR = (signal intensity − background intensity)/background standard deviation). Spots with an SNR of <2.0 and outliers of replicates (>2 standard deviations) were removed. A gene was included in the analysis when a positive hybridization signal was obtained from >34% of the spots (generally, 9 spots for each gene) on the arrays in triplicate hybridizations.
Statistical analysis.
Cluster analysis was performed using the pairwise average-linkage hierarchical clustering algorithm (6) in CLUSTER (http://rana.stanford.edu/), and the results of hierarchical clustering were visualized using TREEVIEW (http://rana.lbl.gov/EisenSoftware.htm). The Bio-Env procedure was used to select environmental variables to find the best subset of environmental variables with maximum (rank) correlation with community dissimilarities (4) in R version 2.11.1 with the vegan package. Canonical correspondence analysis (CCA) was performed to identify the relationship between geochemical parameters and microbial functional genes using CANOCO for Windows version 4.5 (25). Monte Carlo tests were used to assess the significance of the environmental variables with 999 permutations. The Mantel test was performed to infer the correlation between geochemistry and functional genes based on Euclidean distance measurement with PC-ORD (MjM Software, Gleneden Beach, OR). The P value of the standardized Mantel statistic (r) was calculated from 999 Monte Carlo randomizations. All analyses of variances (14) were performed with SPSS 13.0 (SPSS, Inc., Chicago, IL) with Kruskal-Wallis test and variance homogeneity by Levene's test first.
Microarray data accession number.
The microarray data presented in this article are available at http://ieg.ou.edu/4download/.
RESULTS AND DISCUSSION
Field geochemical changes.
Geochemical analysis of the eight samples from four wells with different time points were selected for this study: acetate, U(VI), sulfate, sulfide, Fe(II), DO, pH, conductivity, and redox potential (Table 1). U(VI) concentrations in the background well and the two experimental plots were as high as 1.0 to ∼1.5 μM. There was a continuing decrease of U(VI) with acetate injection in the 2004 experimental plot, and the concentration was lowered to 0.19 μM on 19 September. In the 2005 experimental plot (M21 and M24), the U(VI) concentration decreased first, while there was a rebound of U(VI) with a shift of subsurface redox condition from Fe-reducing to sulfate-reducing conditions (5). In the same wells, there was a loss of sulfate and an accumulation of sulfide when they were dominated by sulfate-reducing conditions.
Table 1.
Groundwater geochemical data of samples selected for GeoChip analysis
| Sample | Concn of: |
pH | Conductivity (μS/cm) | Eh (mV) | |||||
|---|---|---|---|---|---|---|---|---|---|
| Acetate (mM) | U(VI) (μM) | Sulfate (mM) | Sulfide (mM) | Fe(II) (mM) | DO (mg/liter) | ||||
| B05 7/27/06 | 0.00 | 0.91 | 6.62 | 0.000 | 0.063 | 0.33 | 7.04 | 2,047 | 148 |
| B05 9/19/06 | 0.00 | 1.00 | 7.52 | 0.000 | 0.018 | 0.32 | 6.92 | 2,190 | 94 |
| M16 9/5/06 | 2.34 | 0.52 | 7.38 | 0.003 | 0.0246 | 0.36 | 6.87 | 2,431 | 84 |
| M16 9/19/06 | 4.44 | 0.19 | 7.34 | 0.010 | 0.034 | 0.33 | 6.92 | 2,659 | 11 |
| M21 7/27/06 | 6.87 | 0.44 | 7.21 | 0.009 | 0.124 | 0.16 | 7.08 | 2,936 | 33 |
| M21 8/10/06 | 2.30 | 0.36 | 6.54 | 0.114 | 0.038 | 0.11 | 7.21 | 2,465 | −129 |
| M24 7/27/06 | 2.10 | 0.24 | 7.05 | 0.004 | 0.127 | 0.1 | 7.02 | 2,383 | 40 |
| M24 8/10/06 | 0.83 | 0.50 | 5.80 | 0.096 | 0.013 | 0.2 | 7.19 | 2,429 | −141 |
Acetate additions were used to stimulate U(VI) reduction at the Rifle, CO, site(1, 5, 28, 34). At that site, continuous U(VI) reduction was observed until the dominant microbial communities shifted from Fe-reducing bacteria to sulfate-reducing bacteria, at which point U(VI) reduction slowed, ceased, or rebounded (1, 5). Furthermore, the greatest rate of reduction of uranium was observed under Fe-reducing conditions, consistent with previous work showing that Fe-reducing conditions were more favorable for uranium reduction in a laboratory bioreduction experiment (2).
Overall functional gene diversity pattern.
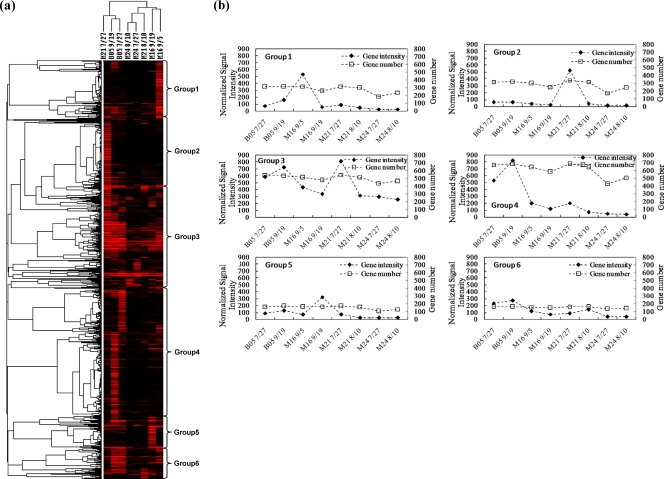
To track the microbial community dynamics during biostimulation, functional genes from selected samples in the wells with unstimulated background (B05), Fe-reducing dominance (M16), and a transition from Fe-reducing to sulfate-reducing conditions (M21 and M24) were analyzed with GeoChip 2.0. More than 1,300 genes showed positive hybridization signals. Hierarchical cluster analysis of all detected functional genes was performed (Fig. 2). The cluster analysis indicated that samples in treatment wells (M16, M21, and M24) and the background well (B05) grouped separately, except for the sample in the early days of acetate injection in well M21 (Fig. 2a). Samples in treatment wells grouped together mainly by Fe- or sulfate-reducing conditions, consistent with the expected correlation of the dominant terminal-electron-accepting process with microbial functional structure (Fig. 2a).
Fig 2.
Hierarchical cluster analysis of all functional genes detected (a). Genes that were present in at least three time points were used for cluster analysis. Results were generated in CLUSTER and visualized using TREEVIEW. Red indicates signal intensities above background, while black indicates signal intensities below background. Brighter red coloring indicates higher signal intensities. A total of 6 major groups were observed (b). The numbers equal groupings found among the hybridization patterns.
From the functional gene cluster results, a total of six major groups were observed (Fig. 2b). Groups 1 and 5 represented genes in high abundance at the beginning and the end of the Fe-reducing conditions, respectively. These groups mainly contained metal resistance and reduction genes such as chromium-, arsenic-, and tellurium-related genes as well as cytochrome genes. Groups 2 and 3 represented genes in high abundance in M21 7/27, which was closer to the injection well and referred to the transition from Fe-reducing to sulfate-reducing conditions, of which many sulfate-reducing genes (dsrA and dsrB) were observed. Groups 3, 4, and 6 represented genes in high abundance in the background wells. These groups of genes were mainly involved in carbon degradation, nitrogen cycling, and metal resistance. These results suggest that the overall functional structure of microbial communities is different with subsurface redox changes and that they were also different from those in the background well.
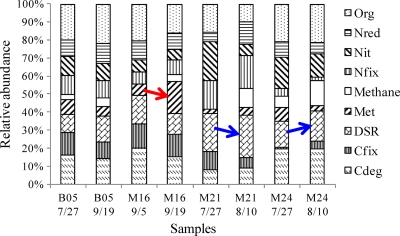
The composition of microbial communities was further analyzed in nine functional categories: carbon degradation, carbon fixation, sulfate reduction, metal reduction and resistance, nitrogen fixation, nitrification, nitrogen reduction, organic contaminant remediation, and methane generation (Fig. 3). Microbial functional gene patterns at the beginning and at the end of the study period in the background well were quite similar (B05 7/27 and B05 9/19). Metal reduction genes increased under Fe-reducing conditions (M16) over time. A previous study also showed that metal-reducing Deltaproteobacteria increased from 5% to nearly 40% by analyzing 16S rRNA gene clone libraries in contaminated subsurface sediments (22). The stimulation of both Fe-reducing bacteria and their metal reduction-related genes plays an important role in enzymatic uranium reduction.
Fig 3.
Functional gene abundance in the background well (B05), the Fe-reducing well (M16), and the shift from Fe-reducing to sulfate-reducing wells (M21 and M24). Abbreviations: Org, organic contaminant degradation; Nred, nitrogen reduction; Nit, nitrification; Nfix, nitrogen fixation; methane, methane generation; Met, metal reduction and resistance; DSR, sulfate reduction; Cfix, carbon fixation; Cdeg, carbon degradation.
When the redox condition shifted from Fe- to sulfate-reducing conditions, an increase in the abundance of sulfate reduction genes (dsrA and dsrB) was observed in M21 and M24, respectively (Fig. 3). Simultaneously, methane generation genes increased during the redox transition period in both M21 and M24 (Fig. 3). This transition was also observed where microbial communities shifted from predominance of metal-reducing Geobacteraceae populations to that of sulfate-reducing Desulfovibrionaceae (12). It was hypothesized that the decrease in U(VI) removal efficiency was due to a loss in the metal-reducing population (i.e., Geobacter) with a depletion of the bioavailable Fe(III) concentration, suggesting that redox conditions should be optimized for continued growth and survival of Geobacter species for long-term in situ uranium reduction (1).
Representatives of functional genes detected under Fe- and sulfate-reducing conditions. (i) Cytochrome genes for metal reduction.
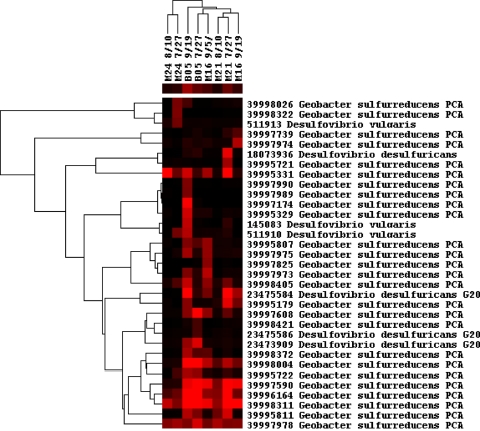
Biogeochemical and genetic studies both indicate that c-type cytochromes are required for U(VI) reduction by a range of microorganisms, including Desulfovibrio sp. (18), Geobacter sp. (23), and Shewanella oneidensis MR-1 (19). Here, we examine the cytochrome genes detected during uranium reduction under Fe-reducing conditions and the transition from Fe-reducing to sulfate-reducing conditions, as visualized by clustering analysis (Fig. 4). The cytochrome genes showed significant correlations (r = 0.71, P = 0.016) with uranium concentrations based on the Mantel test. In addition, cytochrome genes decreased during subsurface redox conditions shifted from Fe-reducing to sulfate-reducing conditions, consistent with the hypotheses that c-type cytochromes are directly involved in U(VI) reduction and that a decrease of U(VI) in groundwater is greatest under Fe-reducing conditions. The c-type cytochrome genes appear to be derived mainly from Geobacter sp. and Desulfovibrio sp., and major groups of cytochrome genes across all samples were from Geobacter sulfurreducens (Fig. 4), whose c-type cytochromes have been shown to be involved in extracellular uranium reduction (23).
Fig 4.
Hierarchical clustering of c-type cytochrome genes. Red indicates signal intensities above background, while black indicates signal intensities below background. Brighter red coloring indicates higher signal intensities. The bar of colors below the sample names indicates the average signal intensities of each sample. Mantel test results indicated significant correlations between the functional gene patterns and the U(VI) concentration (P < 0.05).
(ii) dsrA and -B for sulfate reduction.
To examine the shift of a microbial community composition dominated by sulfate reducers in more detail, total dsr genes were analyzed. In well M16, the number of dsr genes decreased when it was driven to Fe-reducing conditions, while in wells M21 and M24, it increased with the shift to sulfate-reducing conditions (see Table S1 in the supplemental material). The diversity increased in well M21 during sulfate reduction; most of the genes (95%) could be found in samples collected before and after acetate injection (see Table S2 in the supplemental material). Also, dsr genes in M21 showed high similarity to those in the background well (92% to 97%). Hierarchical clustering was performed to assess the dsr gene patterns (see Fig. S1 in the supplemental material). Compared to the Fe-reducing conditions, an obvious increase of dsr genes was detected when the subsurface was dominated by sulfate reduction. The dsr genes came from both cultured bacteria (e.g., Desulfomonile sp., Desulfosporosinus sp., Desulfotomaculum sp., Desulfovibrio sp., Desulfovirga sp., and Chlorobium sp.) and uncultured sulfate-reducing bacteria in well M21 at the late sampling date (8/10/06), when the sulfide concentration was highest. Several dsr genes from uncultured sulfate-reducing bacteria similar to Desulfosporosinus sp. and Pelotomaculum sp. were abundant in all samples.
(iii) mcr for methane generation.
In the background well, mcr genes were mainly derived from an uncultured archaeon and Methanothermobacter sp. (see Fig. S2 in the supplemental material). An increase of total signal intensity of methane generation genes (mcrA, mcrG, mcrC, and mcrD) were observed in well M21 7/27 in comparison to the background well (B05). Methane concentrations would need to be measured during biostimulation to confirm this. The increasing mcr-containing populations detected included primarily Methanoculleus sp., Methanocorpusculum sp., Methanocaldococcus sp., Methanothermobacter sp., Methanopyrus sp., and uncultured archaea.
(iv) nirS and -K and nifH for nitrogen cycling.
Functional genes nirS and -K, and nifH, coding for key enzymes involved in denitrification and nitrogen fixation, respectively, were examined. The nitrogen-reducing genes (nirS and -K) were present under both redox conditions as well as in the background well (see Fig. S3 in the supplemental material). It has been recognized that nitrogen-reducing bacteria are essential for the removal of nitrate to create the low-redox conditions favorable for U(VI) reduction (8, 26). Since only a trace level of nitrate was detected in the Old Rifle site (1, 3), no clear relationship between nitrogen-reducing genes and the redox conditions was observed in this study. These genes were likely derived from populations of Pseudomonas sp., Sinorhizobium sp., Nitrosomonas sp., Ochrobactrum sp., and Paracoccus sp., yet the majority of the nitrogen cycling genes were from uncultured bacteria. Similarly, most of the nifH genes observed were from uncultured bacteria (see Fig. S4 in the supplemental material).
(v) Carbon degradation-related genes.
A variety of carbon degradation genes were detected in the samples (see Fig. S5 in the supplemental material). The functional genes for cellulase from Leuconostoc sp., chitinase from Salinivibrio sp. and Burkholderia sp., and laccase from Trametes sp. were detected across all samples. Additionally, genes for cellulase from Clostridium sp. and Gluconacetobacter sp., chitinase from Serratia sp., Xanthomonas sp., and Burkholderia sp., and polygalacturonase (pgl) from Penicillium sp. were highly abundant in the background well. In the Fe-reducing well, genes for cellulase from Neurospora sp., Reticulitermes sp., Clostridium sp., Xanthomonas sp., and Fusarium sp., chitinase from Aeromonas sp., laccase from Trametes sp. and Basidiomycete sp., and pgl from Xanthomonas sp. were also highly abundant. In the sulfate-reducing well, the cellulase genes from Clostridium sp., Fusarium sp., Halobacterium sp., and Gluconacetobacter sp., chitinase genes from Serratia sp., Xanthomonas sp., and Microbulbifer sp., the mannanase gene from Cellvibrio sp., and pgl from Xanthomonas sp. were in high abundance. Several different species of Clostridium have been shown to reduce U(VI) to U(IV) to various degrees (2, 24), and the hypothesis was proposed that U(VI) reduction occurred through hydrogenases and other enzymes (9). However, hydrogenase genes derived from Clostridium sp. were not contained on the GeoChip, and further work will be required to confirm the mechanism of bioreduction of U(VI) by Clostridium sp. Detrital organic matter that is locally abundant at the Rifle site is probably the principal source of carbon compounds sustaining this part of the microbial community.
Linking geochemistry and microbial community structure.
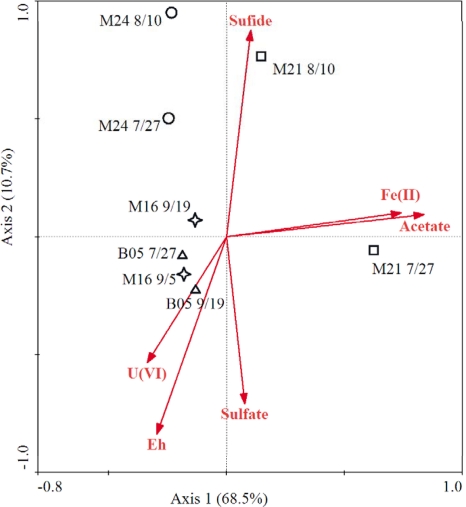
The responses of the microbial community to subsurface redox conditions can be very different based on the local environmental conditions and the biostimulation operations (i.e., exogenous substrate injection) (26). The Bio-Env procedure was used to find the best subset of environmental variables with maximum (rank) correlation with community dissimilarities, and the result indicated that acetate, U(VI), and Eh had the highest correlation coefficient (r = 0.579). Canonical correspondence analysis (CCA) was performed to discern possible linkages between geochemical parameters and microbial community functional structure (Fig. 5). Only geochemical parameters that were significant were included in the CCA biplot [acetate, U(VI), sulfate, sulfide, Fe(II), and Eh], based on a forward selection procedure and variance inflation factors with 999 Monte Carlo permutations. For all functional genes, 37.1% of the total variance could be explained by the first two constrained axes, with the first axis explaining 19.5%. The specified model was significant (first axis, P = 0.019; all axes, P = 0.040). The CCA results reflected the microbial functional distributions among the environmental variables such as U(VI), acetate, Eh, sulfate, sulfide, and Fe(II) (Fig. 5). B05 samples distributed in the area with highest U(VI). In the Fe-reducing well (M16), the change of microbial structure was correlated with Eh. With continuous decrease of Eh and accumulation of sulfide, a clear shift of microbial structure was observed with transition from Fe-reducing to sulfate-reducing conditions (wells M21 and M24, from 7/27 to 8/10). The results here indicated that changes of microbial functional diversity were strongly related to the dominant terminal-electron-accepting process following acetate addition.
Fig 5.
Canonical correspondence analysis (CCA) of total functional genes and geochemical data.
In summary, this study characterized the functional structure and dynamics of microbial communities under Fe-reducing and sulfate-reducing conditions during in situ U(VI) bioreduction. Analysis of key functional genes indicated a shift of functional potential of microbial communities in response to the transition from Fe reduction to sulfate reduction as the dominant terminal-electron-accepting process. As a result, an increase of dsr and mcr genes was detected when the subsurface was dominated by sulfate-reducing conditions. Also, the c-type cytochrome genes detected were primarily from Geobacter sp. and Desulfovibrio sp. and decreased when subsurface redox conditions shifted from Fe-reducing to sulfate-reducing conditions. Overall, microbial functional structure is sensitive to changes in subsurface redox conditions, indicating that tracking microbial functional genes could be very useful for tracking microbial community structure and dynamics during bioremediation.
Supplementary Material
ACKNOWLEDGMENTS
This research was supported by the Office of Science, Office of Biological and Environmental Research, of the U.S. Department of Energy under contract no. DE-AC02-05CH11231 as part of ENIGMA, the Oklahoma Center for the Advancement of Science and Technology under the Oklahoma Applied Research Support Program, the State Key Joint Laboratory of Environment Simulation and Pollution Control (grant 11Z03ESPCT) at Tsinghua University, National Natural Scientific Foundation of China (no. 40730738 and 41101233), and the Natural Scientific Foundation of Jiangsu province (no. BK2011233). The field experiment and biogeochemical sample analyses were supported by the U.S. Department of Energy under the Environmental Remediation Sciences Program.
Footnotes
Published ahead of print 10 February 2012
Supplemental material for this article may be found at http://aem.asm.org/.
REFERENCES
- 1. Anderson R, et al. 2003. Stimulating the in situ activity of Geobacter species to remove uranium from the groundwater of a uranium-contaminated aquifer. Appl. Environ. Microbiol. 69:5884–5891 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Boonchayaanant B, Nayak D, Xin D, Criddle C. 2009. Uranium reduction and resistance to reoxidation under iron-reducing and sulfate-reducing conditions. Water Res. 43:4652–4664 [DOI] [PubMed] [Google Scholar]
- 3. Chang Y, et al. 2005. Microbial incorporation of C-13-labeled acetate at the field scale: detection of microbes responsible for reduction of U(VI). Environ. Sci. Technol. 39:9039–9048 [DOI] [PubMed] [Google Scholar]
- 4. Clarke KR, Ainsworth M. 1993. A method of linking multivariate community structure to environmental variables. Mar. Ecol. Prog. Ser. 92:205–219 [Google Scholar]
- 5. Druhan J, et al. 2008. Sulfur isotopes as indicators of amended bacterial sulfate reduction processes influencing field scale uranium bioremediation. Environ. Sci. Technol. 42:7842–7849 [DOI] [PubMed] [Google Scholar]
- 6. Eisen M, Spellman P, Brown P, Botstein D. 1998. Cluster analysis and display of genome-wide expression patterns. Proc. Natl. Acad. Sci. U. S. A. 95:14863–14868 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Elias D, Suflita J, McInerney M, Krumholz L. 2004. Periplasmic cytochrome c3 of Desulfovibrio vulgaris is directly involved in H2-mediated metal but not sulfate reduction. Appl. Environ. Microbiol. 70:413–420 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Finneran KT, Housewright M, Lovley DR. 2002. Multiple influences of nitrate on uranium solubility during bioremediation of uranium contaminated subsurface sediments. Environ. Microbiol. 4:510–516 [DOI] [PubMed] [Google Scholar]
- 9. Gao W, Francis AJ. 2008. Reduction of uranium(VI) to uranium(IV) by clostridia. Appl. Environ. Microbiol. 74:4580–4584 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. He Z, et al. 2007. GeoChip: a comprehensive microarray for investigating biogeochemical, ecological and environmental processes. ISME J. 1:67–77 [DOI] [PubMed] [Google Scholar]
- 11. Holmes D, et al. 2007. Subsurface clade of Geobacteraceae that predominates in a diversity of Fe(III)-reducing subsurface environments. ISME J. 1:663–677 [DOI] [PubMed] [Google Scholar]
- 12. Hwang C, et al. 2009. Bacterial community succession during in situ uranium bioremediation: spatial similarities along controlled flow paths. ISME J. 3:47–64 [DOI] [PubMed] [Google Scholar]
- 13. Istok J, et al. 2004. In situ bioreduction of technetium and uranium in a nitrate-contaminated aquifer. Environ. Sci. Technol. 38:468–475 [DOI] [PubMed] [Google Scholar]
- 14. Ivanova I, et al. 2000. A survey of 16S rRNA and amoA genes related to autotrophic ammonia-oxidizing bacteria of the beta-subdivision of the class Proteobacteria in contaminated groundwater. Can. J. Microbiol. 46:1012–1020 [DOI] [PubMed] [Google Scholar]
- 15. Komlos J, Moon HS, Jaffe PR. 2008. Effect of sulfate on the simultaneous bioreduction of iron and uranium. J. Environ. Qual. 37:2058–2062 [DOI] [PubMed] [Google Scholar]
- 16. Liang Y, et al. 2011. Functional gene diversity of soil microbial communities from five oil-contaminated fields in China. ISME J. 5:403–413 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Lovley D, Phillips E, Gorby Y, Landa E. 1991. Microbial reduction of uranium. Nature 350:413–416 [Google Scholar]
- 18. Lovley D, Widman P, Woodward J, Phillips E. 1993. Reduction of uranium by cytochrome c3 of Desulfovibrio vulgaris. Appl. Environ. Microbiol. 59:3572–3576 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Marshall M, et al. 2006. c-type cytochrome-dependent formation of U(IV) nanoparticles by Shewanella oneidensis. PLoS Biol. 4:1324–1333 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. N′Guessan A, Vrionis H, Resch C, Long P, Lovley D. 2008. Sustained removal of uranium from contaminated groundwater following stimulation of dissimilatory metal reduction. Environ. Sci. Technol. 42:2999–3004 [DOI] [PubMed] [Google Scholar]
- 21. N′Guessan AL, et al. 2010. Molecular analysis of phosphate limitation in Geobacteraceae during the bioremediation of a uranium-contaminated aquifer. ISME J. 4:253–266 [DOI] [PubMed] [Google Scholar]
- 22. North N, et al. 2004. Change in bacterial community structure during in situ biostimulation of subsurface sediment cocontaminated with uranium and nitrate. Appl. Environ. Microbiol. 70:4911–4920 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Shelobolina E, et al. 2007. Importance of c-type cytochromes for U(VI) reduction by Geobacter sulfurreducens. BMC Microbiol. 7:16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Tebo B, Obraztsova A. 1998. Sulfate-reducing bacterium grows with Cr(VI), U(VI), Mn(IV), and Fe(III) as electron acceptors. FEMS Microbiol. Lett. 162:193–198 [Google Scholar]
- 25. ter Braak CJF, Smilauer P. 1998. CANOCO reference manual and user's guide to CANOCO for windows: software for canonical community ordination, version 4ed. Microcomputer Power, New York, NY [Google Scholar]
- 26. Van Nostrand JD, et al. 2011. Dynamics of microbial community composition and function during in situ bioremediation of a uranium-contaminated aquifer. Appl. Environ. Microbiol. 77:3860–3869 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Van Nostrand JD, et al. 2009. GeoChip-based analysis of functional microbial communities during the reoxidation of a bioreduced uranium-contaminated aquifer. Environ. Microbiol. 11:2611–2626 [DOI] [PubMed] [Google Scholar]
- 28. Vrionis H, et al. 2005. Microbiological and geochemical heterogeneity in an in situ uranium bioremediation field site. Appl. Environ. Microbiol. 71:6308–6318 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Wang F, et al. 2009. GeoChip-based analysis of metabolic diversity of microbial communities at the Juan de Fuca Ridge hydrothermal vent. Proc. Natl. Acad. Sci. U. S. A. 106:4840–4845 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Wilkins MJ, et al. 2009. Proteogenomic monitoring of Geobacter physiology during stimulated uranium bioremediation. Appl. Environ. Microbiol. 75:6591–6599 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Wu L, Kellogg L, Devol A, Tiedje J, Zhou J. 2008. Microarray-based characterization of microbial community functional structure and heterogeneity in marine sediments from the Gulf of Mexico. Appl. Environ. Microbiol. 74:4516–4529 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Wu LY, Liu X, Schadt CW, Zhou JZ. 2006. Microarray-based analysis of subnanogram quantities of microbial community DNAs by using whole-community genome amplification. Appl. Environ. Microbiol. 72:4931–4941 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Xu M, et al. 2010. Responses of microbial community functional structures to pilot-scale uranium in situ bioremediation. ISME J. 4:1060–1070 [DOI] [PubMed] [Google Scholar]
- 34. Yabusaki SB, et al. 2007. Uranium removal from groundwater via in situ biostimulation: field-scale modeling of transport and biological processes. J. Contam. Hydrol. 93:216–235 [DOI] [PubMed] [Google Scholar]
- 35. Zhou J, Bruns M, Tiedje J. 1996. DNA recovery from soils of diverse composition. Appl. Environ. Microbiol. 62:316–322 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Zhou J, Kang S, Schadt C, Garten C. 2008. Spatial scaling of functional gene diversity across various microbial taxa. Proc. Natl. Acad. Sci. U. S. A. 105:7768–7773 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.