Abstract
Analysis of the 16S rRNA gene sequences generated from Xerocomus pruinatus and Scleroderma citrinum ectomycorrhizospheres revealed that similar bacterial communities inhabited the two ectomycorrhizospheres in terms of phyla and genera, with an enrichment of the Burkholderia genus. Compared to the bulk soil habitat, ectomycorrhizospheres hosted significantly more Alpha-, Beta-, and Gammaproteobacteria.
TEXT
Ectomycorrhizal fungi are important actors of nutrient cycling in the forest ecosystems. They enhance the nutrient uptake capacity of plants due to their ability to mobilize carbon from organic matter and access chemical elements with low mobility in the soil, such as nutritive cations, phosphorus, and nitrogen (5, 15, 32). They also connect tree roots to the surrounding soil and form a very specific ecological environment, the ectomycorrhizosphere (19). Apart from its physical and chemical characteristics, which differ from those of the rhizosphere (26, 28), the ectomycorrhizosphere is characterized by diverse fungal and bacterial communities that inhabit the same environment. Consequently, the functioning of the ectomycorrhizal symbiosis is influenced by each partner of the ectomycorrhizal (ECM) complex (6, 8, 9).
Cultivation-dependent and -independent studies have demonstrated the structuring effect of the mycorrhizal fungi on the soil bacterial communities. They revealed that the bacterial communities colonizing the ectomycorrhizal roots differed from those of uncolonized roots and that the ectomycorrhizal species differentially impacted the structure of ectomycorrhizosphere bacterial communities (2, 11, 12, 22, 35, 37). One mycorrhizal species could be colonized by either very similar or contrastingly different bacterial communities (2, 11, 13). Certain ectomycorrhizal species associated with Betula pubescens, such as Piloderma fallax or Pseudotomentella tristis, were colonized by distinct bacterial communities, whereas the bacterial communities colonizing the ectomycorrhizosphere of Tomentellopsis submollis or Lactarius torminosus were more similar (13), thus suggesting that the taxonomic classification of the host mycorrhizal fungi was not always the main structuring parameter of the bacterial communities. Within this context, a comprehensive description of the ectomycorrhizosphere bacterial communities using a pyrosequencing-based approach would detail the structure and diversity of the bacterial communities coexisting in this specific ecological habitat as well as impart access to the rare taxonomic groups.
In a recent study, Uroz et al. (36) used pyrosequencing of 16S rRNA fragments to compare the composition of bacterial communities inhabiting the oak (Quercus petraea) rhizosphere and surrounding bulk soil. In the study presented here, we investigated in the same soil core the composition and structure of bacterial communities inhabiting the ectomycorrhizosphere, which is a specific subhabitat of the rhizosphere. Pyrosequencing tags spanning the V5 to V6 hypervariable regions of the 16S rRNA gene were used to compare the bacterial communities colonizing the Scleroderma citrinum and Xerocomus pruinatus ectomycorrhizospheres. The species richness and the numbers of operational taxonomic units (OTUs) in the ectomycorrhizosphere bacterial communities were compared between the two ectomycorrhizospheres. They were also compared to the data obtained for the rhizosphere and the surrounding bulk soil.
Soil samples were recovered from three independent soil cores in an oak (Quercus petraea) forest located in Breuil-Chenue, France. Dominant ectomycorrhizal morphotypes in each soil core were collected in separated tubes to avoid contaminations between morphotypes. These morphotypes were identified as Scleroderma citrinum (samples C3B, C4A, and C4B) and Xerocomus pruinatus (samples C1B and C3A) according to Agerer's (1987 to 1998) descriptions and the sequencing of the fungal internal transcribed spacer. The mycorrhizal tips, which are a combination of the soil-ectomycorrhiza interface and the symbiotic fungal mantle, were considered ectomycorrhizosphere samples in this study. Samples analyzed in this study were compared to the surrounding bulk soil (BS) and rhizosphere (R) samples described previously (36). DNA was extracted from three mycorrhizal tips of S. citrinum and two mycorrhizal tips of X. pruinatus using the PowerSoil DNA isolation kit (MO BIO Laboratories, Inc.). Amplicon libraries were generated as recommended for 454 pyrosequencing using a combination of two tagged primers targeting the V5 and V6 variable regions of the 16S rRNA gene, using the primers 787r (5′-AxxxATTAGATACCYTGTAGTCC-3′) (23) and 1073f (5′-B-ACGAGCTGACGACARCCATG-3′) (25) to generate PCR 16S rRNA fragments of ca. 250 bp, where A and B represent the linkers CCATCTCATCCCTGCGTGTCTCCGACTCAG and CCTATCCCCTGTGTGCCTTGGCAGTCTCAG and xxx represents the sample identification bar coding key (tag). Pyrosequencing resulted in 158,690 reads (average size, 286 bp) which passed the length and quality criteria (7). MOTHUR was used to trim, denoise, and align the reads and to generate the operational taxonomic units (OTUs; 97% sequence similarity) as well as to perform the nonparametric analyses (31). Taxonomic assignments were obtained with the metagenomics RAST server (MG-RAST) (21) using an 80% confidence threshold according to the methods of Santelli et al. (30). The impact of the fungal species on the relative distributions of the phyla and genera was determined by analysis of variance (one-factor ANOVA) using the SuperANOVA software (Abacus Concepts, Inc., Berkeley, CA).
Nonparametric analyses revealed that the total numbers of OTUs observed were not significantly different between the ectomycorrhizospheres (see Table S1 in the supplemental material). However, for a similar number of 16S rRNA tag sequences corresponding to sample C1B (n = 28,516 reads), a higher number of OTUs was obtained for the Scleroderma citrinum ectomycorrhizosphere (12,523 OTUs) (see Table S1 in the supplemental material). As for other soil studies (27, 36), the rarefaction curves generated for each ectomycorrhizosphere did not reach a plateau (see Fig. S1 in the supplemental material). The Chao1 index estimated that the ectomycorrhizosphere samples contained between 31,000 and 39,000 OTUs (see Table S1 in the supplemental material). Altogether, these analyses highlighted the richness of the ectomycorrhizosphere bacterial communities and the relative overlap existing between the different ectomycorrhizospheres analyzed.
Taxonomic assignments demonstrated that the same 13 phyla were present in each sample, regardless of their ecological origin (Xerocomus pruinatus or Scleroderma citrinum ectomycorrhizosphere) and sampling location (Table 1). In each ectomycorrhizosphere, four major phyla were dominant, the Proteobacteria (mean value, 54.89% ± 3.22%; n = 5), the Acidobacteria (mean value, 19.35% ± 2.87%; n = 5), the Bacteroidetes (mean value, 4.75% ± 0.79%; n = 5), and the Actinobacteria (mean value, 4.39% ± 0.30%; n = 5) (Table 1). Similarly, the same genera were detected in both ectomycorrhizospheres (see Fig. S2 in the supplemental material). Analysis of the 10 most abundant genera showed that the genera Acidobacterium (mean value, 19.35% ± 2.88%; n = 5), Burkholderia (mean value, 6.46% ± 1.11%; n = 5), Rhodoplanes (mean value, 4.70% ± 0.38%; n = 5), Chitinophaga (mean value, 4.28% ± 0.77%; n = 5), and Bradyrhizobium (mean value, 4.37% ± 0.22%; n = 5) were dominant (Table 2), regardless of the type of ectomycorrhizosphere (S. citrinum or X. pruinatus). For both levels (phyla or genera), no significant differences were observed between the X. pruinatus and S. citrinum ectomycorrhizospheres (P > 0.05) (see Fig. S3A in the supplemental material).
Table 1.
Relative abundances of the taxonomic groups present in the oak ectomycorrhizosphere, rhizosphere, and surrounding bulk soila
| Taxonomic group | Relative abundance (%) in each soil source |
Statistics (P value) | ||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| BS |
R |
Myc Xp |
Myc Sc |
|||||||||
| BS1 | BS2 | BS3 | R1 | R2 | R3 | C1B | C3A | C3B | C4A | C4B | ||
| Proteobacteria | 35.57 | 35.85 | 37.76 | 41.03 | 37.81 | 40.04 | 64.32 | 58 | 56.8 | 48.40 | 46.95 | Myc > BS = R (0.004) |
| Acidobacteria | 26.45 | 25.98 | 21.95 | 23.10 | 26.59 | 19.52 | 16.85 | 13.36 | 14.06 | 25.66 | 26.82 | NS |
| Unclassified bacteria | 21.00 | 20.44 | 22.16 | 18.27 | 18.59 | 19.99 | 10.09 | 8.64 | 8.31 | 14.19 | 12.42 | Myc < R < BS (0.0007) |
| Actinobacteria | 11.29 | 12.72 | 11.43 | 11.38 | 11.52 | 10.79 | 3.85 | 3.86 | 3.99 | 5.01 | 5.24 | Myc < BS = R (0.0001) |
| Bacteroidetes | 1.13 | 1.17 | 1.88 | 2.30 | 1.91 | 2.09 | 2.47 | 6 | 6.23 | 3.18 | 5.87 | Myc Sc > BS (0.02) |
| Gemmatimonadetes | 1.14 | 0.74 | 1.58 | 1.09 | 0.88 | 0.92 | 0.18 | 0.28 | 0.26 | 0.34 | 0.17 | Myc < BS = R (0.0013) |
| Verrucomicrobia | 1.18 | 1.07 | 1.09 | 1.01 | 1.28 | 2.43 | 0.60 | 0.93 | 0.86 | 1.02 | 0.85 | NS |
| Planctomycetes | 0.66 | 0.63 | 0.54 | 0.47 | 0.52 | 2.29 | 0.25 | 0.51 | 0.52 | 0.38 | 0.31 | NS |
| Firmicutes | 0.54 | 0.50 | 0.40 | 0.44 | 0.29 | 0.78 | 0.29 | 0.24 | 0.27 | 0.29 | 0.20 | NS |
| Chlamydiae | 0.46 | 0.57 | 0.75 | 0.40 | 0.31 | 0.72 | 0.16 | 0.13 | 0.12 | 0.09 | 0.14 | Myc < BS = R (0.002) |
| Nitrospira | 0.31 | 0.15 | 0.23 | 0.22 | 0.07 | 0.13 | 0.01 | 0.02 | 0 | 0.06 | 0.02 | Myc < BS = R (0.008) |
| Genera incertae sedis OP10 | 0.10 | 0.06 | 0.12 | 0.14 | 0.10 | 0.12 | 0.04 | 0.05 | 0.04 | 0.05 | 0.10 | NS |
| Genera incertae sedis TM7 | 0.06 | 0.06 | 0.07 | 0.08 | 0.06 | 0.13 | 0.01 | 0.01 | 0.01 | 0.03 | 0.03 | NS |
The data generated in this study are presented in bold in the table. These data are compared with the data published by Uroz et al. (36) on the distribution of the bacterial communities in the rhizosphere and in the surrounding bulk soil. Because all the samples have been collected at the same time and treated using the same methods, a one-factor (niche) ANOVA at a threshold level of P = 0.05 and a Bonferroni-Dunn test were applied on the relative distribution values after an arcsine transformation. The results are presented in the column entitled “Statistics.” The taxonomic groups for which a significant or nearly significant effect of the niche was found are presented. NS, nonsignificant differences; Myc, both Xerocomus pruinatus and Scleroderma citrinum ectomycorrhizospheres; Myc Xp, Xerocomus pruinatus ectomycorrhizosphere; Myc Sc, Scleroderma citrinum ectomycorrhizosphere; BS, bulk soil; R, rhizosphere; >, significantly more present; <, significantly less present.
Table 2.
Relative abundances of the 10 most abundant genera in each of the ectomycorrhizospheresa
| Rank | Most abundant genera (relative abundance [%]) in each soil source |
||||
|---|---|---|---|---|---|
|
Xerocomus pruinatus mycorrhizosphere |
Scleroderma citrinum mycorrhizosphere |
||||
| C1B | C3A | C3B | C4A | C4B | |
| 1 | Acidobacterium (16.85) | Acidobacterium (13.36) | Acidobacterium (14.06) | Acidobacterium (25.66) | Acidobacterium (26.82) |
| 2 | Burkholderia (7.99) | Burkholderia (8.33) | Burkholderia (8.44) | Burkholderia (3.30) | Burkholderia (4.24) |
| 3 | Chitinophaga (2.03) | Chitinophaga (5.61) | Chitinophaga (5.82) | Chitinophaga (2.86) | Chitinophaga (5.09) |
| 4 | Rhodoplanes (3.42) | Rhodoplanes (5.14) | Rhodoplanes (4.82) | Rhodoplanes (5.72) | Rhodoplanes (4.42) |
| 5 | Bradyrhizobium (3.90) | Bradyrhizobium (4.97) | Bradyrhizobium (4.75) | Bradyrhizobium (3.87) | Bradyrhizobium (4.37) |
| 6 | Caulobacter (6.40) | Caulobacter (1.95) | Caulobacter (1.87) | Caulobacter (0.46) | Caulobacter (0.40) |
| 7 | Curtobacterium (1.58) | Curtobacterium (0.92) | Curtobacterium (1.02) | Curtobacterium (1.61) | Curtobacterium (2.02) |
| 8 | Alterococcus (2.70) | Alterococcus (0.95) | Alterococcus (0.86) | Alterococcus (1.34) | Alterococcus (0.74) |
| 9 | Phenylobacterium (0.35) | Planctomyces (0.38) | Planctomyces (0.39) | Gemmatimonas (0.34) | Pedobacter (0.58) |
| 10 | Brevundimonas (0.34) | Conexibacter (0.32) | Rhodanobacter (0.31) | Acidimicrobium (0.26) | Nocardia (0.29) |
Shading indicates the position of the genus Burkholderia.
Overall, the 158,690 read sequences generated in this study defined 47,292 OTUs. The 10 most abundant OTUs detected in the ectomycorrhizosphere, regardless of the type of ectomycorrhizal fungus, belonged to Rhizobiales, Gammaproteobacteria (Steroidobacter spp.), Burkholderia, Acidobacteria, Bacteroidetes, and Actinobacteria (data not shown). The detailed list of the 10 most abundant OTUs for each ectomycorrhizosphere is presented in Table 3. An in-depth analysis revealed that 42% of the total sequences (66,951 sequences distributed in 1,266 OTUs) were common to all the ectomycorrhizosphere samples. This result suggests a common bacterial core between the two mycorrhizal species but also a relative heterogeneity between the different samples regardless of the fungal species. Around 1% of the total sequences appeared specific to the X. pruinatus ectomycorrhizospheres, corresponding to 584 OTUs (1,694 sequences). Similarly, 0.85% of the total sequences were specific to the S. citrinum ectomycorrhizospheres, corresponding to 265 OTUs (1,367 sequences). Both ectomycorrhizospheres appeared dominated by OTUs related to Proteobacteria and Acidobacteria. These relatively low proportions of phylotypes specific to the X. pruinatus or S. citrinum ectomycorrhizospheres suggest a low impact of the fungal species on the associated bacterial communities, a finding which is in line with those of previous studies which have suggested that other environmental factors, such as soil characteristics, strongly influence bacterial distribution (2, 13).
Table 3.
Relative abundances of the 10 most abundant phylotypes in each of the ectomycorrhizospheresa
| Rank | Most abundant phylotypes (relative abundance [%]) in each soil source |
||||
|---|---|---|---|---|---|
|
Xerocomus pruinatus mycorrhizosphere |
Scleroderma citrinum mycorrhizosphere |
||||
| C1B | C3A | C3B | C4A | C4B | |
| 1 | Steroidobacter sp. (1.14) | Burkholderia sp. (0.93) | Rhizobiales (0.72) | Steroidobacter sp. (0.65) | Bradyrhizobium sp. (0.74) |
| 2 | Bradyrhizobium elkanii (0.92) | Steroidobacter sp. (0.64) | Burkholderia sp. (0.57) | Bradyrhizobium sp. (0.58) | Flavosolibacter sp. (0.52) |
| 3 | Steroidobacter sp. (0.88) | Rhizobium miluonense (0.64) | Burkholderia sp. (0.54) | Mesorhizobium sp. (0.57) | Bradyrhizobium sp. (0.50) |
| 4 | Burkholderia glathei (0.63) | Rhizobiales (0.60) | Bradyrhizobium sp. (0.50) | Bradyrhizobium sp. (0.55) | Steroidobacter sp. (0.49) |
| 5 | Burkholderia glathei (0.55) | Burkholderia sp. (0.59) | Bradyrhizobium sp. (0.49) | Mesorhizobium sp. (0.52) | Acidobacteria (0.46) |
| 6 | Steroidobacter sp. (0.55) | Steroidobacter sp. (0.54) | Acidobacteriaceae (0.47) | Steroidobacter sp. (0.52) | Acidobacteria (0.46) |
| 7 | Steroidobacter sp. (0.49) | Bradyrhizobium elkanii (0.51) | Rhizobiales (0.46) | Acidobacteria bacterium (0.49) | Bradyrhizobium sp. (0.45) |
| 8 | Caulobacter sp. (0.46) | Pseudolabrys taiwanensis (0.50) | Steroidobacter sp. (0.46) | Steroidobacter sp. (0.46) | Rhizobiales (0.42) |
| 9 | Curtobacterium flaccumfaciens (0.45) | Steroidobacter sp. (0.49) | Bradyrhizobium sp. (0.43) | Bradyrhizobium elkanii (0.44) | Steroidobacter sp. (0.41) |
| 10 | Steroidobacter sp. (0.45) | Bradyrhizobium sp. (0.42) | Bradyrhizobium sp. (0.42) | Bradyrhizobium sp. (0.36) | Flavosolibacter sp. (0.40) |
Based on the OTUs generated with a threshold of 97%.
The most abundant bacterial genera detected in the ectomycorrhizospheres of X. pruinatus and S. citrinum, including Acidobacterium, Burkholderia, Chitinophaga, Rhodoplanes, and Bradyrhizobium, have also been reported among the most abundant genera in different agricultural or forest soils (10, 27, 36), as well as in association with ectomycorrhizal roots (13, 14, 17, 33) (Table 2). Our knowledge on some of them, such as the Acidobacterium or Chitinophaga genus, is limited due to the fact that bacteria from these genera remain hitherto unculturable (10, 24, 29). In contrast, culturable approaches have revealed that the Burkholderia and Bradyrhizobium genera were frequently detected in mycorrhizospheres (1, 16, 20, 22, 34, 35). Notably, the Burkholderia OTUs defined in the present study showed high similarity (99 to 100%) with sequences of Burkholderia glathei strains coming from the same experimental site and characterized for their ability to weather minerals, a process of high importance in nutrient-poor forest soils (3, 4, 35). The pyrosequencing approach has also permitted the detection of rare phylotypes related to genera, such as Nitrospira, Collimonas, or Streptomyces, that are known to be involved in nitrogen and nutrient cycling or antibiotic production (18) (see Table S2 in the supplemental material). Our results thus reinforce the interest in performing a pyrosequencing approach to detect rare taxonomic groups.
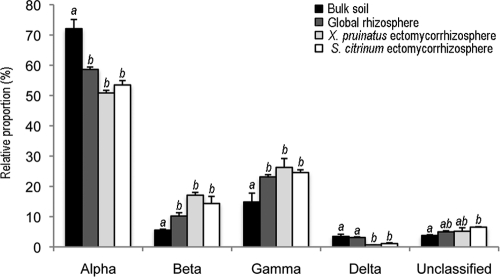
Comparative analysis with the 16S rRNA sequences generated from the same soil cores (36) but from the rhizosphere and the surrounding bulk soil revealed specificities of the mycorrhizosphere bacterial communities. For the same number of sequences, the ectomycorrhizosphere bacterial communities were characterized by a higher number of OTUs than those of the rhizosphere and the surrounding bulk soil. Although similar taxonomic groups were detected in all the soil habitats considered (ectomycorrhizosphere, rhizosphere, or bulk soil), the ectomycorrhizosphere was significantly enriched in Proteobacteria (P = 0.004) (Table 1) and depleted in Actinobacteria (P = 0.0001), Gemmatimonadetes (P = 0.001), Chlamydia (P = 0.002), Nitrospira (P = 0.008), and unclassified bacteria (P = 0.0007). Closer investigation of proteobacterial sequences showed that the ectomycorrhizosphere was significantly dominated by Betaproteobacteria (P = 0.002) and Gammaproteobacteria (P = 0.003), in contrast to the surrounding bulk soil environment (Fig. 1). Significantly fewer Alphaproteobacteria (P = 0.0006) and Deltaproteobacteria (P = 0.002) were detected in the ectomycorrhizosphere than in the surrounding bulk soil environment. At the genus level, our analysis also revealed a significant predominance of sequences related to Burkholderia in ectomycorrhizosphere compared to that of the rhizosphere and the surrounding bulk soil (P = 0.0018), thus suggesting selection of this bacterial genus in the ectomycorrhizosphere. On the contrary, significantly fewer sequences related to Rhodoplanes were detected in the ectomycorrhizosphere than in the surrounding bulk soil (P = 0.02). Detailed comparisons at the phylotype (OTU) level were not as clear, highlighting a high heterogeneity between samples. The ectomycorrhizosphere did not cluster with the rhizosphere or the bulk soil (see Fig. S3B in the supplemental material), demonstrating once again that it is a specific ecological habitat of the soil. In this sense, the ectomycorrhizosphere OTUs represented 45% of the total number of OTUs (64,492 reads). A detailed analysis revealed that slightly more OTUs were common with the surrounding bulk soil (4.5%; 16,136 reads) than with the rhizosphere (3%; 17,054 reads) (see Fig. S4 in the supplemental material).
Fig 1.
Relative abundances of Proteobacteria in the ectomycorrhizosphere, the rhizosphere, and the surrounding bulk soil. A one-factor (habitat) ANOVA was applied on the relative distribution values after an arcsine square root transformation for each proteobacterial group. Different letters for each proteobacterial group indicate that the values are significantly different.
In conclusion, this study highlights for the first time, through 454 pyrosequencing, the richness and diversity of the ectomycorrhizosphere bacterial communities. We show that the bacterial communities inhabiting the ECM complex of two ectomycorrhizal fungi associated with oak, S. citrinum and X. pruinatus, are very similar at the phylum or genus level but clearly different at the OTU (phylotype) level. Our analysis demonstrates that the ectomycorrhizosphere bacterial communities qualitatively resemble those of the rhizosphere or bulk soil environments at the phylum and genus levels but present significant quantitative differences, illustrating the specificity of the ectomycorrhizosphere.
Nucleotide sequence accession numbers.
The sequences determined in this study have been deposited in the Sequence Read Archive (SRA) service of the GenBank database under the accession numbers SRA029325.1 (mycorrhizosphere samples) and SRA029106.2 (for the rhizosphere and bulk soil samples of the study of Uroz et al. [36]).
Supplementary Material
ACKNOWLEDGMENTS
This work was made possible by grants from EC2CO, BRG, and Région Lorraine. All of these are gratefully acknowledged.
We thank S. Antony-Babu, M. Buée, P.-E. Courty, and J. Garbaye for helpful discussions on the manuscript. We are grateful to Pat Schloss's bioinformatics platform (http://www.mothur.org/) for providing help and support.
Footnotes
Published ahead of print 3 February 2012
Supplemental material for this article may be found at http://aem.asm.org/.
REFERENCES
- 1. Bianciotto V, et al. 2000. Detection and identification of bacterial endosymbionts in arbuscular mycorrhizal fungi belonging to the family Gigasporaceae. Appl. Environ. Microbiol. 66:4503–4509 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Burke DJ, Dunham SM, Kretzer AM. 2008. Molecular analysis of bacterial communities associated with the roots of Douglas fir (Pseudotsuga menziesii) colonized by different ECM fungi. FEMS Microbiol. Ecol. 65:299–309 [DOI] [PubMed] [Google Scholar]
- 3. Calvaruso C, Turpault M-P, Leclerc E, Frey-Klett P. 2007. Impact of ectomycorrhizosphere on the functional diversity of soil bacterial and fungal communities from a forest stand in relation to nutrient mobilization processes. Microb. Ecol. 54:567–577 [DOI] [PubMed] [Google Scholar]
- 4. Calvaruso C, et al. 2010. Influence of forest trees on the distribution of mineral weathering-associated bacterial communities of the Scleroderma citrinum mycorrhizosphere. Appl. Environ. Microbiol. 76:4780–4787 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Courty P-E, et al. 2010. The role of ectomycorrhizal communities in forest ecosystem processes: new perspectives and emerging concepts. Soil Biol. Biochem. 42:679–698 [Google Scholar]
- 6. Cusano AM, et al. 2010. Pseudomonas fluorescens BBc6R8 type III secretion mutants no longer promote ectomycorrhizal symbiosis. Environ. Microbiol. Rep. 3:203–210 [DOI] [PubMed] [Google Scholar]
- 7. Droege M, Hill BB. 2008. The genome sequencer FLXTM system—longer reads, more applications, straight forward bioinformatics and more complete data sets. J. Biotechnol. 136:3–10 [DOI] [PubMed] [Google Scholar]
- 8. Frey-Klett P, et al. 2005. Ectomycorrhizal symbiosis affects functional diversity of rhizosphere fluorescent pseudomonads. New Phytol. 165:317–328 [DOI] [PubMed] [Google Scholar]
- 9. Frey-Klett P, Tarkka M, Garbaye J. 2007. The mycorrhiza helper bacteria revisited. New Phytol. 176:22–36 [DOI] [PubMed] [Google Scholar]
- 10. Fulthorpe RR, Roesch LF, Riva A, Triplett EW. 2008. Distantly sampled soils carry few species in common. ISME J. 2:901–910 [DOI] [PubMed] [Google Scholar]
- 11. Izumi H, Moore ERB, Killham K, Alexander IJ, Anderson IC. 2007. Characterisation of endobacterial communities in ectomycorrhizas by DNA- and RNA-based molecular methods. Soil Biol. Biochem. 39:891–899 [Google Scholar]
- 12. Izumi H, et al. 2008. Bacteria associated with ectomycorrhizas of slash pine (Pinus elliottii) in south-eastern Queensland, Australia. FEMS Microbiol. Lett. 282:196–204 [DOI] [PubMed] [Google Scholar]
- 13. Izumi H, Finlay RD. 2011. Ectomycorrhizal roots select distinctive bacterial and ascomycete communities in Swedish subarctic forests. Env. Microbiol. 74:645–650 [DOI] [PubMed] [Google Scholar]
- 14. Janssen PH. 2006. Identifying the dominant soil bacterial taxa in libraries of 16S rRNA and 16S rRNA genes. Appl. Environ. Microbiol. 72:1719–1728 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Johansson JF, Paul LR, Finlay RD. 2004. Microbial interactions in the mycorrhizosphere and their significance for sustainable agriculture. FEMS Microbiol. Ecol. 48:1–13 [DOI] [PubMed] [Google Scholar]
- 16. Kataoka R, Taniguchi T, Ooshima H, Futai K. 2008. Comparison of the bacterial communities established on the mycorrhizae formed on Pinus thunbergii root tips by eight species of fungi. Plant Soil 304:267–275 [Google Scholar]
- 17. Khetmalas MB, Egger KN, Massicotte HB, Tackaberry LE, Clapperton MJ. 2002. Bacterial diversity associated with subalpine fir (Abies lasiocarpa) ectomycorrhizae following wildfire and salvage-logging in central British Columbia. Can. J. Microbiol. 48:611–625 [DOI] [PubMed] [Google Scholar]
- 18. Leveau JHJ, Uroz S, de Boer W. 2010. The bacterial genus Collimonas: mycophagy, weathering and other adaptive solutions to life in oligotrophic soil environments. Environ. Microbiol. 12:281–292 [DOI] [PubMed] [Google Scholar]
- 19. Linderman RG. 1988. Mycorrhizal interactions with the rhizosphere microflora: the mycorrhizosphere effect. Phytopathology 78:366–371 [Google Scholar]
- 20. Mansfeld-Giese K, Larsen J, Bødker L. 2002. Bacterial populations associated with mycelium of the arbuscular mycorrhizal fungus Glomus intraradices. FEMS Microbiol. Ecol. 41:133–140 [DOI] [PubMed] [Google Scholar]
- 21. Meyer F, et al. 2008. The metagenomics RAST server—a public resource for the automatic phylogenetic and functional analysis of metagenomes. BMC Bioinformatics 9:386. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Mogge B, Loferer C, Agerer R, Hutzler P, Hartmann A. 2000. Bacterial community structure and colonization patterns of Fagus sylvatica L-ectomycorrhizospheres as determined by fluorescence in situ hybridization and confocal laser scanning microscopy. Mycorrhiza 9:271–278 [Google Scholar]
- 23. Nadkarni MA, Martin FE, Jacques NA, Hunter N. 2002. Determination of bacterial load by real-time PCR using a broad-range (universal) probe and primers set. Microbiology 148:257–266 [DOI] [PubMed] [Google Scholar]
- 24. Nunes da Rocha U, van Overbeek L, van Elsas JD. 2009. Exploration of hitherto-uncultured bacteria from the rhizosphere. FEMS Microbiol. Ecol. 69:313–328 [DOI] [PubMed] [Google Scholar]
- 25. On SLW, Atabay HI, Corry JEL, Harrington CS, Vandamme P. 1998. Emended description of Campylobacter sputorum and revision of its infrasubspecific (biovar) divisions, including C. sputorum biovar paraureolyticus, a urease-producing variant from cattle and humans. Int. J. Syst. Bacteriol. 48:195–206 [DOI] [PubMed] [Google Scholar]
- 26. Rambelli A. 1973. The rhizosphere of mycorrhizae, p 299–349 In Marks GC, Kozlowski TT. (ed), Ectomycorrhizae, their ecology and physiology. Academic Press, New York, NY [Google Scholar]
- 27. Roesch LF, et al. 2007. Pyrosequencing enumerates and contrasts soil microbial diversity. ISME J. 1:283–290 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Rygiewicz PT, Andersen CP. 1994. Mycorrhizae alter quality and quantity of carbon allocated below ground. Nature 369:58–60 [Google Scholar]
- 29. Sangkhobol V, Skerman VBD. 1981. Chitinophaga, a new genus of chitinolytic myxobacteria. Int. J. Syst. Bacteriol. 31:285–293 [Google Scholar]
- 30. Santelli CM, Edgcomb VP, Bach W, Edwards KJ. 2009. The diversity and abundance of bacteria inhabiting seafloor lavas positively correlate with rock alteration. Environ. Microbiol. 11:86–98 [DOI] [PubMed] [Google Scholar]
- 31. Schloss PD, et al. 2009. Introducing mothur: open-source, platform-independent, community-supported software for describing and comparing microbial communities. Appl. Environ. Microbiol. 75:7537–7541 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Smith SE, Read DJ. 2008. Mycorrhizal symbiosis, 3rd ed Academic Press, London, United Kingdom [Google Scholar]
- 33. Timonen S, Jorgensen KS, Haahtela K, Sen R. 1998. Bacterial community structure at defined locations of Pinus sylvestris-Suillus bovinus and Pinus sylvestris-Paxillus involutus mycorrhizospheres in dry pine forest humus and nursery peat. Can. J. Microbiol. 44:499–513 [Google Scholar]
- 34. Timonen S, Hurek T. 2006. Characterization of culturable bacterial populations associating with Pinus sylvestris-Suillus bovinus mycorrhizospheres. Can. J. Microbiol. 52:769–778 [DOI] [PubMed] [Google Scholar]
- 35. Uroz S, et al. 2007. Effect of the mycorrhizosphere on the genotypic and metabolic diversity of the soil bacterial communities involved in mineral weathering in a forest soil. Appl. Environ. Microbiol. 73:3019–3027 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Uroz S, Buée M, Murat C, Frey-Klett P, Martin F. 2010. Pyrosequencing highlights the contrasted bacterial diversity in forest soil. Environ. Microbiol. Rep. 2:281–288 [DOI] [PubMed] [Google Scholar]
- 37. Viollet A, et al. 2011. Fluorescent pseudomonads harboring type III secretion genes are enriched in the mycorrhizosphere of Medicago truncatula. FEMS Microbiol. Ecol. 75:457–467 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.