Abstract
The type strain of Vibrio owensii (DY05) was isolated during an epizootic of aquaculture-reared larvae (phyllosomas) of the ornate spiny lobster (Panulirus ornatus). V. owensii DY05 was formally demonstrated to be the etiological agent of a disease causing rapid and reproducible larval mortality with pathologies similar to those seen during disease epizootics. Vectored challenge via the aquaculture live feed organism Artemia (brine shrimp) caused consistent cumulative mortality rates of 84 to 89% after 72 h, in contrast to variable mortality rates seen after immersion challenge. Histopathological examination of vector-challenged phyllosomas revealed bacterial proliferation in the midgut gland (hepatopancreas) concomitant with epithelial cell necrosis. A fluorescent-protein-labeled V. owensii DY05 transconjugant showed dispersal of single cells in the foregut and hepatopancreas 6 h postexposure, leading to colonization of the entire hepatopancreas within 18 h and eventually systemic infection. V. owensii DY05 is a marine enteropathogen highly virulent to P. ornatus phyllosoma that uses vector-mediated transmission and release from host association to a planktonic existence to perpetuate transfer. This understanding of the infection process will improve targeted biocontrol strategies and enhance the prospects of commercially viable larviculture for this valuable spiny lobster species.
INTRODUCTION
Major steps toward the commercialization of closed life cycle aquaculture production of the ornate spiny lobster (Panulirus ornatus) have been reported recently (36), yet nutritional deficits (41) and mortality caused by bacterial disease (5) remain major constraints to hatchery productivity. Mass mortalities of larvae (phyllosomas) are often associated with enteric vibriosis, an infection of the midgut gland (hepatopancreas) caused by Vibrio species (6, 52).
Members of the genus Vibrio are natural marine inhabitants, playing important roles in nutrient cycling and forming associations with zooplankton (49). Microhabitat preferences and ecological selection may be key factors in the speciation of vibrios (20), and intensive aquaculture systems are thought to select for bacterial virulence, including traits that enhance infectivity and transmission (29, 33). Accordingly, many Vibrio species are pathogenic to cultured crustacean zooplanktonic larval forms, including the three closely related species Vibrio harveyi (32, 35), V. campbellii (17, 44), and the recently described V. owensii (8).
It is of paramount importance to the development of efficient disease management strategies that pathogens be identified by using experimental infection models that provide information on infection routes and infection dynamics (38). Recently, microorganisms engineered to express fluorescent proteins (FP) have significantly increased the understanding of invasive pathways and infection dynamics of pathogens, including V. anguillarum, Aeromonas hydrophila, and Edwardsiella tarda in fish models (10, 24, 30) and V. harveyi in abalone (50). Panulirus sp. phyllosoma (from Greek: leaf-like body) larvae are dorsoventrally flattened and transparent, which makes them excellent candidates for live, nondestructive direct microscopic observations of bacterium-host symbioses in situ.
This study describes the development of a robust experimental infection model to evaluate the pathogenicity of the V. owensii type strain (DY05) toward P. ornatus phyllosomas. The study included (i) comparison of immersion and vector challenges as natural routes of infection, (ii) verification of Koch's postulates, (iii) pathological assessment of experimentally infected animals, and (iv) visualization of the spatiotemporal localization and infection process using FP-tagged strains.
MATERIALS AND METHODS
Larviculture.
P. ornatus phyllosomas were sourced from broodstock held at the Tropical Aquaculture Facility of the Australian Institute of Marine Science (AIMS), Townsville, Australia. Maintenance of broodstock, production of phyllosomas, and larviculture were performed according to the methods of Smith et al. (41). Only apparently healthy individuals as assessed by positive phototaxis were used for experiments.
Isolation of the pathogen.
Vibrio spp. were isolated from moribund stage 3 phyllosomas during an epizootic in the AIMS larval rearing system. Briefly, phyllosomas were washed in sterile artificial seawater (ASW) (Instant Ocean; Spectrum Brands, Madison, WI) to remove loosely attached epibionts and excess detritus, homogenized in ASW, and plated on thiosulfate citrate bile sucrose agar (TCBS) (BD, Franklin Lakes, NJ). The 3 numerically dominant and 3 unique morphotypes were cultured to purity on TCBS and cryopreserved (−80°C). In preliminary infection experiments (described below), one of the dominant isolates (DY05) caused 85% mortality of stage 3 phyllosomas 72 h after vectored challenge (data not shown). DY05 was subsequently characterized by exhaustive phenotypical, biochemical, and phylogenetic analyses and determined to belong to the novel species V. owensii, of which DY05 is the type strain (8).
Bacterial strains and culture conditions.
Cryopreserved stocks of V. owensii DY05 were revived and cultured in 5 ml marine broth (MB) (BD; 28°C; 170 rpm) for 18 to 24 h. Transconjugations used the helper strain Escherichia coli CC118λpir harboring pEVS104 (45) and the green fluorescent protein (GFP) donor strain E. coli DH5αλpir carrying the V. fischeri pES213-derived plasmid pVSV102 (13), both encoding kanamycin resistance. The helper and donor strains were revived and cultured in LB broth (5 g liter−1 yeast extract, 10 g liter−1 neutralized peptone; Oxoid, United Kingdom) supplemented with 40 μg ml−1 kanamycin (30°C; 170 rpm).
Inoculum preparation.
Bacterial cells were washed 3 times by centrifugation (10 min at 5,200 × g; 10°C) and resuspended in 0.22-μm-filtered seawater (FSW). The final suspension was adjusted to an optical density at 600 nm (OD600) of 0.1 (Nanodrop ND1000; Nanodrop Products, Wilmington, DE). Corresponding total viable counts were determined by spiral plating (Eddy Jet; IUL, Spain) on marine agar (MA) (BD) and enumeration by an automatic colony counter (Flash and Grow v1.2; IUL).
Experimental infection by immersion.
Phyllosomas (stage 1) were washed twice in FSW and distributed into 12-well cell culture plates (NUN150628; Nunc, Denmark) at 1 larva well−1 with 3 ml FSW per well. The animals were acclimated in darkness (28°C; 45 rpm) for 2 h before adding V. owensii DY05 inoculum directly into the wells at low (1 × 103 CFU ml−1), moderate (1 × 105 CFU ml−1), or high (1 × 107 CFU ml−1) doses. Experimental controls were not exposed to V. owensii DY05. Each treatment was performed in separate plates and run in quadruplicate (n = 48). Larval mortality was assessed every 24 h for 5 days, with phyllosomas not displaying any active movement after prolonged inspection being recorded as dead. No feeding or water changes were performed during the experiment. The immersion challenge experiment was repeated three times using progeny produced by different broodstocks and genetic lineages.
Experimental infection by vectored challenge.
Phyllosomas (stage 1 and stage 3) were exposed to V. owensii DY05 using live Artemia stage II (nauplii; INVE, Belgium) as vectors. Formalin-disinfected nauplii (200 nauplii ml−1) were enriched through filter feeding with V. owensii DY05 (1 × 106 CFU ml−1) in tissue culture flasks (Sarstedt, Germany) for 2 h (28°C; 45 rpm). Control nauplii were treated similarly, except no bacteria were added. Both control and enriched nauplii were homogenized and plated on TCBS.
Phyllosomas were distributed to cell culture plates and acclimated (28°C; 45 rpm) for 2 h (stage 1) or 24 h (stage 3) prior to experimental infection. On day 0, phyllosomas were fed with live enriched or nonenriched Artemia nauplii (control) at approximately 3 nauplii ml−1. Each treatment was performed in separate plates and run in quadruplicate (n = 48). One stage 3 experiment included a control with phyllosomas treated with an antibiotic cocktail (25 mg liter−1 erythromycin, 25 mg liter−1 oxytetracycline, 10 mg liter−1 streptomycin, 40 mg liter−1 ciprofloxacin) for 24 h prior to collection and then rinsed, acclimated, and challenged with control nauplii as outlined above. This experiment also included additional replicate plates for sacrificial sampling for analysis of culturable vibrios (see below).
Larval mortality was assessed every 24 h for 5 days as described above. Water changes and additional feeds were not performed during the experiment. For each stage, three replicate experiments were performed using progeny from different broodstocks and genetic lineages.
Reisolation of Vibrio.
Vibrios were recovered from Artemia cultures and experimentally infected phyllosomas (stage 3) in sacrificial plates. Briefly, six live and six dead (within 4 h postmortem) phyllosomas were sampled within the first 3 days, homogenized in ASW, and plated on TCBS. Dominant colony morphotypes were cultured to purity and cryopreserved.
For each isolate, colony PCR with universal bacterial 16S rRNA gene primers 27F and 1492R (22) was performed under standard amplification conditions. The PCR products were purified and sequenced in both directions by Macrogen (Seoul, South Korea). The sequences were edited using 4Peaks (Mekentosj) and analyzed using the BLAST algorithm (http://www.ncbi.nlm.nih.gov/BLAST/) to determine nucleotide-nucleotide similarity with sequences in the nr/nt database. Isolates with high 16S rRNA gene sequence identity (>99%) to V. owensii were analyzed further by sequencing topA (topoisomerase I; DNA replication and repair) and mreB (rod-shaping protein gene B subunit) using primers VtopA400f/VtopA1200R or VmreB12F/VmreB999R, respectively (39), as described by Thompson et al. (48). Sequences were generated and analyzed as described above.
Histopathological analysis.
Phyllosomas (stage 3) were sampled prior to and 18, 24, and 42 h after vector challenge. Phyllosomas with limbs removed were fixed in Bouin's fixative (75 ml saturated picric acid, 25 ml formaldehyde [40%], 5 ml glacial acetic acid) for 20 h at room temperature with gentle shaking. Fixed samples were washed and stored in 70% ethanol at 4°C until further processing. The samples were dehydrated in an ethanol series (70%, 96%, and 100%; 30 min), followed by preinfiltration and embedding in 2-hydroxyethyl-methacrylate resin (Technovit 7100; Heraeus Kulzer, Germany) according to the manufacturer's protocol. The phyllosomas were sagittally sectioned (2 μm) using a carbide tungsten blade (Delaware Diamond Knives, Wilmington, DE) on a rotary microtome (HM360; Microm International, Germany). Sections were progressively stained with Gill's No. 2 hematoxylin and aqueous eosin (1%) (ProSciTech, Australia) and examined using light microscopy (AxioSkop 2 mot plus; Carl Zeiss, Germany). Images were captured using an AxioCam MRc5 camera (Carl Zeiss) and processed using AxioVision release 4.8 software (Carl Zeiss). Special attention was given to the hepatopancreas tubules.
In situ visualization of the infection process.
The GFP-carrying plasmid (pVSV102) was transferred to V. owensii DY05 cells by triparental conjugation (see File S1 in the supplemental material for detailed protocol). The transconjugant V. owensii DY05[GFP] was assessed for stable expression of GFP, and its growth profile was compared with that of the V. owensii DY05 wild type (see File S1 in the supplemental material for detailed protocol). Vectored challenge of phyllosomas (stage 1) with DY05[GFP] was performed as outlined above. Enriched Artemia nauplii and four vector-challenged phyllosomas were live mounted in FSW and viewed using differential inference contrast (DIC) and fluorescence microscopy (AxioSkop 2 mot plus; Carl Zeiss). Fluorescence was detected using a dual-band filter set (59004; Chroma Technology Corp., Bellows Falls, VT), and images were captured by an AxioCam MRc5 camera (Carl Zeiss) directed by the multidimensional acquisition module of the AxioVision release 4.8 software (Carl Zeiss).
Statistical analysis.
Differences between survival curves were determined using the product limit (Kaplan-Meier) estimator, employing log rank and Wilcoxon chi-square statistics, with confirmation by analysis of variance (ANOVA). A post hoc Dunnett's test was used to compare treatments to defined control groups. Student's t test, assuming two-tailed distribution and equal variance, was used to test statistical significance between survival means on individual days. Statistical significance was standardized at an α value of 0.05. Analyses were performed using the statistical software package JMP7 (SAS, Cary, NC).
RESULTS
Pathogenicity of V. owensii DY05 toward P. ornatus phyllosomas.
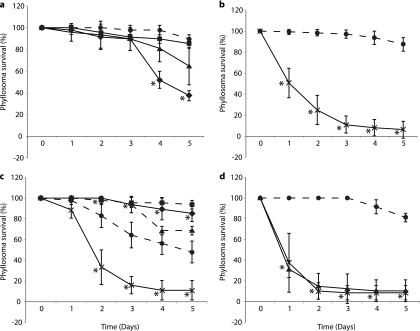
Immersion challenge experiments were performed with nonfeeding phyllosomas and low, moderate, or high concentrations of V. owensii DY05 (Fig. 1a). Significant interexperimental differences were seen for groups exposed to the same pathogen dose (ANOVA, P < 0.05), but not for control phyllosoma (ANOVA, P > 0.05), so data for the controls were pooled before further analyses. In two of the three replicate experiments, mortalities were significantly increased relative to the control for moderate (Dunnett's test, P < 0.05) and high (Dunnett's test, P < 0.0005) pathogen concentrations.
Fig 1.
Survival of P. ornatus phyllosomas experimentally infected with V. owensii DY05 by immersion (a) or vectored challenge via Artemia nauplii (b to d). (a) Stage 1 phyllosoma survival after immersion with V. owensii DY05 at concentrations of 0 (control; ●), 1 × 103 (■), 1 × 105 (▲), and 1 × 107 (♦) CFU ml−1. Data from one of three independent experiments are presented, as high interexperimental variability prevented pooling of data. (b) Stage 1 phyllosoma survival after vectored challenge with nonenriched nauplii (control; ●) or nauplii enriched with V. owensii DY05 (×). Pooled data from three independent experiments are presented. (c) Stage 3 phyllosoma survival after vectored challenge with nonenriched nauplii (controls; ●, ■, and ▲) or nauplii enriched in V. owensii DY05 (×). Data from three independent experiments are shown; data for controls were not pooled due to high interexperimental variability, while data for V. owensii-challenged phyllosomas were pooled. The first experiment (control; ●) included a second control treated with an antibiotic cocktail (♦) prior to feeding with nauplii. (d) Stage 1 phyllosoma survival after vectored challenge with nonenriched nauplii (control; ●) or nauplii enriched with V. owensii DY05 (×) or DY05[GFP] (▲). The asterisks indicate data points significantly different (Student's t test) from the corresponding control. Means ± standard deviations (SD) are shown.
Vectored challenge (via Artemia) of stage 1 phyllosomas resulted in reproducible mortality rates, with no significant interexperimental differences for challenged or control phyllosomas (ANOVA, P > 0.05); hence, data from the three experiments were pooled (Fig. 1b). Vector-mediated exposure to V. owensii DY05 had a significant (ANOVA, P < 0.0001) detrimental effect on phyllosoma survival. The highest number of phyllosoma deaths occurred within the first 24 h (49% of the total), and cumulative mortality after 72 h was 89% (Fig. 1b).
Vectored challenge experiments of stage 3 phyllosomas showed significant interexperimental differences for control phyllosomas (ANOVA, P < 0.0001), but not for V. owensii DY05-challenged phyllosomas (ANOVA, P > 0.05), so the latter data were pooled (Fig. 1c). Vector-challenged phyllosomas showed decreased survival relative to each control group (Dunnett's test, P < 0.0001). The highest number of phyllosoma deaths occurred between 24 and 48 h (56% of the total), and cumulative mortality after 72 h was 84% (Fig. 1c). A control group treated with antibiotics was included in one experiment to evaluate the contribution of opportunistic infections by autochthonous bacteria to stage 3 phyllosoma survival. Antibiotic treatment increased survival significantly relative to the corresponding control (Dunnett's test, P < 0.0001).
Confirmation of Koch's postulates and identification of phyllosoma-associated vibrios.
Isolates recovered from enriched Artemia and vector-challenged stage 3 phyllosomas are listed in Table S1 in the supplemental material. No vibrios were recovered on TCBS from control Artemia cultures. In contrast, a strain showing 16S rRNA, mreB, and topA gene sequence identity to V. owensii DY05 was recovered from enriched Artemia nauplii. For five out of six dead vector-challenged phyllosomas, the dominant bacterial morphotypes were identical to V. owensii DY05 as determined by sequencing of the 16S rRNA, mreB, and topA genes, thereby satisfying Koch's postulates. It should be noted that for the sixth vector-challenged individual, a V. neptunis-like strain (A37) was the dominant member of the culturable community. Importantly, V. owensii DY05 was not recovered from apparently healthy live phyllosoma or dead control individuals, for which recovered strains were affiliated with V. neptunis, V. parahaemolyticus, and V. harveyi.
Histology.
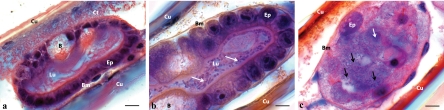
Prior to experimental infection with V. owensii DY05, stage 3 phyllosomas showed structural integrity of lateral hepatopancreas tubules, characterized by a dilated lumen lined by cuboidal epithelial cells anchored to an intact basement membrane (Fig. 2a). Eighteen hours after infection, most tubule sections showed pathologies similar to those of control phyllosomas; however, some exhibited dissociation of intercellular junctions. Proliferation of rod-shaped bacteria in the tubule lumen was observed 24 h postexposure, concomitant with epithelial cell rounding (Fig. 2b) and, in some specimens, detachment of epithelial cells from the basement membrane. After 42 h, the hepatopancreas tubules of dead phyllosomas were necrotic (Fig. 2c) and in some instances had completely disintegrated, with the tissue remnants and intertubular spaces inhabited by masses of bacterial cells. Examination of eyes and thoracic musculature also indicated bacterial infiltration, suggesting progression to systemic infection in the late stages of moribundity or postmortem.
Fig 2.
Histopathological analysis of stage 3 P. ornatus phyllosoma hepatopancreas tubules during vectored challenge with V. owensii DY05. (a) Distal end of tubule before exposure to V. owensii DY05, showing no visible bacterial cells, cuboidal epithelial cells, large lumen, and B-cell vacuoles. (b) Tubules 24 h after exposure, demonstrating bacterial proliferation (white arrows) within the lumen and rounding of epithelial cells. (c) Necrotic tubule 42 h after exposure, characterized by disintegration of epithelial cells (white arrow), cell sloughing, absence of defined lumen, and proliferation of bacteria (black arrows). B, B-cell vacuole; Bm, basement membrane; Ct, connective tissue; Cu, cuticle; Ep, epithelial cells; Lu, tubular lumen. All scale bars, 10 μm.
In situ visualization of the V. owensii DY05 infection process.
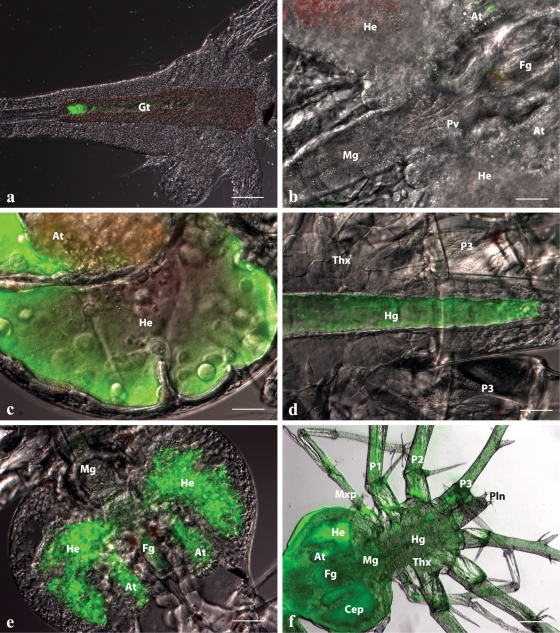
The transconjugant DY05[GFP] stably expressed GFP after continuous subculture (see Fig. S1a in the supplemental material) and showed a growth profile similar to that of the wild type (see Fig. S1b in the supplemental material). The survival curves of stage 1 phyllosomas after vectored challenge with DY05[GFP] (Fig. 1d) did not differ significantly from those of the wild type (ANOVA, P > 0.05). These observations strongly indicate that FP expression had a low bioenergetic cost and no detrimental effect on virulence, making DY05[GFP] a suitable biomarker to visualize the infection process of V. owensii DY05. The spatiotemporal dynamics of DY05[GFP] through the infection cycle were monitored during vectored challenge of stage 1 phyllosomas. Following a 2-h enrichment of the vector organism, Artemia, fluorescent cells were detected along the length of the Artemia gastrointestinal tract, with colonization typically more concentrated toward the gut posterior (Fig. 3a). The degree of colonization varied between Artemia individuals, suggesting bioaccumulation was not uniform. Small numbers of fluorescent cells were observed on appendages, indicating incidental entanglement.
Fig 3.
In situ spatiotemporal localization of V. owensii DY05[GFP] in Artemia nauplius after 2-h enrichment (a) and during the infection process of vector-challenged stage 1 P. ornatus phyllosomas (b to f). (a) Artemia nauplius showing colonization of DY05[GFP] in the gastrointestinal tract. Scale bar, 100 μm. (b) Hepatopancreas and midgut region of phyllosoma after 6-h exposure, demonstrating the presence of single cells or small aggregations of DY05[GFP] cells in the foregut, the midgut, and the anterior and lateral lobes of the hepatopancreas. Scale bar, 50 μm. (c) Proliferation of DY05[GFP] in the distal ends of the lateral and anterior hepatopancreas lobes 12 h after exposure. Scale bar, 50 μm. (d) Hindgut 12 h after exposure showing trafficking of DY05[GFP] cells followed by evacuation. Scale bar, 50 μm. (e) Hepatopancreas 18 h after exposure, showing illumination of the entire organ with fluorescent DY05[GFP] cells concomitant with tissue granulation and loss of architecture. Scale bar, 100 μm. (f) Dead phyllosoma 24 h after exposure, showing colonization of the entire body (systemic infection) by DY05[GFP] cells associated with loss of internal organ structural integrity. Scale bar, 200 μm. At, hepatopancreas anterior lobe; Cep, cephalic shield; Fg, foregut; Gt, gastrointestinal tract; He, hepatopancreas lateral lobe; Hg, hindgut; Mg, midgut; Mxp, maxilliped; P, pereiopod (1 to 3); Pln, pleon; Pv, pyloric valve; Thx, thorax.
After vectored challenge, the in situ localization of DY05[GFP] in stage 1 phyllosomas was monitored at 6-h intervals for 24 h. After 6 h, monodispersed and small aggregates of fluorescent cells were visualized within the foregut and in the hepatopancreas, and cells were seen transiting through the midgut (Fig. 3b). At this stage, phyllosomas were still active, hemocytes were circulating, and the structural integrity of tissues and organs was retained. After 12 h, mass proliferation of fluorescent bacteria was visualized in the distal ends of the hepatopancreas lobes (Fig. 3c). Most bacteria in the lobes appeared to be pulsating, indicating motility. During this phase, many bacteria were actively expelled from the hepatopancreas through the pyloric valve into the midgut, and through a series of rhythmic convulsions, bacteria were pushed through the hindgut and evacuated from the anus (Fig. 3d). After 18 h, the entire hepatopancreas was illuminated by fluorescent cells, and this was associated with tissue granulation, loss of architecture, and detachment of the hepatopancreas gland from the cuticle epithelia (Fig. 3e). At this stage, phyllosomas showed symptoms of lethargy, and hepatopancreas opaqueness could be discerned macroscopically. After 24 h, when >63% of the population had died (Fig. 1d), the entire dead phyllosoma was fluorescent, suggestive of systemic infection (Fig. 3f). The vast majority of fluorescent cells were still motile while inhabiting internal tissues, which appeared to lose structural integrity, with the exception of the posterior hindgut. Bacteria transited through the vascular systems of the limbs, antennae, antennules, and eyes, indicating translocation pathways.
In general, external colonization was minimal during the infection process. Individual fluorescent cells, rather than clusters, were observed infrequently on the cephalic shield, thorax, and limbs. However, incidental entanglement of fluorescent cells in the plumose natatory setae (feathery appendages of the limbs used for propulsion) was noted and seemed to increase over time in some individuals. Interestingly, in one individual, a severed leg (pereiopod) had extensive colonization of cells around the lesion, which could indicate an alternative route of infection.
DISCUSSION
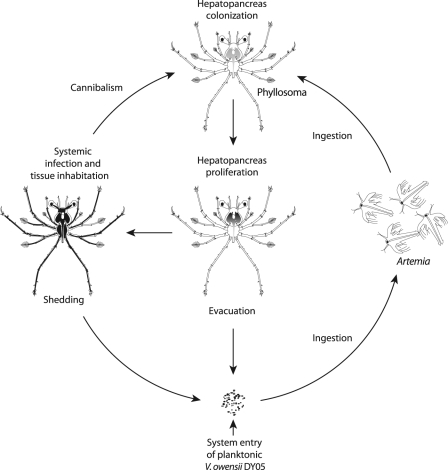
The present study provides the first description of the pathogenicity, pathology, and infection dynamics of the V. owensii type strain (DY05) toward phyllosomas of the ornate spiny lobster, P. ornatus. Infection of early-stage phyllosomas appears to be a multifaceted process, involving (i) vector-facilitated transmission, (ii) targeted colonization and proliferation in the hepatopancreas, (iii) evacuation of cells into the ambient environment, and (iv) systemic infection and acute mortality. In addition, occasional cannibalism of moribund and dead phyllosomas by vigorous individuals (our personal observation) may serve as an additional infection route. A conceptualized model to describe and summarize the infection cycle in the larviculture ecosystem is proposed in Fig. 4.
Fig 4.
Infection cycle of V. owensii DY05 in the larviculture ecosystem of early-stage P. ornatus phyllosomas. The pathogen enters the larviculture environment through an unascertained source as a free-living planktonic form. Live prey Artemia nauplii actively ingest the bacteria, resulting in bioaccumulation in the gut. Early-stage phyllosomas (stages 1 to 3) capture and masticate the Artemia nauplii, triturating food particulates along with the pathogen through the foregut into the hepatopancreas. The pathogen proliferates in the distal ends of the hepatopancreas, eventually colonizing the entire organ, with planktonic cells dispelled from the midgut and evacuated back into the ambient environment. These planktonic cells are ingested by Artemia nauplii, which perpetuates invasion of the phyllosomas through the oral route. Eventually, the condition progresses to systemic infection in association with acute mortality. By utilizing and inhabiting the tissues of moribund and dead hosts, the pathogen can also be transmitted to new hosts through occasional cannibalism. Continuous shedding of bacteria from dead hosts likely contributes to the ambient bacterioplankton, again reactivating the oral infection pathway.
The causative effect of V. owensii DY05 infection on phyllosoma mortality was demonstrated by reisolating the agent from moribund experimentally infected phyllosomas, thereby fulfilling Koch's postulates. Identification of the reisolated strains was performed by sequencing multiple loci (16S rRNA, topA, and mreB genes) in order to enable discrimination between V. harveyi-like species at the strain level, which cannot reliably be achieved by sequencing the 16S rRNA gene alone (8, 9).
Members of the genus Vibrio form major constituents of the microbiota of healthy and diseased cultured P. ornatus phyllosomas (31, 52). We isolated strains affiliated with V. neptunis, V. harveyi, and V. parahaemolyticus from both apparently healthy stage 3 phyllosomas and dead control individuals. These species include strains pathogenic to cultured aquatic invertebrates and fish (3, 4, 7, 25, 35, 46), and it is possible they, together with other autochthonous microbiota, contributed to the interexperimental variability for control stage 3 phyllosomas (Fig. 1c). This is supported by the enhanced survival of the control group exposed to antibiotic treatment (Fig. 1c). Another factor likely contributing to the differences in survival between stage 3 control cohorts is larval batch variability arising from underlying differences in genotype and physiological condition. For one of six experimentally infected individuals, a V. neptunis strain was dominant in the cultured community, introducing the possibility of synergistic interaction between V. owensii and other vibrios and the involvement of multiple etiological agents in disease epizootics (15). Despite the use of xenic hosts in our infection model, the consistent and reproducible mortality patterns of vector-challenged phyllosomas and the visualization of mass hepatopancreatic colonization by a GFP-tagged transconjugant in moribund individuals suggest a singular effect imposed by V. owensii DY05.
Clear pathological changes were visualized in experimentally infected phyllosomas, including bacterial proliferation in the hepatopancreas tubule lumen, rounding of epithelial cells, dissociation of intercellular junctions, detachment and necrosis of epithelial cells from the basement membrane, and, eventually, complete disintegration of tubules associated with systemic infection. Similar pathologies have been reported previously in cultured P. ornatus phyllosomas during disease epizootics (5, 6), reinforcing the epizootiological relevance of our experiments. Detachment and rounding of epithelial cells were also reported by Martin et al. (28) in the gut of ridgeback rock shrimp (Sicyonia ingentis) exposed to V. harveyi and V. parahaemolyticus. Comparable pathologies concomitant with Vibrio infections have also been described in cultured phyllosomas of packhorse rock lobster [Sagmariasus (Jasus) verreauxi] (12), southern rock lobster (Jasus edwardsii) (18), and various life stages of penaeid shrimp (23, 42).
Mortality was more rapid within the first 24 h in stage 1 than in stage 3 phyllosomas, suggesting that newly hatched phyllosomas are more susceptible to invasion by V. owensii DY05. Increased sensitivity to pathogens in early developmental stages of crustaceans has been reported previously (2, 32). Factors that could contribute to delayed onset of disease in more developed phyllosomas include differential expression of immune-related genes (21), a more complex hepatopancreas structure (40), and potential antagonistic activity from resident gut microbiota (14).
A key determinant of a reliable infection model is the delivery of putative pathogens to the host using natural infection routes (38), which can significantly influence the onset and pathology of disease (26, 27). In this study, vectored challenge via the aquaculture live feed organism Artemia was tested as a possible infection route, as it is a demonstrated carrier of Vibrio spp. in our system (19). Vectored challenge with V. owensii DY05 via Artemia consistently caused 84 to 89% cumulative mortality in stage 1 and 3 phyllosomas within 72 h after exposure. Mass mortalities of comparable magnitude and across similar temporal scales have been reported previously in larval rearing runs of P. ornatus, usually coinciding with molting (5). In contrast, mortality patterns after immersion challenge were more erratic and variable between experiments, suggestive of a less reliable model. Artemia organisms have long been recognized as important disease vectors in marine larviculture (19). They ingest and bioaccumulate bacterioplankton at 102 to 104 CFU nauplius−1 (43), thereby increasing the chance of delivery into the phyllosoma digestive tract. Furthermore, association with ingested Artemia may protect the pathogen from a potentially unfavorable gastrointestinal microenvironment, as previously suggested for V. anguillarum in turbot (Scophthalmus maximus) larvae (16). The results from the present study underscore the importance of Artemia in the infection process of V. owensii DY05 in the larviculture ecosystem (Fig. 4).
Incidental ingestion of V. owensii DY05 by nonfeeding phyllosomas likely occurred during the immersion challenge at moderate and high concentrations (≥1 × 105 CFU ml−1), when mortality was significantly increased relative to controls. Entry through breakages in the cuticle is another possible infection route, given that colonization of a severed pereiopod was observed with FP-tagged V. owensii DY05. However, external colonization was generally lacking, suggesting that the cuticle is not a preferred microenvironment for V. owensii DY05 and that unaided invasion through the cuticle is a less likely portal of entry. Diggles et al. (12) demonstrated that experimentally injured S. verreauxi phyllosomas were more susceptible to V. jasicida (formerly V. harveyi [53]) in similar immersion experiments and suggested that inadvertent injuries acquired during experimental processing could have contributed to some inconsistent mortality patterns (12).
Bacterial proliferation in the hepatopancreas has been reported for pathogenic vibrios in spiny lobster phyllosomas (6, 12, 18, 52), penaeid shrimp (1, 23, 35), and molluscs (54). In the present study, proliferation of the pathogen occurred in the distal ends of the hepatopancreas 12 h after exposure. The distal region harbors a high proportion of digestive cells (40), which could indicate a potential nutrient source for bacterial proliferation. Evacuation of pathogen cells from the phyllosoma hepatopancreas (12 h after exposure) and reversion to planktonic forms may reflect a behavioral mechanism evolved to enhance the prospects of colonizing other hosts (47). Reversion to a planktonic existence may be facilitated by quorum sensing, whereby cell density-dependent downregulation of virulence factors constitutes a switch from epithelium-attached forms to free-living lifestyles (11, 56). Shedding of large numbers of planktonic forms into the aquaculture environment increases the probability of uptake by Artemia, thus possibly perpetuating vector-mediated transmission through a feedback loop (Fig. 4). This phase of the infection process may represent the survival strategy of a pathogen exquisitely adapted to an ecosystem where the availability of hosts is high (29, 33).
Progression from hepatopancreas colonization to systemic infection was likely facilitated by the observed epithelial cell detachment and necrosis. Destruction of gut epithelial cells could allow bacteria unrestricted access to the basal lamina, ultimately leading to translocation to other tissues and organs (34). V. proteolyticus is known to interfere with gut epithelial cell junctions of Artemia, enabling penetration through the intercellular spaces and eventual invasion of the body cavity (51). While the mechanism by which V. owensii DY05 caused cell detachment and necrosis is unknown, it can be hypothesized that it was related to production of toxins. Preliminary studies have demonstrated that V. owensii DY05 produces hemolysins, proteases, and phospholipases (data not shown), previously identified as virulence factors in Harveyi clade vibrios (4, 25, 43, 46, 55). It is likely that the pathogenicity of V. owensii DY05 is a multifactorial process, given that some virulence genes, including those encoding hemolysins and proteases, may be linked or controlled by the same regulatory mechanism (37).
V. owensii DY05 represents a specialist marine enteropathogen posing a serious threat to the development of viable hatchery technologies for P. ornatus. The present study provides a conceptualized snapshot of the adaptive strategy used by the pathogen to enhance infectivity in the larviculture ecosystem, including vector-mediated transmission and release from host association to a planktonic existence to perpetuate transfer. Importantly, identification of the bacterium's host-associated ecological niches will facilitate the development of targeted biocontrol strategies for P. ornatus larviculture. As new pathogens emerge in larviculture settings, researchers are encouraged to take advantage of transparent zooplanktonic forms to elucidate bacterial-host symbioses in situ. It is anticipated that the robust infection model described here will be used as an important diagnostic tool to identify pathogens in Panulirus and Jasus sp. hatcheries.
Supplementary Material
ACKNOWLEDGMENTS
We thank Eric Stabb (University of Georgia) for kindly donating the helper and donor strains used to construct the fluorescent-protein-expressing transconjugants of V. owensii. We also acknowledge the zootechnical assistance of Greg Smith, Matt Kenway, Matt Salmon, Grant Milton, Justin Hochen, Katie Holroyd, Jane Gioffre, and Michael Clarkson and thank Rochelle Soo and Rose Cobb for their technical assistance, Jean-Baptiste Raina for help with statistical analyses, and Tim Simmonds for help with preparation of figures (all AIMS).
E.F.G. was supported by an Australian Postgraduate Award Ph.D. scholarship.
Footnotes
Published ahead of print 3 February 2012
Supplemental material for this article may be found at http://aem.asm.org/.
REFERENCES
- 1. Aguirre-Guzmán A, et al. 2010. Pathogenicity and infection route of Vibrio parahaemolyticus in American white shrimp, Litopenaeus vannamei. J. World Aquacult. Soc. 41:464–470 [Google Scholar]
- 2. Aguirre-Guzmán A, Vázquez-Juárez R, Ascencio F. 2001. Differences in the susceptibility of American white shrimp larval substages (Litopenaeus vannamei) to four Vibrio species. J. Invertebr. Pathol. 78:215–219 [DOI] [PubMed] [Google Scholar]
- 3. Austin B, Zhang X-H. 2006. Vibrio harveyi: a significant pathogen of marine vertebrates and invertebrates. Lett. Appl. Microbiol. 43:119–124 [DOI] [PubMed] [Google Scholar]
- 4. Austin B, Austin D, Sutherland R, Thompson F, Swings J. 2005. Pathogenicity of vibrios to rainbow trout (Oncorhynchus mykiss, Walbaum) and Artemia nauplii. Environ. Microbiol. 7:1488–1495 [DOI] [PubMed] [Google Scholar]
- 5. Bourne DG, et al. 2004. Microbial community dynamics in a larval aquaculture system of the tropical rock lobster, Panulirus ornatus. Aquaculture 242:31–51 [Google Scholar]
- 6. Bourne D, et al. 2007. Microbiological aspects of phyllosoma rearing of the ornate rock lobster Panulirus ornatus. Aquaculture 268:274–287 [Google Scholar]
- 7. Cai J, Han Y, Wang Z. 2006. Isolation of Vibrio parahaemolyticus from abalone (Haliotis diversicolor supertexta L.) postlarvae associated with mass mortalities. Aquaculture 257:161–166 [Google Scholar]
- 8. Cano-Gómez A, Goulden EF, Owens L, Høj L. 2010. Vibrio owensii sp. nov., isolated from cultured crustaceans in Australia. FEMS Microbiol. Lett. 302:175–181 [DOI] [PubMed] [Google Scholar]
- 9. Cano-Gómez A, Høj L, Owens L, Andreakis N. 2011. Multilocus sequence analysis provides basis for fast and reliable identification of Vibrio harveyi-related species and reveals previous misidentification of important marine pathogens. Syst. Appl. Microbiol. 34:561–565 [DOI] [PubMed] [Google Scholar]
- 10. Chu WH, Lu C-P. 2008. In vivo fish models for visualizing Aeromonas hydrophila invasion pathway using GFP as a biomarker. Aquaculture 277:152–155 [Google Scholar]
- 11. Defoirdt T, Ruwandeepika HAD, Karunaasagar I, Boon N, Bossier P. 2010. Quorum sensing negatively regulates chitinase in Vibrio harveyi. Environ. Microbiol. Rep. 2:44–49 [DOI] [PubMed] [Google Scholar]
- 12. Diggles BK, Moss GA, Carson J, Anderson CD. 2000. Luminous vibriosis in rock lobster Jasus verreauxi (Decapoda: Palinuridae) phyllosoma larvae associated with infection by Vibrio harveyi. Dis. Aquat. Org. 43:127–137 [DOI] [PubMed] [Google Scholar]
- 13. Dunn AK, Millikan DS, Adin DM, Bose JL, Stabb EV. 2006. New rfp- and pES213-derived tools for analyzing symbiotic Vibrio fischeri reveal patterns of infection and lux expression in situ. Appl. Environ. Microbiol. 72:802–810 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Fjellheim AJ, Playfoot KJ, Skjermo J, Vadstein O. 2007. Vibrionaceae dominates the microflora antagonistic towards Listonella anguillarum in the intestine of cultured Atlantic cod (Gadus morhua L.) larvae. Aquaculture 269:98–106 [Google Scholar]
- 15. Gay M, Berthe FCJ, Le Roux F. 2004. Screening of Vibrio isolates to develop an experimental infection model in the Pacific oyster Crassostrea gigas. Dis. Aquat. Org. 59:49–56 [DOI] [PubMed] [Google Scholar]
- 16. Grisez L, Chair M, Sorgeloos P, Ollevier F. 1996. Mode of infection and spread of Vibrio anguillarum in turbot Scophthalmus maximus larvae after oral challenge through live feed. Dis. Aquat. Org. 26:181–187 [Google Scholar]
- 17. Hameed ASS, Rao PV, Farmer JJ, Hickman-Brenner FW, Fanning GR. 1996. Characteristics and pathogenicity of a Vibrio campbelli-like bacterium affecting hatchery-reared Penaeus indicus (Milne Edwards, 1837) larvae. Aquacult. Res. 27:853–863 [Google Scholar]
- 18. Handlinger J, et al. 2001. Disease conditions of cultured phyllosoma larvae and juveniles of the southern rock lobster (Jasus edwardsii, Decapoda; Palinuridae), p 75–87 In Evans LH, Jones JB. (ed), Proceedings of the International Symposium on Lobster Health Management, Adelaide, Australia, 19 to 21 September 1999 Curtin University of Technology, Perth, Australia [Google Scholar]
- 19. Høj L, Bourne DG, Hall MR. 2009. Localization, abundance and community structure of bacteria associated with Artemia: effects of nauplii enrichment and antimicrobial treatment. Aquaculture 293:278–285 [Google Scholar]
- 20. Hunt DE, et al. 2008. Resource partitioning and sympatric differentiation among closely related bacterioplankton. Science 320:1081–1085 [DOI] [PubMed] [Google Scholar]
- 21. Jiravanichpaisal P, et al. 2007. Expression of immune-related genes in larval stages of the giant tiger shrimp, Penaeus monodon. Fish Shellfish Immunol. 23:815–824 [DOI] [PubMed] [Google Scholar]
- 22. Lane DJ. 1991. 16S/23S rRNA sequencing, p 115–175 In Stackebrandt E, Goodfellow M. (ed), Nucleic acid techniques in bacterial systematics. John Wiley and Sons, New York, NY [Google Scholar]
- 23. Lavilla-Pitogo CR, Leaño EM, Paner MG. 1998. Mortalities of pond-cultured juvenile shrimp, Penaeus monodon, associated with dominance of luminescent vibrios in the rearing environment. Aquaculture 164:337–349 [Google Scholar]
- 24. Ling SHM, Wang XH, Lim TM, Leung KY. 2001. Green fluorescent protein-tagged Edwardsiella tarda reveals portal of entry in fish. FEMS Microbiol. Lett. 194:239–243 [DOI] [PubMed] [Google Scholar]
- 25. Liu P-C, Lee K-K, Chen S-N. 1996. Pathogenicity of different isolates of Vibrio harveyi in tiger prawn, Penaeus monodon. Lett. Appl. Microbiol. 22:413–416 [Google Scholar]
- 26. Magi GE, et al. 2009. Experimental Pseudomonas anguilliseptica infection in turbot Psetta maxima (L.): a histopathological and immunohistochemical study. Eur. J. Histochem. 53:73–80 [DOI] [PubMed] [Google Scholar]
- 27. Magnadóttir B, Bambir SH, Gudmundsdóttir BK, Pilström L, Helgason S. 2002. Atypical Aeromonas salmonicida infection in naturally and experimentally infected cod, Gadus morhua L. J. Fish Dis. 25:583–597 [Google Scholar]
- 28. Martin GG, Rubin N, Swanson E. 2004. Vibrio parahaemolyticus and V. harveyi cause detachment of the epithelium from the midgut trunk of the penaeid shrimp Sicyonia ingentis. Dis. Aquat. Org. 60:21–29 [DOI] [PubMed] [Google Scholar]
- 29. Mennerat A, Nilsen F, Ebert D, Skorping A. 2010. Intensive farming: evolutionary implications for parasites and pathogens. Evol. Biol. 37:59–67 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. O'Toole R, von Hofsten J, Rosqvist R, Olsson P-E, Wolf-Watz H. 2004. Visualisation of Zebrafish infection by GFP-labelled Vibrio anguillarum. Microb. Pathog. 37:41–46 [DOI] [PubMed] [Google Scholar]
- 31. Payne MS, Hall MR, Sly L, Bourne DG. 2007. Microbial diversity within early-stage cultured Panulirus ornatus phyllosomas. Appl. Environ. Microbiol. 73:1940–1951 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Prayitno SB, Latchford JW. 1995. Experimental infections of crustaceans with luminous bacteria related to Photobacterium and Vibrio. Effect of salinity and pH on infectiosity. Aquaculture 132:105–l12 [Google Scholar]
- 33. Pulkkinen K, et al. 2010. Intensive fish farming and the evolution of pathogen virulence: the case of columnaris disease in Finland. Proc. R. Soc. B 277:593–600 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Ringø E, Myklebust R, Mayhew TM, Olsen RE. 2007. Bacterial translocation and pathogenesis in the digestive tract of larvae and fry. Aquaculture 268:251–264 [Google Scholar]
- 35. Robertson PAW, et al. 1998. Experimental Vibrio harveyi infections in Penaeus vannamei larvae. Dis. Aquat. Org. 32:151–155 [Google Scholar]
- 36. Rogers PP, Barnard R, Johnston MM. 2010. Lobster aquaculture a commercial reality: a review. J. Mar. Biol. Assoc. India 52:327–335 [Google Scholar]
- 37. Ruwandeepika HAD, et al. 2011. In vitro and in vivo expression of virulence genes in Vibrio isolates belonging to the Harveyi clade in relation to their virulence towards gnotobiotic brine shrimp (Artemia franciscana). Environ. Microbiol. 13:506–517 [DOI] [PubMed] [Google Scholar]
- 38. Saulnier D, Haffner P, Goarant C, Levy P, Ansquer D. 2000. Experimental infection models for shrimp vibriosis studies: a review. Aquaculture 191:133–144 [Google Scholar]
- 39. Sawabe T, Kita-Tsukamoto K, Thompson FL. 2007. Inferring the evolutionary history of vibrios by means of multilocus sequence analysis. J. Bacteriol. 189:7932–7936 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Smith GG, Hall MR, Salmon M. 2009. Use of microspheres, fresh and microbound diets to ascertain dietary path, component size, and digestive gland functioning in phyllosoma of the spiny lobster Panulirus ornatus. N. Z. J. Mar. Fresh. Res. 43:205–215 [Google Scholar]
- 41. Smith G, Salmon M, Kenway M, Hall M. 2009. Description of the larval morphology of captive reared Panulirus ornatus spiny lobsters, benchmarked against wild-caught specimens. Aquaculture 295:76–88 [Google Scholar]
- 42. Soonthornchai W, et al. 2010. Expression of immune-related genes in the digestive organ of shrimp, Penaeus monodon, after an oral infection by Vibrio harveyi. Dev. Comp. Immunol. 34:19–28 [DOI] [PubMed] [Google Scholar]
- 43. Soto-Rodriguez SA, Roque A, Lizarraga-Partida ML, Guerra-Flores AL, Gomez-Gil B. 2003. Virulence of luminous vibrios to Artemia franciscana nauplii. Dis. Aquat. Org. 53:231–240 [DOI] [PubMed] [Google Scholar]
- 44. Soto-Rodriguez SA, Simoes N, Roque A, Gomez-Gil B. 2006. Pathogenicity and colonisation of Litopenaeus vannamei larvae by luminescent vibrios. Aquaculture 258:109–115 [Google Scholar]
- 45. Stabb EV, Ruby EG. 2002. RP4-based plasmids for conjugation between Escherichia coli and members of the Vibrionaceae. Methods Enzymol. 358:413–426 [DOI] [PubMed] [Google Scholar]
- 46. Sudheesh PS, Xu H-S. 2001. Pathogenicity of Vibrio parahaemolyticus in tiger prawn Penaeus monodon Fabricius: possible role of extracellular proteases. Aquaculture 196:37–46 [Google Scholar]
- 47. Tang KW, Turk V, Grossart H-P. 2010. Linkage between crustacean zooplankton and aquatic bacteria. Aquat. Microb. Ecol. 61:261–277 [Google Scholar]
- 48. Thompson FL, et al. 2005. Phylogeny and molecular identification of Vibrios on the basis of multilocus sequence analysis. Appl. Environ. Microbiol. 71:5107–5115 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Thompson FL, Iida T, Swings J. 2004. Biodiversity of Vibrios. Microbiol. Mol. Biol. Rev. 68:403–431 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Travers M-A, et al. 2008. Construction of a stable GFP-tagged Vibrio harveyi strain for bacterial dynamics analysis of abalone infection. FEMS Microbiol. Lett. 289:34–40 [DOI] [PubMed] [Google Scholar]
- 51. Verschuere L, Heang H, Criel G, Sorgeloos P, Verstraete W. 2000. Selected bacterial strains protect Artemia spp. from the pathogenic effects of Vibrio proteolyticus CW8T2. Appl. Environ. Microbiol. 66:1139–1146 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Webster NS, Bourne DG, Hall M. 2006. Vibrionaceae infection in phyllosomas of the tropical rock lobster Panulirus ornatus as detected by fluorescence in situ hybridisation. Aquaculture 255:173–178 [Google Scholar]
- 53. Yoshizawa S, et al. 2011. Vibrio jasicida sp. nov., a member of the Harveyi clade, from marine animals (packhorse lobster, abalone, and Atlantic salmon). Int. J. Syst. Evol. Microbiol. doi:10.1099/ijs.0.025916-0 [DOI] [PubMed] [Google Scholar]
- 54. Yue X, Liu B, Sun L. 2011. Isolation and characterization of a virulent Vibrio sp. bacterium from clams (Meretrix meretrix) with mass mortality. J. Invertebr. Pathol. 106:242–249 [DOI] [PubMed] [Google Scholar]
- 55. Zhang X-H, Austin B. 2000. Pathogenicity of Vibrio harveyi to salmonids. J. Fish Dis. 23:93–102 [Google Scholar]
- 56. Zhu J, Mekalanos JJ. 2003. Quorum sensing-dependent biofilms enhance colonization in Vibrio cholerae. Dev. Cell 5:647–665 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.