Abstract
Flavonoids, secondary plant metabolites which mainly have a polyphenolic structure, play an important role in plant-microbe communications for nitrogen-fixing symbiosis. Among 10 polyphenolic compounds isolated from soybean roots in our previous study, coumestrol showed the highest antioxidant activity. In this study, its effect on the soybean nodulation was tested. The soybean symbiont Bradyrhizobium japonicum USDA110 pretreated with 20 μM coumestrol enhanced soybean nodulation by increasing the number of nodules 1.7-fold compared to the control. We also tested the effect of coumestrol on B. japonicum biofilm formation. At a concentration of 2 μM, coumestrol caused a higher degree of biofilm formation than two major soybean isoflavonoids, genistein and daidzein, although no biofilm formation was observed at a concentration of 20 μM each compound. A genome-wide transcriptional analysis was performed to obtain a comprehensive snapshot of the B. japonicum response to coumestrol. When the bacterium was incubated in 20 μM coumestrol for 24 h, a total of 371 genes (139 upregulated and 232 downregulated) were differentially expressed at a 2-fold cutoff with a q value of less than 5%. No common nod gene induction was found in the microarray data. However, quantitative reverse transcription-PCR (qRT-PCR) data showed that incubation for 12 h resulted in a moderate induction (ca. 2-fold) of nodD1 and nodABC, indicating that soybean coumestrol is a weak inducer of common nod genes. In addition, disruption of nfeD (bll4952) affected the soybean nodulation by an approximate 30% reduction in the average number of nodules.
INTRODUCTION
Flavonoids are a diverse class of secondary plant metabolites which typically have polyphenolic structures (44). Flavonoids, classified into several subgroups such as chalcones, flavanones, flavones, flavonols, and isoflavonoids, are accumulated in plant root tips and/or released into the rhizosphere (34). They play an important role in plant-microbe symbioses, specifically between legume plants and nitrogen-fixing soil bacteria, called rhizobia, as molecular signals to trigger the nodule development process (40, 41). It has been well known that the legume-rhizobium symbiosis is initiated by the secretion of flavonoids, which induce the rhizobial nodulation (nod) genes, resulting in the production of Nod factor (8, 39, 47). In turn, plants recognize the Nod factor and initiate a series of molecular events for nodulation.
Bradyrhizobium japonicum is a nitrogen-fixing soil bacterium that has a symbiotic relationship with the host legume soybean (Glycine max). Various exudates from soybean roots or seeds have been studied for their effects on nod gene expression in B. japonicum (1, 2, 25, 26, 28). Genistein and daidzein are major compounds among the soybean exudates that induce nod genes. In particular, genistein has been known to induce all nod box-associated genes, including the nodYABCSUIJ operon (30).
Ten polyphenolic compounds (three isoflavones, five pterocarpans, one flavonol, one anthocyanidin) were isolated from soybean roots in our previous study (31). The antioxidant activity of the isolated compounds was evaluated by measuring their inhibitory activity in copper-induced low-density lipoprotein (LDL) oxidation. Among them was a pterocarpan, identified as coumestrol, which turned out to be the strongest antioxidant, even compared to daidzein and genistein. Coumestrol, which was first identified from alfalfa in 1957, is a kind of pterocarpan possessing benzofuran and benzopyran rings (5). The original study with the isolated compounds was carried out from a medical perspective, focusing on their antioxidant activity. It has been reported that various (iso)flavonoids have antibacterial, antiviral, and antifungal activities that suit medical applications for treatments of a number of infections (9, 23, 52).
It is likely that coumestrol is produced by soybeans as one of the plant's responses to exogenous invaders. It has been reported that coumestrol is accumulated in soybean leaves inoculated with Xanthomonas campestris, a pathogen causing pustule disease of soybeans (13). Coumestrol also showed antimicrobial activities against strains of Bacillus subtilis, Bacillus licheniformis, Staphylococcus aureus, Streptococcus thermophilus, and Xanthomonas campestris pv. glycines (14). In addition to accumulation by bacteria, coumestrol is also involved in the early recognition process of the symbiotic relationship between soybean and its symbionts, B. japonicum and Rhizobium sp. NGR234 (48). Coumestrol is likely a weaker inducer of nod genes of B. japonicum compared to the other two major isoflavonoids genistein and daidzein (28).
Because the effects of coumestrol on nodulation and associated rhizobial physiology are relatively little known compared to those of genistein, the most studied isoflavonoid, we attempted to demonstrate how soybean-derived coumestrol influences soybean nodulation by B. japonicum. We also investigated the biofilm-forming ability and transcriptome-level changes of B. japonicum to obtain a comprehensive snapshot of the gene expression in response to coumestrol.
MATERIALS AND METHODS
Bacterial strains, growth, and chemicals.
B. japonicum USDA110 was cultivated at 30°C with shaking at 200 rpm in arabinose-gluconate (AG) medium (46) containing 125 mg Na2HPO4, 250 mg Na2SO4, 320 mg NH4Cl, 180 mg MgSO4 · 7H2O, 10 mg CaCl2, 4 mg FeCl3, 1.3 g HEPES, 1.1 g MES (morpholineethanesulfonic acid), 1.0 g yeast extract, 1.0 g l-arabinose, and 1.0 g Na-gluconate per liter (pH 6.8). The NtrC mutant and its complementary strains were generously provided by Gregory B. Martin (33) and William Franck (unpublished results). All mutant strains were also routinely maintained in AG medium. Antibiotics were used at the following concentrations when needed: kanamycin (150 μg/ml) and streptomycin (50 μg/ml).
For soybean nodulation assay and RNA isolation, cells were grown in a 1-liter-flask containing 250 ml of AG medium until mid-log phase. The cells were harvested by centrifugation at 8,000 × g and 4°C for 20 min, and the supernatant was removed. The cell pellets were washed with half-strength Broughton and Dilworth (B&D) medium, which does not contain a nitrogen source: 500 μM CaCl2, 250 μM KH2PO4, 250 μM K2HPO4, 5 μM Fe-citrate, 125 μM MgSO4 · 7H2O, 125 μM K2SO4, 0.5 μM MnSO4 · H2O, 1 μM H3BO3, 0.25 μM ZnSO4 · 7H2O, 1 μM CuSO4 · 5H2O, 0.05 μM CoSO4 · 7H2O, and 0.05 μM Na2MoO4 · 2H2O (pH 6.8). The cell suspension was centrifuged again under the same conditions, and the supernatant was removed. The cell pellets were resuspended in half-strength B&D to adjust to an optical density at 600 nm (OD600) of 1 (ca. 1 × 109 CFU/ml) in a 200-ml volume. After the addition of dimethyl sulfoxide (DMSO) (solvent for coumestrol) or coumestrol solution, the cell suspension was incubated at 30°C for 24 h without shaking. Coumestrol (molecular weight [MW] = 268) was made as 100 mM stock solution in DSMO, and an appropriate volume of the stock solution was added to each culture. For example, 20 μl of the 100 mM stock solution was added into 200 ml culture to make a final concentration of 10 μM. A corresponding volume of DMSO was also added to each control sample to ensure that the amounts of DMSO used did not affect bacterial growth. Coumestrol was obtained from soybean root exudates as described in our previous study (31), and DMSO, daidzein, and genistein were purchased from Sigma (St. Louis, MO).
Construction of nfeD mutant.
The 2.9-kb-sized fragment including the nfeD gene (bll4952, 1.4 kb) and its upstream and downstream regions was amplified by PCR with the primers PN1 (5′-AGA ACT GCG AGC TTC ATG GGA TCT-3′) and PN2 (5′-AAT GGC AAG CAT GGA TGT TCG ACG-3′). The PCR product was restricted by BamHI and ApaI and subsequently ligated with pJQ200SK (43) digested by the same restriction enzymes. The correct ligation product was named pHL4952. pHL4952 was introduced into the strain DH5α harboring pKD78, which expresses λ Red recombination genes under the control of the araBAD promoter. The kanamycin-resistant gene was amplified from pKD4 (10) using the primers PS1 (5′-GTG TAG GCT GGA GCT GCT TC-3′) and PS2 (5′-CAT ATG AAT ATC CTC CTT AG-3′). Using the PCR product as a template, the kanamycin-resistant gene was amplified again with the 60-bp primers PSS1 and PSS2, which contain 40-bp regions homologous to both bordering sequences of nfeD at the 5′ end of PS1 and PS2, respectively, by Phusion Taq polymerase under the following conditions: one cycle of 98°C for 30 s; two cycles of 98°C for 10 s, 54°C for 30 s, and 72°C for 45 s; 30 cycles of 98°C for 10 s, 68°C for 30 s, and 72°C for 45 s; and then 72°C for 5 min for final extension of the amplification. The sequences of PSS1 and PSS2 are 5′-CTC CTT CCC AGT TCA GCA GAG GAG AAC GGC CGT CTT GCA CGT GTA GGC TGG AGC TGC TTC-3′ and 5′-TTT GCG ACT TCG ACC GTC TCT CCG GGC TTG AAC GTT TCG GCA TAT GAA TAT CCT CCT TAG-3′, respectively. To induce λ Red recombinase genes in pKD78, the competent cells were prepared through the following steps: (i) an overnight culture of Escherichia coli harboring pHL4952 and pKD78 was subcultured in 30 ml of SOB (20 g tryptone, 5 g yeast extract, 2 ml of 5 M NaCl, and 2.5 ml of 1 M KCl per liter); (ii) l-arabinose was added (300 μl of 1 M stock solution) in the culture medium to induce the araBAD promoter; (iii) the subculture was grown to OD600 = 0.5 at 30°C, washed twice with 10% glycerol, and resuspended in 2 ml of 10% glycerol; and (iv) the 100-μl aliquots of the competent cells were flash frozen and stored at −80°C until use. Then, the mixture of the competent cells and the kanamycin-resistant gene containing nfeD-homologous regions (∼200 ng) was electroporated (22 kV/cm), incubated at 37°C for 1 h after the addition of 800 μl of LB, and selected on LB agar with gentamicin (15 μg/ml) and kanamycin (50 μg/ml). The created plasmid was named pHL4952::Km. pHL4952::Km was transferred from E. coli DH5α to B. japonicum USDA110 by triparental mating with the helper plasmid pRK2073 (11). The nfeD mutant strain created by double-homologous recombination was selected on AG agar plates containing chloroamphenicol (30 μg/ml), kanamycin (150 μg/ml), and 8% (wt/vol) sucrose, and disruption of the gene by the kanamycin resistance gene was confirmed by colony PCR (data not shown). The addition of sucrose allowed us to positively select double-crossover homologous recombination events against single-crossover recombinants, which are not replicable under these conditions (5 to 10% sucrose) due to the insertion of the sacB gene from pJQ200SK into the chromosome.
Biofilm assay and growth kinetics.
For the biofilm assay, we partially modified a method described in a previous study (12). Cells were harvested from 10-ml cultures at mid-log phase. The cell pellets were washed with 10 ml of Bergersen minimal medium (4) supplemented with 0.4% (vol/vol) glycerol (BMM) and resuspended in BMM to adjust to an OD600 of 1. The cell suspension was 3% (vol/vol) diluted (OD600 was approximately 0.03; ca. 3 × 107 cells/ml) into BMM supplemented with coumestrol, genistein, daidzein, or their solvent DMSO. After vortexing, 150-μl volumes were transferred into 6 wells of a polystyrene 96-well microtiter plate per sample. A total of 8 plates were identically prepared and incubated at 30°C without shaking. Each plate was used for the analysis of biofilm-forming abilities at each time point.
The cell density at every time period was measured by reading the OD600 on a microtiter plate reader (Synergy 2; BioTek). The medium and planktonic cells in every well were aseptically removed by vacuum aspiration, and each well was washed three times with 200 μl of sterile double-distilled water (ddH2O). Then, the plate was dried in air aseptically for 30 min. Every well was stained with 150 μl of 1% (wt/vol) crystal violet (CV) for 45 min in the dark. The CV was removed by vacuum suction, and each well was washed three times with 400 μl of sterile ddH2O. To quantify the amount of biofilm, the CV was destained with 200 μl of 100% ethanol. The absorbance of 100 μl of the destained CV which was transferred into a new microtiter plate was measured at 595 nm. All experiments were repeated independently three times.
Nodulation assay.
A nodulation assay was performed on a laboratory scale using small plastic pouches (Mega International). Soybean seeds were washed with 20% (vol/vol) bleach for 10 min, rinsed three times with sterile ddH2O, washed again with 0.01 M HCl for 10 min, and rinsed three times with sterile ddH2O. After being allowed to dry at room temperature, the surface-sterilized soybean seeds were placed on presoaked Whatman paper in petri dishes and were germinated in the dark at room temperature for 2 or 3 days until the root length was between 2 and 3 cm. The germinated soybean seeds were aseptically placed in pouches and inoculated with 1 ml of the cell suspension (OD600 = 0.1; ca. 1 × 108 CFU/ml) of B. japonicum USDA110 pretreated with various concentrations of coumestrol. The position of the root tip was marked on the surface of the pouch. The seeds were grown in a plant chamber at 26°C with 15 h of day and 9 h of night. The pouches were watered with half-strength B&D when needed. After 2 weeks, the number of nodules was counted. In addition, the nodule distribution was analyzed by measuring the distance (in millimeters) of all nodules on the primary root from the root tip mark (21). A total of nine plants were used to measure the parameters in each experiment, and all experiments were repeated three times.
RNA isolation, cDNA synthesis, and hybridization.
Total RNA was isolated by the hot phenol extraction method as described previously (6). DNase treatment and RNA purification were performed using a Qiagen RNeasy minikit (Qiagen Sciences) and RNase-free DNase set (Qiagen Sciences) as recommended in the manufacturer's instructions.
For the genome-wide transcriptional profiling of B. japonicum, we used the whole-genome 70-mer oligonucleotide microarrays developed previously (7), which contain probes specific to 8,453 predicted open reading frames. Every step of cDNA synthesis, labeling of Cy3 and Cy5, hybridization, and scanning was performed as previously described (7). Three biological replicates of the cell cultures were applied, and hybridization was conducted in three replications with dye swaps.
Microarray data analysis.
The microarray data were analyzed as described in a previous study (7) through microarray analysis of variance (MAANOVA) (27) and significance analysis of microarray (SAM) (53) with a false-discovery rate lower than 5%.
qRT-PCR analysis.
Quantitative reverse transcription-PCR (qRT-PCR) was performed in order to (i) confirm microarray data for the selected genes and (ii) examine the expression of common nod genes and some of the significantly induced genes in the microarray data. Each RNA sample for the first qRT-PCR was obtained from the same batch for microarray. On the other hand, RNAs for the latter experiment were isolated from different batches after treatment of the cells with 20 μM coumestrol for 3, 12, and 24 h. The primers for the selected genes are listed in Table S1 in the supplemental material. The whole process was performed as described in the previous study (16). Histidyl-tRNA synthetase hisS (bll7457) was used as the reference gene for data normalization in both experiments.
Microarray data accession numbers.
The microarray data from this study are compiled in the NCBI Gene Expression Omnibus (GEO) database (http://www.ncbi.nlm.nih.gov/geo/). The GEO series accession number is GSE26380.
RESULTS
Pretreatment of B. japonicum with the high concentration of coumestrol enhances the soybean nodulation.
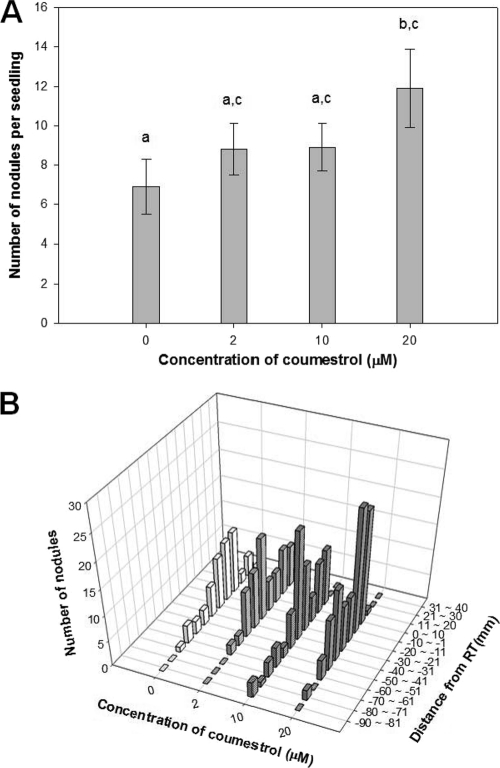
It has been known that preincubation of rhizobia with isoflavonoids accelerates nodule development (57). Therefore, we hypothesized that pretreatment of B. japonicum with coumestrol may enhance the soybean nodulation. The B. japonicum cultures were treated with coumestrol at three different concentrations (2, 10, and 20 μM) prior to inoculation of soybean seeds. At the concentrations of 2 and 10 μM, the nodulation was increased to some degree but with no statistical significance (Fig. 1A). However, when inoculated with B. japonicum treated with 20 μM, the soybean plants formed 1.7 times more nodules than the control (Fig. 1A).
Fig 1.
Effect of pretreatment of B. japonicum with different concentrations of coumestrol on soybean nodulation. (A) Numbers of nodules. Values are means ± standard errors of the means from nine replications. The different letters above the bars indicate that the data are significantly different from each other (P < 0.05; by t test using JMP8 statistical software). (B) Distribution of nodules. Positive and negative (−) values on the “Distance from RT (mm)” axis indicate measurements above and below root tips, respectively. The values on the “Number of nodules” axis are the total nodule numbers from nine replications.
The distribution of nodules was also analyzed by measuring the distance from the initial root tip point to the position of each nodule to determine whether any enhanced or delayed nodulation occurred. The overall pattern of the nodule distribution on the primary root is similar among all samples, and the most nodules have been formed near the initial root tip position, suggesting that there is no difference in timing and duration of nodule development by pretreatment of B. japonicum with coumestrol, even at the high concentration. However, the overall increased nodule numbers at 20 μM were due to the increased number of nodules located at ±10 mm from the root tip (Fig. 1B).
Coumestrol inhibits the biofilm formation and growth of B. japonicum.
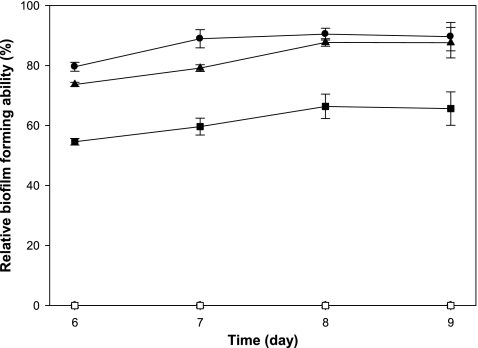
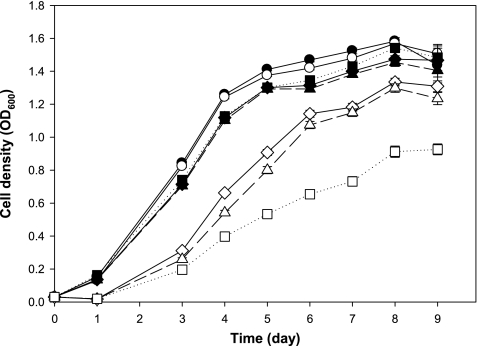
Enhanced nodulation by coumestrol pretreatment could be due to increased biofilm formation by coumestrol-treated B. japonicum. It can be postulated that biofilm formation may be advantageous to rhizobial colonization on legume root surfaces (45). Luteolin, a flavonoid isolated from alfalfa, promoted biofilm formation by its symbiotic partner Sinorhizobium meliloti (17). To investigate the effect of coumestrol on B. japonicum biofilm formation, the quantitative biofilm assay was performed at various concentrations of coumestrol. We also included two other major soybean isoflavonoids, genistein and daidzein, and their relative abilities of biofilm formation were monitored from 6 to 9 days. At a concentration of 2 μM, coumestrol showed the highest biofilm formation, although no biofilm formation was observed at 20 μM for any of the three compounds (Fig. 2). Given the fact that a 6-to-9-day incubation time indicates the stationary phase, the lack of biofilm formation at the high concentration of 20 μM led us to examine whether it is related to growth rate. The growth of B. japonicum in the presence of coumestrol, genistein, or daidzein was monitored at the concentrations of 2 and 20 μM each. As shown in Fig. 3, the bacterial growth was inhibited by all of the compounds tested, although the degree of inhibition was different among the compounds. At a concentration of 20 μM, genistein showed the highest inhibitory effect on the bacterial growth, while coumestrol showed the lowest effect (Fig. 3). However, at a concentration of 2 μM, there was no difference of growth rates among cells treated with each compound (Fig. 3). Therefore, we conclude that the enhanced biofilm formation in the case of coumestrol compared to that in genistein and daidzein is not mediated by growth rate (Fig. 2 and 3).
Fig 2.
Effect of coumestrol, daidzein, and genistein on the biofilm formation ability of B. japonicum. Circles (● and ○), triangles (▲ and △), and squares (■ and □) indicate coumestrol, daidzein, and genistein, respectively. Solid and open symbols are 2 and 20 μM, respectively. All of the open symbols are overlapped because B. japonicum did not form biofilms in 20 μM each compound. To quantify the biofilm formation ability, the specific biofilm-forming ability was normalized by dividing the total biofilm OD595 value by the cell growth OD600 value. The relative biofilm-forming ability was then calculated by dividing the specific biofilm-forming ability value of each treatment sample by that of the control DMSO. Values are means ± standard errors of the mean for six replications.
Fig 3.
Growth of B. japonicum in the presence of coumestrol, daidzein, and genistein. H2O and DMSO were used as the general and vehicle controls, respectively. Circle symbols indicate controls [solid (●) for H2O and open (○) for DMSO]. Diamond (♦ and ♢), triangle (▲ and △), and square (■ and □) symbols indicate coumestrol, daidzein, and genistein, respectively. Solid (♦, ▲, and ■) and open (♢, △, and □) symbols indicate 2 and 20 μM, respectively. Values are means ± standard errors of the mean for six replications.
Transcriptome analysis.
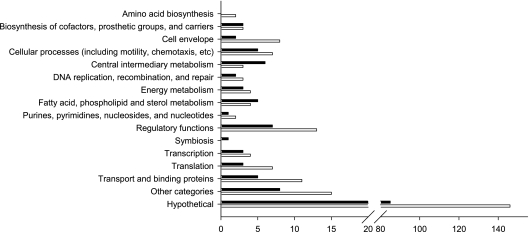
In order to investigate genes responsible for coumestrol-mediated nodulation, we employed global gene expression profiling using the oligonucleotide microarray technology that has been previously described and validated (7). A total of 371 genes (139 upregulated and 232 downregulated) were differentially expressed at a 2-fold cutoff with a q value of <0.05 (Fig. 4). Substantial numbers of differentially expressed genes were observed in the functional categories of cell envelope, cellular process, including motility and chemotaxis genes, regulatory proteins, translation, and transport and binding proteins (Fig. 4). Of 10 genes differentially expressed in the cell envelope group, 5 genes related to outer membrane were downregulated (see Table S2 in the supplemental material), which probably is linked to the antimicrobial effect of coumestrol. Coumestrol did not induce any gene involved in motility and chemotaxis groups. Instead, pilA (bsl6587), ctpA (bsl7141), and bll7954 (putative methyl-accepting chemotaxis protein) were downregulated (see Table S2), indicating that coumestrol may not enhance soybean nodulation via chemotaxis and motility functions of B. japonicum.
Fig 4.
Functional group assignment of genes differentially expressed in the presence of 20 μM coumestrol at a 2-fold cutoff with q value less than 5%. Black and white bars indicate up- and downregulated genes, respectively. Functional classifications were derived from B. japonicum USDA110 genome annotations available through Rhizobase (http://genome.kazusa.or.jp/rhizobase/).
A total of 20 regulatory protein genes were differentially expressed: 7 genes upregulated and 13 downregulated (see Table S2). The upregulated genes include a two-component response regulator (bll2758), a putative monoamine oxidase regulator (bll3297), one LuxR family regulator (blr6291), two TetR family regulators (bll7024 and blr4080), and two Crp family regulators (bll6061 and blr7084). The induction of the two Crp family regulators, named fixK1 (bll6061) and nnrR, seems to be related to the expression of fixK2 (bll2757), since they are regulated by FixK2 in the FixLJ-FixK2 regulatory system under symbiotic and denitrification conditions (22, 35).
Validation of microarray data using qRT-PCR.
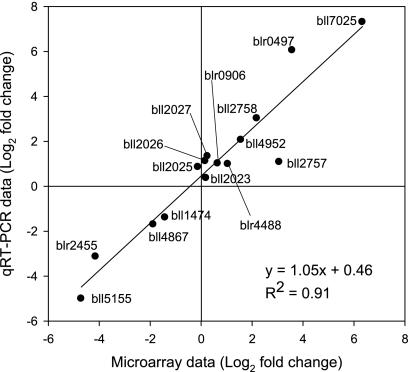
To confirm the microarray data, qRT-PCR was employed for the genes listed in Table S1 in the supplemental material. Fifteen genes were selected based on their functional categories and fold change values in the microarray data. The relationship between microarray and qRT-PCR data denotes a good accordance of expression values (R2 = 0.91) (Fig. 5).
Fig 5.
Correlation between microarray and qRT-PCR data for the control versus the 20 μM coumestrol treatment for 15 genes listed in Table S1 in the supplemental material. The genes were selected based on the fold induction and functional categories. Expression differences were log2 transformed.
Coumestrol is a weak inducer of B. japonicum nod genes.
From the microarray data, the result that coumestrol did not induce common nod genes was surprising since other isoflavonoids such as genistein and daidzein are known to induce canonical nod genes (28, 30). We performed qRT-PCR with RNAs harvested at various time points, since the conditions of the transcriptome snapshot may not have been sufficient enough to detect expression of nod genes (nodD1 and nodABC). The higher sensitivity of qRT-PCR over microarray is the other reason for this confirmation (3, 7). All of the nod genes tested were induced at low levels (1.75- to 2.40-fold) after 12 h, whereas after 24 h only nodB and nodC remained induced (Table 1). This is consistent with the previous study that coumestrol weakly induced B. japonicum common nod genes compared to genistein and daidzein (28).
Table 1.
Expression of nod genes and the genes selected from the microarray data
| Gene ID | Fold change in microarray (24-h treatment) | Fold change in qRT-PCRa (mean ± SE) |
||
|---|---|---|---|---|
| 3 h | 12 h | 24 h | ||
| bll2023 (nodD1) | 1.14 | 1.43 ± 0.58 | 2.40 ± 0.84 | 1.29 ± 0.28 |
| blr2025 (nodA) | −1.09 | −1.16 ± 0.11 | 1.79 ± 0.35 | 1.81 ± 1.42 |
| blr2026 (nodB) | 1.12 | −1.10 ± 0.11 | 2.33 ± 0.14 | 2.17 ± 0.33 |
| blr2027 (nodC) | 1.19 | −1.13 ± 0.08 | 1.75 ± 0.11 | 2.52 ± 0.35 |
| bll4952 (nfeD) | 2.95 | 8.74 ± 4.17 | 6.87 ± 3.64 | 4.18 ± 2.45 |
| bll2757 (fixK2) | 8.37 | 30.94 ± 21.91 | 12.80 ± 7.03 | 2.11 ± 0.92 |
| blr4488 (ntrC) | 2.07 | 1.27 ± 0.10 | 2.31 ± 0.47 | 1.98 ± 0.78 |
Different RNA samples were used for microarray and qRT-PCR experiments, specifically 24-h samples. Each time indicates incubation time (in hours) with 20 μM coumestrol before cells were harvested for RNA isolation.
Expression of ntrC is not responsible for the coumestrol-mediated nodulation enhancement.
Among the nitrogen fixation-related genes expressed, ntrC (blr4488) is of interest because NtrC is a key regulator in nitrogen metabolism (32) and regulates the glutamine synthetase II gene (glnII) in B. japonicum (33). It has been also reported that the ntrC mutant of Azorhizobium caulinodans exhibits delayed nodulation and reduced nitrogen-fixing activity but is able to form effective nodules on the semiaquatic legume Sesbania rostrata (38). An ntrC mutant was tested for its nodulation ability to determine whether ntrC expression is responsible for the enhanced nodulation by 20 μM coumestrol. We could not find significantly decreased nodulation by the ntrC mutant compared to that of its wild type and the complemented strain (data not shown).
NfeD-like protein is responsible for the soybean nodulation.
Among the genes found to be induced by coumestrol and categorized in the symbiosis group was an NfeD protein homolog (Bll4952) (see Table S2 in the supplemental material). In B. japonicum, there has been no report on the function of nfeD to date. Thus, we mutagenized the gene to examine its effect on soybean nodulation. The physiological properties, such as colony morphology and growth rate, were not different between USDA110 and ΔnfeD (data not shown). However, the disruption of nfeD affected the soybean nodulation. The number of nodules formed by ΔnfeD was reduced compared to those in the wild type (wild type, 8.3 ± 0.7; ΔnfeD, 5.7 ± 0.7 per seedling; P < 0.05). In addition, coumestrol treatment did not cause an increase of the number of nodules formed by ΔnfeD (4.8 ± 0.8 per seedling), indicating that nfeD plays an important role in the enhancement of soybean nodulation by coumestrol-treated B. japonicum.
DISCUSSION
We showed that soybean coumestrol enhanced the nodulating ability of B. japonicum on soybean roots, and we attempted to find out the molecular mechanisms of the nodulation enhancement at the transcriptional level using an oligonucleotide microarray platform developed previously (7). Since other studies have demonstrated not only that common nod genes are essential for nodule development (40, 41) but also that (iso)flavonoids induce the nod genes in rhizobia (8, 39, 47), we, of course, expected that the enhancement of soybean nodulation was also mediated by the expression of nod genes. Although our initial transcriptomic data showed no induction of nod genes, we were able to confirm the low-level induction of nodD1 and nodABC by qRT-PCR (Table 1), which could be the most sensitive method for the detection of low levels of mRNA. In contrast to genistein and daidzein, well-known strong inducers of the nod genes (28, 29), coumestrol has not been attractable due to its weak effects, as shown in several previous studies (25, 28). Moreover, there have been some contradictory roles of coumestrol in nod gene induction. Coumestrol, on the one hand, may be a weak inducer of common nod genes (42), but on the other hand it may be considered to be a weak inhibitor (29). Our qRT-PCR data are in accordance with the former perspective (Table 1). Alternatively, it appears somewhat confusing to define coumestrol as a weak inhibitor in the latter study, as coumestrol merely reduced the nod gene induction by daidzein or genistein when combined with them but itself showed approximately 65% induction compared to daidzein (29). Therefore, coumestrol might be weakly involved in the induction of nod genes at the initial step of soybean nodule development by B. japonicum.
Among the notable genes induced by pretreatment of coumestrol from the microarray data is nfeD (bll4952; NfeD protein homolog), which is the only gene categorized in the symbiosis group. Based on the protein sequence analysis (data not shown), it contains an N-terminal membrane-bound ClpP-class protease domain and a C-terminal NfeD domain (PFAM01957). NfeD-like protein genes are located in many prokaryotic genomes, and they frequently accompany genes encoding stomatin-like proteins in an operon (20). Like many other species, in the B. japonicum genome the nfeD gene forms an operon with a stomatin-like protein (bll4951). Correctly speaking, the name nfeD for this gene might be erroneous, presumably led by an incorrect annotation of the protein domain PFAM01957 (19, 20). Moreover, this gene is entirely unrelated to the nfeD (nodulation formation efficiency) gene described in previous studies (18, 50), which showed delayed nodulation by the S. meliloti nfeD mutant strain. The product of the gene is rather similar to an ornithine cyclodeaminase from Agrobacterium tumefaciens pTiAch5 (50). Because of this confusion, the NfeD protein homolog gene (bll4952) of B. japonicum needs to be further studied for its role(s) in B. japonicum-soybean symbiosis. Therefore, our finding that the lack of NfeD protein homolog gene in B. japonicum affects the soybean nodulation is noteworthy. However, we cannot rule out the possibility that the insertion of a kanamycin resistance gene into the nfeD gene might influence transcription and/or translation of the next gene, stomatin-like protein (bll4951), although bll4951 has not been differentially expressed by a coumestrol pretreatment. It has been reported that stomatin-like protein in Rhizobium etli plays an important role in nodulation competitiveness on the common bean (56); that report is, to our knowledge, the only study of the function of a stomatin-like protein in the nodulation process.
The induction of several nitrogen fixation genes, such as fixK2, nifN, fixQ, nnrR, and fixK1, is remarkable. In particular, fixK2, like nfeD, was induced from an early time point and continued to be induced through 24 h, although the induction level is decreased over incubation time (Table 1). Since the induction of those nitrogen fixation genes is known to be triggered by micro-oxic culture conditions (15, 22, 35), we checked growth conditions in our study. Although aerobic culture conditions were used, the high-density culture incubated without shaking after coumestrol treatment might cause a partially micro-oxic condition in the culture. Nevertheless, the culture conditions of the control and the treatment samples were exactly the same with the only exception being the treatment compound (DMSO versus coumestrol dissolved in DMSO). Therefore, it would be interesting to reveal a molecular mechanism by which the weaker inducer coumestrol affects nitrogen fixation genes in B. japonicum.
The antimicrobial activity of coumestrol on B. japonicum and coumestrol-mediated nodulation enhancement is likely a general phenomenon of (iso)flavonoids. In this study, high concentrations of genistein and daidzein that induce nod genes also revealed antimicrobial activity. In addition, flavonoids isolated from other legume plants possess antimicrobial activities (9). Expression of heat shock protein genes like hspH (bll0729) and hsp20 (blr4637) (see Table S2 in the supplemental material) is a line of evidence that coumestrol is an environmental cue causing the growth inhibition. Six ribosomal protein genes (blr0420, bll5378, bsr8096, bsr5117, bsr7117, and blr0365) were also downregulated (see Table S2). It has been reported that a substantial number of ribosomal protein genes were repressed by cultivation in minimal medium, in which the bacteria grow slowly, for E. coli (51) and B. japonicum (7). The repression of those genes by coumestrol is likely to be related to the inhibition of B. japonicum growth.
There are three mechanisms suggested for flavonoids as an antimicrobial agent at the gene expression level: (i) inhibition of nucleic acid synthesis, (ii) inhibition of cytoplasmic membrane function, and (iii) inhibition of energy metabolism (9). Based on these criteria, we sorted out our microarray data for the antimicrobial activity of coumestrol. Regarding nucleic acid synthesis, several genes, which are related to DNA replication, recombination, and repair, including bll4517 (DNA polymerase III), bll6773 (probable DNA ligase), and uvrA (excinuclease, blr8051), were downregulated (see Table S2 in the supplemental material). For the membrane function, the genes related to outer membrane proteins such as bll4853, bll4867, bll7469, blr4701, and blr7695 were downregulated (see Table S2). Among the energy metabolism-related genes, aceA (isocitrate lyase, blr2455) was most highly repressed (17.65-fold) (see Table S2). In addition, genes (bll5913 and blr7544) related to respiration were also downregulated. Hence, growth inhibition and reduced biofilm formation might be due to the combined effects of those downregulated genes in DNA synthesis, membrane integrity, and energy metabolism.
Flagella are known to be necessary for the initial attachment stage during nodule development (36). Although Lang et al. reported that genistein induced a large number of genes in a flagellar cluster (30), coumestrol did not induce any chemotaxis-related genes in this study. Coumestrol even repressed the expression of pilA (bsl6587), ctpA (bsl7141), and putative methyl-accepting chemotaxis protein (bll7954). This could be consistent with the previous study results that isoflavones are not principal chemoattractants to B. japonicum (2). In addition, repression of both pilA- and ctpA-encoding components of type IV pilus pilin subunit might be involved in the lack of biofilm formation at the high concentration of coumestrol, since it has been demonstrated that type IV pili contribute to biofilm formation in many bacteria (24, 37, 49).
Taken together, our studies examined the antimicrobial activities of coumestrol isolated from soybean roots and its effects on the nodulation, biofilm formation, and transcriptome of B. japonicum. The two major properties of coumestrol found in this study include the growth inhibition and the enhancement of nodulation. These findings seem to be contradictory to each other. Moreover, these properties were found only at a high concentration (20 μM) of coumestrol and not at a low concentration (2 μM). Thus, we could postulate that B. japonicum cells that have adapted to a high concentration of coumestrol may become more resistant to plant defense mechanisms, thereby enhancing the probability of nodulation. This might be the nature of phenolic compounds isolated from legume plants, since genistein has similar characteristics (9, 54, 55, 57). In addition, our transcriptomic and mutation studies indicate that NfeD-like protein is involved in coumestrol-mediated soybean nodulation.
Supplementary Material
ACKNOWLEDGMENTS
We thank Gregory B. Martin and William L. Franck for the ntrC mutant and its complementary strains. We also thank Andrew Donati for his critical reading of the manuscript.
This research was supported by a Research Enhancement Program (REP) grant from the University of Texas—Arlington.
Footnotes
Published ahead of print 3 February 2012
Supplemental material for this article may be found at http://aem.asm.org/.
REFERENCES
- 1. Banfalvi Z, Nieuwkoop A, Schell M, Besl L, Stacey G. 1988. Regulation of nod gene expression in Bradyrhizobium japonicum. Mol. Gen. Genet. 214:420–424 [DOI] [PubMed] [Google Scholar]
- 2. Barbour WM, Hattermann DR, Stacey G. 1991. Chemotaxis of Bradyrhizobium japonicum to soybean exudates. Appl. Environ. Microbiol. 57:2635–2639 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Barnett MJ, Toman CJ, Fisher RF, Long SR. 2004. A dual-genome symbiosis chip for coordinate study of signal exchange and development in a prokaryote-host interaction. Proc. Natl. Acad. Sci. U. S. A. 101:16636–16641 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Bergersen FJ. 1961. The growth of rhizobium in synthetic media. Aust. J. Biol. Sci. 14:349–360 [Google Scholar]
- 5. Bickoff EM, et al. 1957. Coumestrol, a new estrogen isolated from forage crops. Science 126:969–970 [DOI] [PubMed] [Google Scholar]
- 6. Bittner M, et al. 2003. Expression analysis of RNA, p 101–288 In Bowtell D, Sambrook J. (ed), DNA microarrays: a molecular cloning manual. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY [Google Scholar]
- 7. Chang WS, et al. 2007. An oligonucleotide microarray resource for transcriptional profiling of Bradyrhizobium japonicum. Mol. Plant Microbe Interact. 20:1298–1307 [DOI] [PubMed] [Google Scholar]
- 8. Cooper JE. 2004. Multiple responses of rhizobia to flavonoids during legume root infection. Adv. Bot. Res. 41:1–62 [Google Scholar]
- 9. Cushnie TPT, Lamb AJ. 2005. Antimicrobial activity of flavonoids. Int. J. Antimicrob. Agents 26:343–356 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Datsenko KA, Wanner BL. 2000. One-step inactivation of chromosomal genes in Escherichia coli K-12 using PCR products. Proc. Natl. Acad. Sci. U. S. A. 97:6640–6645 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Ditta G, Stanfield S, Corbin D, Helinski DR. 1980. Broad host range DNA cloning system for gram-negative bacteria: construction of a gene bank of Rhizobium meliloti. Proc. Natl. Acad. Sci. U. S. A. 77:7347–7351 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Djordjevic D, Wiedmann M, McLandsborough LA. 2002. Microtiter plate assay for assessment of Listeria monocytogenes biofilm formation. Appl. Environ. Microbiol. 68:2950–2958 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Fett WF. 1984. Accumulation of isoflavonoids and isoflavone glucosides after inoculation of soybean leaves with Xanthomonas campestris pv. glycines and pv. campestris and a study of their role in resistance. Physiol. Plant Pathol. 24:303–320 [Google Scholar]
- 14. Fett WF, Osman SF. 1982. Inhibition of bacteria by the soybean isoflavonoids glyceollin and coumestrol. Phytopathology 72:755–760 [Google Scholar]
- 15. Fischer HM, et al. 2001. One of two hemN genes in Bradyrhizobium japonicum is functional during anaerobic growth and in symbiosis. J. Bacteriol. 183:1300–1311 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Franck WL, et al. 2008. Whole-genome transcriptional profiling of Bradyrhizobium japonicum during chemoautotrophic growth. J. Bacteriol. 190:6697–6705 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Fujishige NA, et al. 2008. Rhizobium common nod genes are required for biofilm formation. Mol. Microbiol. 67:504–515 [DOI] [PubMed] [Google Scholar]
- 18. Garcia-Rodriguez FM, Toro N. 2000. Sinorhizobium meliloti nfe (nodulation formation efficiency) genes exhibit temporal and spatial expression patterns similar to those of genes involved in symbiotic nitrogen fixation. Mol. Plant Microbe Interact. 13:583–591 [DOI] [PubMed] [Google Scholar]
- 19. Green JB, et al. 2004. Eukaryotic and prokaryotic stomatins: the proteolytic link. Blood Cells Mol. Dis. 32:411–422 [DOI] [PubMed] [Google Scholar]
- 20. Green JB, Lower RP, Young JP. 2009. The NfeD protein family and its conserved gene neighbours throughout prokaryotes: functional implications for stomatin-like proteins. J. Mol. Evol. 69:657–667 [DOI] [PubMed] [Google Scholar]
- 21. Halverson LJ, Stacey G. 1986. Effect of lectin on nodulation by wild-type Bradyrhizobium japonicum and a nodulation-defective mutant. Appl. Environ. Microbiol. 51:753–760 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Hauser F, et al. 2006. Design and validation of a partial-genome microarray for transcriptional profiling of the Bradyrhizobium japonicum symbiotic gene region. Mol. Genet. Genomics 275:55–67 [DOI] [PubMed] [Google Scholar]
- 23. Havsteen BH. 2002. The biochemistry and medical significance of the flavonoids. Pharmacol. Ther. 96:67–202 [DOI] [PubMed] [Google Scholar]
- 24. Jurcisek JA, Bakaletz LO. 2007. Biofilms formed by nontypeable Haemophilus influenzae in vivo contain both double-stranded DNA and type IV pilin protein. J. Bacteriol. 189:3868–3875 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Kape R, Parniske M, Brandt S, Werner D. 1992. Isoliquiritigenin, a strong nod gene- and glyceollin resistance-inducing flavonoid from soybean root exudate. Appl. Environ. Microbiol. 58:1705–1710 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Kape R, Parniske M, Werner D. 1991. Chemotaxis and nod gene activity of Bradyrhizobium japonicum in response to hydroxycinnamic acids and isoflavonoids. Appl. Environ. Microbiol. 57:316–319 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Kerr MK, Martin M, Churchill GA. 2000. Analysis of variance for gene expression microarray data. J. Comput. Biol. 7:819–837 [DOI] [PubMed] [Google Scholar]
- 28. Kosslak RM, Bookland R, Barkei J, Paaren HE, Appelbaum ER. 1987. Induction of Bradyrhizobium japonicum common nod genes by isoflavones isolated from Glycine max. Proc. Natl. Acad. Sci. U. S. A. 84:7428–7432 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Kosslak RM, Joshi RS, Bowen BA, Paaren HE, Appelbaum ER. 1990. Strain-specific inhibition of nod gene induction in Bradyrhizobium japonicum by flavonoid compounds. Appl. Environ. Microbiol. 56:1333–1341 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Lang K, Lindemann A, Hauser F, Gottfert M. 2008. The genistein stimulon of Bradyrhizobium japonicum. Mol. Genet. Genomics 279:203–211 [DOI] [PubMed] [Google Scholar]
- 31. Lee JH, et al. 2006. LDL-antioxidant pterocarpans from roots of Glycine max (L.) Merr. J. Agric. Food Chem. 54:2057–2063 [DOI] [PubMed] [Google Scholar]
- 32. Magasanik B. 1982. Genetic control of nitrogen assimilation in bacteria. Annu. Rev. Genet. 16:135–168 [DOI] [PubMed] [Google Scholar]
- 33. Martin GB, Chapman KA, Chelm BK. 1988. Role of the Bradyrhizobium japonicum ntrC gene product in differential regulation of the glutamine synthetase II gene (glnII). J. Bacteriol. 170:5452–5459 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Maxwell CA, Phillips DA. 1990. Concurrent synthesis and release of nod-gene-inducing flavonoids from alfalfa roots. Plant Physiol. 93:1552–1558 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Mesa S, Bedmar EJ, Chanfon A, Hennecke H, Fischer HM. 2003. Bradyrhizobium japonicum NnrR, a denitrification regulator, expands the FixLJ-FixK2 regulatory cascade. J. Bacteriol. 185:3978–3982 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. O'Toole GA, Kolter R. 1998. Initiation of biofilm formation in Pseudomonas fluorescens WCS365 proceeds via multiple, convergent signalling pathways: a genetic analysis. Mol. Microbiol. 28:449–461 [DOI] [PubMed] [Google Scholar]
- 37. Paranjpye RN, Strom MS. 2005. A Vibrio vulnificus type IV pilin contributes to biofilm formation, adherence to epithelial cells, and virulence. Infect. Immun. 73:1411–1422 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Pawlowski K, Ratet P, Schell J, de Bruijn FJ. 1987. Cloning and characterization of nifA and ntrC genes of the stem nodulating bacterium ORS571, the nitrogen fixing symbiont of Sesbania rostrata: regulation of nitrogen fixation (nif) genes in the free living versus symbiotic state. Mol. Gen. Genet. 206:207–219 [Google Scholar]
- 39. Peters NK, Frost JW, Long SR. 1986. A plant flavone, luteolin, induces expression of Rhizobium meliloti nodulation genes. Science 233:977–980 [DOI] [PubMed] [Google Scholar]
- 40. Peters NK, Verma DP. 1990. Phenolic compounds as regulators of gene expression in plant-microbe relations. Mol. Plant Microbe Interact. 3:4–8 [DOI] [PubMed] [Google Scholar]
- 41. Phillips DA. 1992. Flavonoids: plant signals to soil microbes, p 201–231 In Stafford HA, Ibrahim RK. (ed), Recent advances in phytochemistry, vol 26. Phenolic metabolism in plants. Plenum Press, New York, NY [Google Scholar]
- 42. Phillips DA, Kapulnik Y. 1995. Plant isoflavonoids, pathogens and symbionts. Trends Microbiol. 3:58–64 [DOI] [PubMed] [Google Scholar]
- 43. Quandt J, Hynes MF. 1993. Versatile suicide vectors which allow direct selection for gene replacement in Gram-negative bacteria. Gene 127:15–21 [DOI] [PubMed] [Google Scholar]
- 44. Rao JR, Cooper JE. 1994. Rhizobia catabolize nod gene-inducing flavonoids via C-ring fission mechanisms. J. Bacteriol. 176:5409–5413 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Rinaudi LV, Giordano W. 2010. Bacterial biofilms: role in Rhizobium-legume symbiosis, p 325–335 In Khan MS, Musarrat J, Zaidi A. (ed), Microbes for legume improvement. Springer, Berlin, Germany [Google Scholar]
- 46. Sadowsky MJ, Tully RE, Cregan PB, Keyser HH. 1987. Genetic diversity in Bradyrhizobium japonicum serogroup 123 and its relation to genotype-specific nodulation of soybean. Appl. Environ. Microbiol. 53:2624–2630 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Schlaman HRM, Okker RJH, Lugtenberg BJJ. 1992. Regulation of nodulation gene-expression by NodD in rhizobia. J. Bacteriol. 174:5177–5182 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Schmidt PE, Broughton WJ, Werner D. 1994. Nod factors of Bradyrhizobium japonicum and Rhizobium sp. NGR234 induce flavonoid accumulation in soybean root exudate. Mol. Plant Microbe Interact. 7:384–390 [Google Scholar]
- 49. Shime-Hattori A, et al. 2006. Two type IV pili of Vibrio parahaemolyticus play different roles in biofilm formation. FEMS Microbiol. Lett. 264:89–97 [DOI] [PubMed] [Google Scholar]
- 50. Soto MJ, et al. 1994. Identification of a novel Rhizobium meliloti nodulation efficiency nfe gene homolog of Agrobacterium ornithine cyclodeaminase. Mol. Plant Microbe Interact. 7:703–707 [DOI] [PubMed] [Google Scholar]
- 51. Tao H, Bausch C, Richmond C, Blattner FR, Conway T. 1999. Functional genomics: expression analysis of Escherichia coli growing on minimal and rich media. J. Bacteriol. 181:6425–6440 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Tripoli E, La Guardia M, Giammanco S, Di Majo D, Giammanco M. 2007. Citrus flavonoids: molecular structure, biological activity and nutritional properties: a review. Food Chem. 104:466–479 [Google Scholar]
- 53. Tusher VG, Tibshirani R, Chu G. 2001. Significance analysis of microarrays applied to the ionizing radiation response. Proc. Natl. Acad. Sci. U. S. A. 98:5116–5121 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Ulanowska K, Majchrzyk A, Moskot M, Jakobkiewicz-Banecka J, Wegrzyn G. 2007. Assessment of antibacterial effects of flavonoids by estimation of generation times in liquid bacterial cultures. Biologia 62:132–135 [Google Scholar]
- 55. Ulanowska K, Tkaczyk A, Konopa G, Wegrzyn G. 2006. Differential antibacterial activity of genistein arising from global inhibition of DNA, RNA and protein synthesis in some bacterial strains. Arch. Microbiol. 184:271–278 [DOI] [PubMed] [Google Scholar]
- 56. You Z, Gao X, Ho MM, Borthakur D. 1998. A stomatin-like protein encoded by the slp gene of Rhizobium etli is required for nodulation competitiveness on the common bean. Microbiology 144:2619–2627 [DOI] [PubMed] [Google Scholar]
- 57. Zhang F, Smith DL. 1995. Preincubation of Bradyrhizobium japonicum with genistein accelerates nodule development of soybean at suboptimal root zone temperatures. Plant Physiol. 108:961–968 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.