Abstract
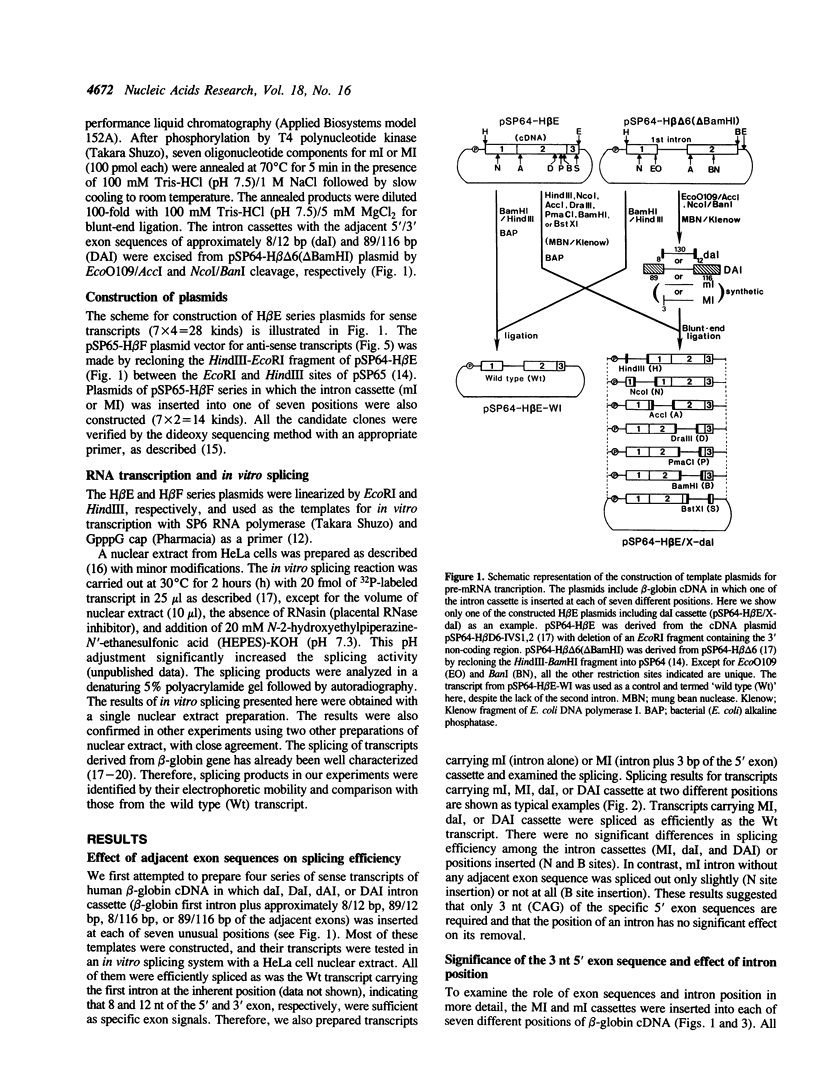
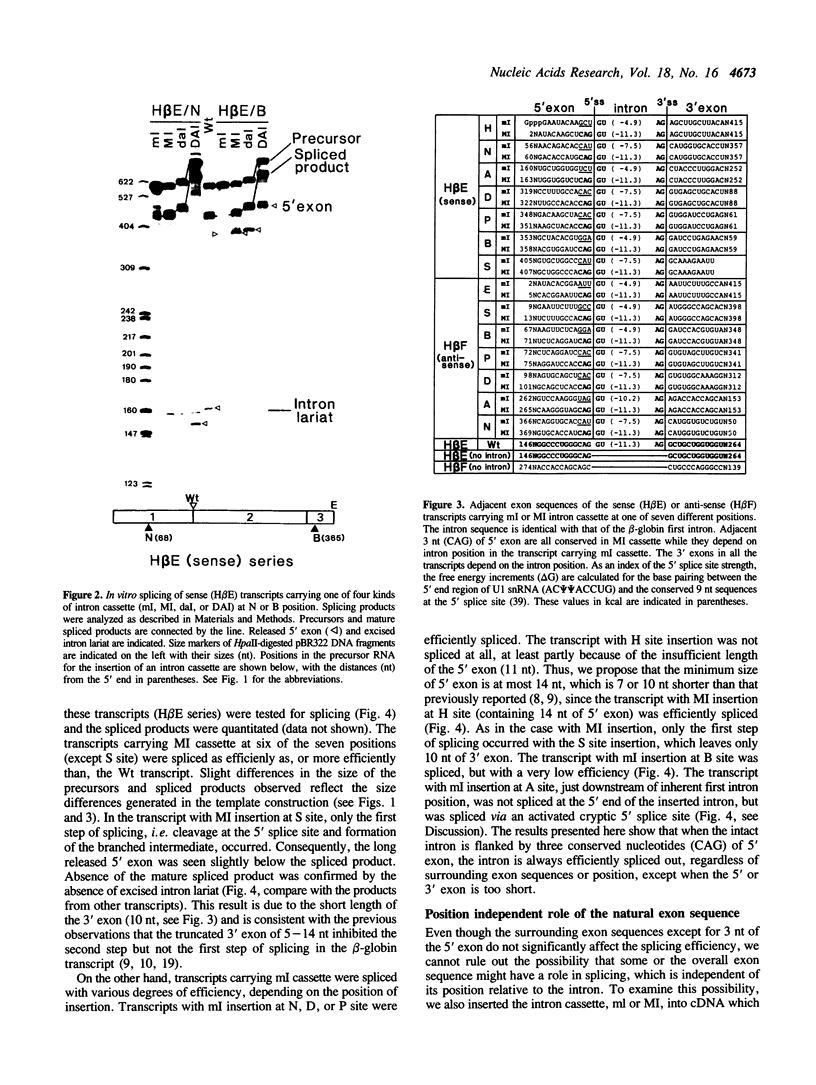
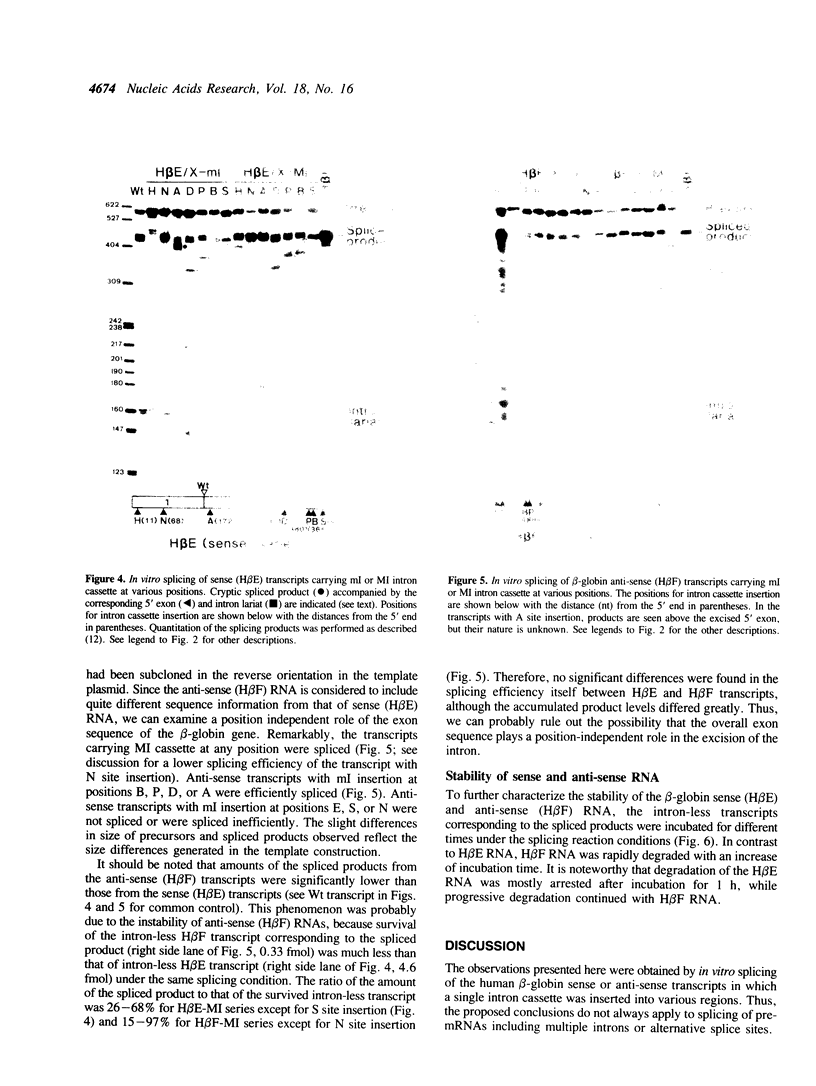
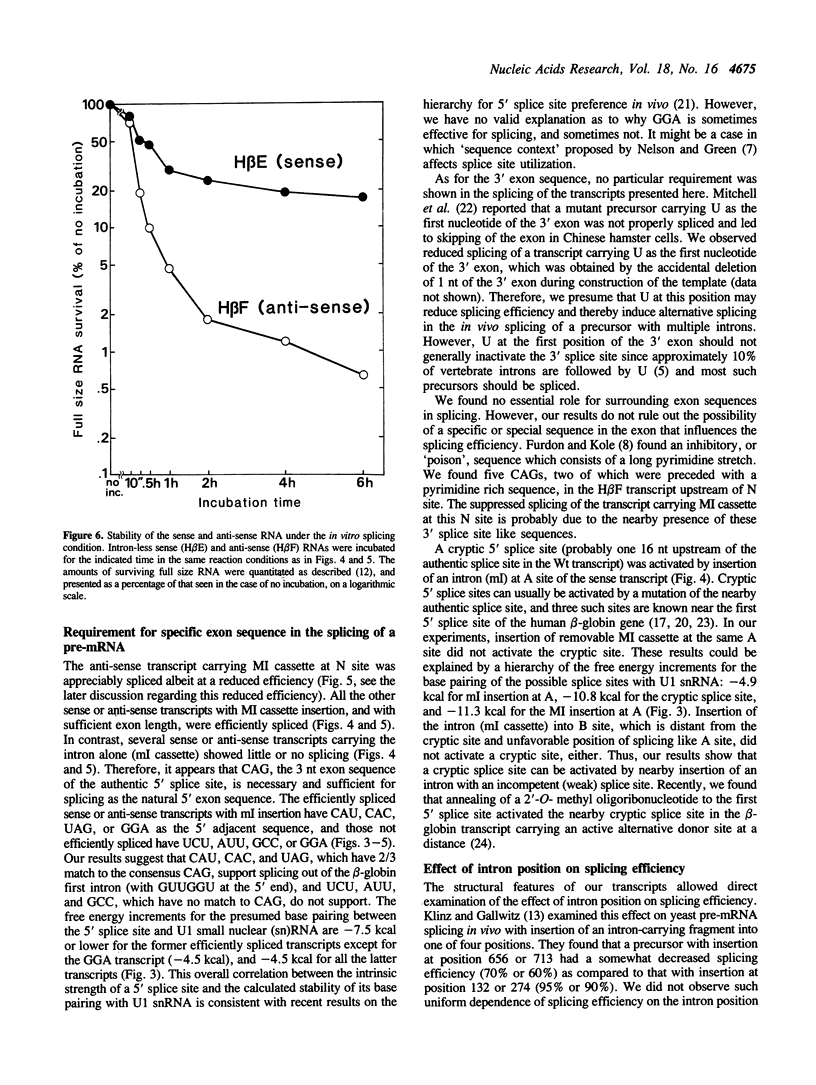
To examine the role of exon sequences and intron position in the splicing of an mRNA precursor, we prepared series of sense or anti-sense transcripts of human beta-globin cDNA in which a cassette containing the beta-globin first intron was inserted into one of seven unusual positions. The intron cassette consisted of the intron alone (ml), the intron with three adjacent base pairs of the 5' exon (MI), or the intron with both 5' and 3' exon sequences. All these transcripts were examined in an in vitro splicing system with a HeLa cell nuclear extract. The sense transcripts carrying MI cassette were spliced efficiently and independently of the intron position, except when the 3' exon was too short. The anti-sense transcripts carrying MI cassette produced significantly less spliced products than did those of the sense transcripts. This was mostly because of the instability of the anti-sense transcripts, and the actual splicing efficiency was similar to that seen in the sense transcripts. Sense or anti-sense transcripts carrying ml cassette were spliced to various extents depending on the surrounding sequences. The results indicate that only three nucleotides of the 5' exon are required as specific exon sequences in the splicing of an mRNA precursor carrying a single intron, and that the intron position does not significantly affect the splicing efficiency in vitro.
Full text
PDF

Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Augustin S., Müller M. W., Schweyen R. J. Reverse self-splicing of group II intron RNAs in vitro. Nature. 1990 Jan 25;343(6256):383–386. doi: 10.1038/343383a0. [DOI] [PubMed] [Google Scholar]
- Dignam J. D., Lebovitz R. M., Roeder R. G. Accurate transcription initiation by RNA polymerase II in a soluble extract from isolated mammalian nuclei. Nucleic Acids Res. 1983 Mar 11;11(5):1475–1489. doi: 10.1093/nar/11.5.1475. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eperon L. P., Estibeiro J. P., Eperon I. C. The role of nucleotide sequences in splice site selection in eukaryotic pre-messenger RNA. Nature. 1986 Nov 20;324(6094):280–282. doi: 10.1038/324280a0. [DOI] [PubMed] [Google Scholar]
- Freier S. M., Kierzek R., Jaeger J. A., Sugimoto N., Caruthers M. H., Neilson T., Turner D. H. Improved free-energy parameters for predictions of RNA duplex stability. Proc Natl Acad Sci U S A. 1986 Dec;83(24):9373–9377. doi: 10.1073/pnas.83.24.9373. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Furdon P. J., Kole R. Inhibition of splicing but not cleavage at the 5' splice site by truncating human beta-globin pre-mRNA. Proc Natl Acad Sci U S A. 1986 Feb;83(4):927–931. doi: 10.1073/pnas.83.4.927. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Furdon P. J., Kole R. The length of the downstream exon and the substitution of specific sequences affect pre-mRNA splicing in vitro. Mol Cell Biol. 1988 Feb;8(2):860–866. doi: 10.1128/mcb.8.2.860. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gatermann K. B., Hoffmann A., Rosenberg G. H., Käufer N. F. Introduction of functional artificial introns into the naturally intronless ura4 gene of Schizosaccharomyces pombe. Mol Cell Biol. 1989 Apr;9(4):1526–1535. doi: 10.1128/mcb.9.4.1526. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gilbert W. Genes-in-pieces revisited. Science. 1985 May 17;228(4701):823–824. doi: 10.1126/science.4001923. [DOI] [PubMed] [Google Scholar]
- Green M. R. Pre-mRNA splicing. Annu Rev Genet. 1986;20:671–708. doi: 10.1146/annurev.ge.20.120186.003323. [DOI] [PubMed] [Google Scholar]
- Greenberg M. E., Ziff E. B. Stimulation of 3T3 cells induces transcription of the c-fos proto-oncogene. Nature. 1984 Oct 4;311(5985):433–438. doi: 10.1038/311433a0. [DOI] [PubMed] [Google Scholar]
- Greenspan D. S., Weissman S. M. Synthesis of predominantly unspliced cytoplasmic RNAs by chimeric herpes simplex virus type 1 thymidine kinase-human beta-globin genes. Mol Cell Biol. 1985 Aug;5(8):1894–1900. doi: 10.1128/mcb.5.8.1894. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hattori M., Sakaki Y. Dideoxy sequencing method using denatured plasmid templates. Anal Biochem. 1986 Feb 1;152(2):232–238. doi: 10.1016/0003-2697(86)90403-3. [DOI] [PubMed] [Google Scholar]
- Klinz F. J., Gallwitz D. Size and position of intervening sequences are critical for the splicing efficiency of pre-mRNA in the yeast Saccharomyces cerevisiae. Nucleic Acids Res. 1985 Jun 11;13(11):3791–3804. doi: 10.1093/nar/13.11.3791. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krainer A. R., Maniatis T., Ruskin B., Green M. R. Normal and mutant human beta-globin pre-mRNAs are faithfully and efficiently spliced in vitro. Cell. 1984 Apr;36(4):993–1005. doi: 10.1016/0092-8674(84)90049-7. [DOI] [PubMed] [Google Scholar]
- Lear A. L., Eperon L. P., Wheatley I. M., Eperon I. C. Hierarchy for 5' splice site preference determined in vivo. J Mol Biol. 1990 Jan 5;211(1):103–115. doi: 10.1016/0022-2836(90)90014-D. [DOI] [PubMed] [Google Scholar]
- Mayeda A., Ohshima Y. Short donor site sequences inserted within the intron of beta-globin pre-mRNA serve for splicing in vitro. Mol Cell Biol. 1988 Oct;8(10):4484–4491. doi: 10.1128/mcb.8.10.4484. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Melton D. A., Krieg P. A., Rebagliati M. R., Maniatis T., Zinn K., Green M. R. Efficient in vitro synthesis of biologically active RNA and RNA hybridization probes from plasmids containing a bacteriophage SP6 promoter. Nucleic Acids Res. 1984 Sep 25;12(18):7035–7056. doi: 10.1093/nar/12.18.7035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mitchell P. J., Urlaub G., Chasin L. Spontaneous splicing mutations at the dihydrofolate reductase locus in Chinese hamster ovary cells. Mol Cell Biol. 1986 Jun;6(6):1926–1935. doi: 10.1128/mcb.6.6.1926. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mount S. M. A catalogue of splice junction sequences. Nucleic Acids Res. 1982 Jan 22;10(2):459–472. doi: 10.1093/nar/10.2.459. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mörl M., Schmelzer C. Integration of group II intron bI1 into a foreign RNA by reversal of the self-splicing reaction in vitro. Cell. 1990 Feb 23;60(4):629–636. doi: 10.1016/0092-8674(90)90666-3. [DOI] [PubMed] [Google Scholar]
- Nelson K. K., Green M. R. Splice site selection and ribonucleoprotein complex assembly during in vitro pre-mRNA splicing. Genes Dev. 1988 Mar;2(3):319–329. doi: 10.1101/gad.2.3.319. [DOI] [PubMed] [Google Scholar]
- Ohshima Y., Gotoh Y. Signals for the selection of a splice site in pre-mRNA. Computer analysis of splice junction sequences and like sequences. J Mol Biol. 1987 May 20;195(2):247–259. doi: 10.1016/0022-2836(87)90647-4. [DOI] [PubMed] [Google Scholar]
- Padgett R. A., Grabowski P. J., Konarska M. M., Seiler S., Sharp P. A. Splicing of messenger RNA precursors. Annu Rev Biochem. 1986;55:1119–1150. doi: 10.1146/annurev.bi.55.070186.005351. [DOI] [PubMed] [Google Scholar]
- Parent A., Zeitlin S., Efstratiadis A. Minimal exon sequence requirements for efficient in vitro splicing of mono-intronic nuclear pre-mRNA. J Biol Chem. 1987 Aug 15;262(23):11284–11291. [PubMed] [Google Scholar]
- Perler F., Efstratiadis A., Lomedico P., Gilbert W., Kolodner R., Dodgson J. The evolution of genes: the chicken preproinsulin gene. Cell. 1980 Jun;20(2):555–566. doi: 10.1016/0092-8674(80)90641-8. [DOI] [PubMed] [Google Scholar]
- Piechaczyk M., Yang J. Q., Blanchard J. M., Jeanteur P., Marcu K. B. Posttranscriptional mechanisms are responsible for accumulation of truncated c-myc RNAs in murine plasma cell tumors. Cell. 1985 Sep;42(2):589–597. doi: 10.1016/0092-8674(85)90116-3. [DOI] [PubMed] [Google Scholar]
- Reed R., Maniatis T. A role for exon sequences and splice-site proximity in splice-site selection. Cell. 1986 Aug 29;46(5):681–690. doi: 10.1016/0092-8674(86)90343-0. [DOI] [PubMed] [Google Scholar]
- Rogers J. H. How were introns inserted into nuclear genes? Trends Genet. 1989 Jul;5(7):213–216. doi: 10.1016/0168-9525(89)90084-x. [DOI] [PubMed] [Google Scholar]
- Ruskin B., Green M. R. Role of the 3' splice site consensus sequence in mammalian pre-mRNA splicing. Nature. 1985 Oct 24;317(6039):732–734. doi: 10.1038/317732a0. [DOI] [PubMed] [Google Scholar]
- Ruskin B., Krainer A. R., Maniatis T., Green M. R. Excision of an intact intron as a novel lariat structure during pre-mRNA splicing in vitro. Cell. 1984 Aug;38(1):317–331. doi: 10.1016/0092-8674(84)90553-1. [DOI] [PubMed] [Google Scholar]
- Sharp P. A. On the origin of RNA splicing and introns. Cell. 1985 Sep;42(2):397–400. doi: 10.1016/0092-8674(85)90092-3. [DOI] [PubMed] [Google Scholar]
- Shaw G., Kamen R. A conserved AU sequence from the 3' untranslated region of GM-CSF mRNA mediates selective mRNA degradation. Cell. 1986 Aug 29;46(5):659–667. doi: 10.1016/0092-8674(86)90341-7. [DOI] [PubMed] [Google Scholar]
- Treisman R., Orkin S. H., Maniatis T. Specific transcription and RNA splicing defects in five cloned beta-thalassaemia genes. Nature. 1983 Apr 14;302(5909):591–596. doi: 10.1038/302591a0. [DOI] [PubMed] [Google Scholar]
- Vidaud M., Gattoni R., Stevenin J., Vidaud D., Amselem S., Chibani J., Rosa J., Goossens M. A 5' splice-region G----C mutation in exon 1 of the human beta-globin gene inhibits pre-mRNA splicing: a mechanism for beta+-thalassemia. Proc Natl Acad Sci U S A. 1989 Feb;86(3):1041–1045. doi: 10.1073/pnas.86.3.1041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Volloch V., Housman D. Stability of globin mRNA in terminally differentiating murine erythroleukemia cells. Cell. 1981 Feb;23(2):509–514. doi: 10.1016/0092-8674(81)90146-x. [DOI] [PubMed] [Google Scholar]
- Yoshimatsu T., Nagawa F. Control of gene expression by artificial introns in Saccharomyces cerevisiae. Science. 1989 Jun 16;244(4910):1346–1348. doi: 10.1126/science.2544026. [DOI] [PubMed] [Google Scholar]
- Zakut R., Shani M., Givol D., Neuman S., Yaffe D., Nudel U. Nucleotide sequence of the rat skeletal muscle actin gene. Nature. 1982 Aug 26;298(5877):857–859. doi: 10.1038/298857a0. [DOI] [PubMed] [Google Scholar]





