SUMMARY
Decremental loss of PTEN results in cancer susceptibility and tumor progression. In turn this raises the possibility that PTEN elevation might be an attractive option for cancer prevention and therapy. We have generated several transgenic mouse lines with variably elevated PTEN expression levels, taking advantage of BAC (Bacterial Artificial Chromosome)-mediated transgenesis. Super-PTEN mutants are viable and show reduced body size due to decreased cell number, with no effect on cell size. Unexpectedly, PTEN elevation at the organism level results in healthy metabolism characterized by increased energy expenditure and reduced body fat accumulation. Cells derived from these mice show reduced glucose and glutamine uptake, increased mitochondrial oxidative phosphorylation, and are resistant to oncogenic transformation. Mechanistically we find that PTEN elevation orchestrates this metabolic switch by regulating PI3K-dependent and independent pathways, and negatively impacts two of the most pronounced metabolic features of tumor cells: glutaminolysis and the Warburg effect.
INTRODUCTION
PTEN, a tumor suppressor frequently mutated or deleted in human cancer, is a main negative regulator of the phosphoinositide 3-kinase (PI3K) signalling pathway by dephosphorylating the 3´ position of phosphatidylinositol-3,4,5-trisphosphate (PIP3) (Maehama and Dixon, 1998). The PI3K pathway transduces intracellular signals for growth, proliferation and cell survival (Leevers et al., 1999). Somatic inactivation of Pten occurs in a wide range of tumors, including glioblastoma, melanoma, prostate and endometrial neoplasia (Bonneau and Longy, 2000; Cantley and Neel, 1999; Simpson and Parsons, 2001). Furthermore, germline mutations of Pten are the underlying genetic cause of three related multiple hamartoma disorders: Cowden disease, characterized by an increased risk of breast and thyroid cancers; Bannayan-Zonana and Proteus syndromes (Eng, 2003). Homozygous deletion of Pten in mice results in embryonic lethality, and Pten heterozygous mutant mice develop dysplasia in a wide spectrum of tissues and have a high incidence of prostate and colon tumors (Di Cristofano et al., 1998; Podsypanina et al., 1999; Suzuki et al., 1998). It has been previously reported that PTEN dose is a key determinant in prostate cancer progression (Trotman et al., 2003). Interestingly, a more recent study shows that even a slight reduction in PTEN levels dictates cancer susceptibility (Alimonti et al., 2010). These studies highlight the crucial dose-dependent role of PTEN in cancer progression.
Studies in Drosophila melanogaster reveal a novel role for PTEN in the control of tissue growth (Gao et al., 2000; Goberdhan et al., 1999; Huang et al., 1999). The phenotypes of flies carrying mutations for various components of the PI3K-PKB/Akt pathway have shown that this pathway positively controls cell number and cell size (Bohni et al., 1999; Scanga et al., 2000; Verdu et al., 1999; Weinkove et al., 1999). Consistent with its role as an antagonist of this pathway, Drosophila PTEN (dPTEN) loss-of-function mutants display increased cell and organ size while overexpression of dPTEN yields the opposite phenotype.
While consequences of gradual PTEN loss have been extensively studied, the consequences and potential benefits of elevating PTEN in the whole organism remain unknown. Of relevance, it has been reported in Drosophila that ubiquitous overexpression of PTEN results in lethality during embryonic and larval stages (Gao et al., 2000; Huang et al., 1999). Importantly, the tumor suppressor PTEN maintains cellular homeostasis through the regulation of biological processes both in the cytoplasm and within the nucleus (Salmena et al., 2008). However, it is currently unknown whether fluctuations in the dose of PTEN (e.g. its elevation) would also impact on its nuclear functions and/or on its ability to modulate metabolic cues at the organismal level. This information is critically needed, as the elevation of PTEN is in principle a desirable objective for tumor prevention and therapy.
Tumor cells have a remarkably different metabolism from normal differentiated cells. Transformed cells uptake and metabolize nutrients such as glucose and glutamine at high levels that support anabolic growth (Tong et al., 2009). In contrast to normal differentiated cells that rely primarily on mitochondrial oxidative phosphorylation to generate the energy needed for cellular processes, most cancer cells instead rely on aerobic glycolysis, a phenomenon termed “the Warburg effect” (Warburg, 1956). The metabolic alterations and adaptations of cancer cells create a phenotype that is essential for tumor cell growth and survival, altering the flux along key metabolic pathways such as glycolysis and glutaminolysis. On the basis of these observations, there is mounting evidence for the therapeutic potential of targeting cancer metabolic reprogramming (Tennant et al., 2010).
Here we unexpectedly report that PTEN elevation is compatible with adult life and triggers a systemic metabolic regrogramming that results in healthy and tumor suppressive anti-Warburg state through the modulation of both PI3K-dependent and independent pathways.
RESULTS
Generation of Super-PTEN mice
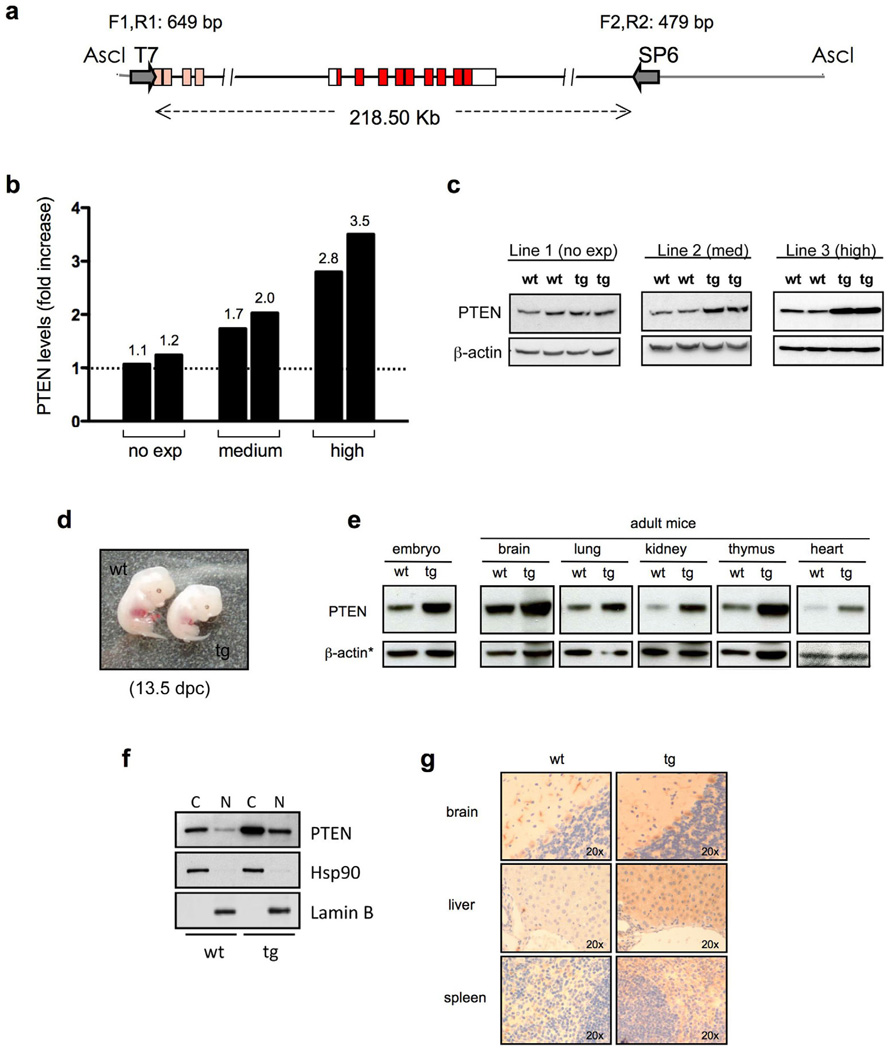
While consequences of gradual PTEN loss have been extensively studied, the potential benefit of elevating PTEN in the whole organism remains unknown. In order to elucidate the pathophysiological impact of PTEN elevation, we attempted to generate transgenic mice carrying additional copies of this critical tumor suppressor gene (referred to as Super-PTEN mice). In order to maintain the regulation properties of the endogenous Pten gene, we made use of large genomic fragments containing the entire Pten locus carried by BACs (Bacterial Artificial Chromosomes). These large genomic fragments protect the gene of interest from chromatin positional effects, preserving in every respect the pattern of expression of the endogenous gene. A genomic insert containing Pten (Fig. 1a) was isolated from a mouse BAC genomic library. We were able to obtain different transgenic lines containing variable copies of the entire Pten locus. We next generated mouse embryonic fibroblasts (MEFs) from these lines to determine the level of expression of PTEN, which we found to vary from 1.1 to 3.5-fold above the endogenous level (Fig. 1b, c), confirming successful overexpression of PTEN by the BAC transgenic system.
Figure 1. Generation of Super-PTEN mice.
a, Map of the genomic insert carried by BAC RP23-215F15 clone (RPCI library, C57BL/6J) used to generate mice with increased gene dosage of PTEN (“Super-PTEN” mice). The genomic insert is 218.50 Kb long and contains the entire Pten locus (red boxes represent PTEN coding sequence). Gray lines indicate the BAC vector (pBACe3.6) sequence. AscI was used to linearize the BAC clone. Primers used for detection of the transgene are shown (T7 side: F1,R1; SP6 side: F2,R2). b, Quantification of PTEN levels in the different BAC-PTEN-transgenic lines generated. MEFs were obtained from each line and total protein lysates were probed with antibodies towards PTEN and β-actin. The graph shows the fold increase in PTEN levels in each transgenic line relative to wild-type littermates. c, Representative immunoblotting of MEFs derived from lines with no expression (line 1), moderate expression (line 2) or high expression of the tg (line 3). d, Representative images of wt and tg embryos harvested at 13.5 days post-coitum (dpc). Note reduced body size in tg embryo compared to wt. e, Western blot showing PTEN expression during embryogenesis and in tissues from adult mice (wt and tg from line 3). *in the case of heart, coomassie is shown as loading control. f, Western blot showing PTEN levels in cytoplasmic (C) and nuclear (N) extracts from wt and tg MEFs (line 3). g, PTEN expression by immunohistochemistry in tissues from wt and tg mice (line 3).
PTEN regulates mammalian body size by controlling cell number but not cell size
Surprisingly, elevation of PTEN levels in the mouse was compatible with life but resulted in reduced body weight and size, a phenotype that was already evident in embryonic developmental stages (Fig. 1d). Increased PTEN levels were observed in all adult tissues analyzed as well as during embryogenesis (Fig. 1e) with no changes in its sub-cellular distribution and expression pattern (Fig. 1f, g). All organs examined from transgenic mice with high PTEN levels weighed below normal, while the ratio of their weight to whole body weight was indistinguishable from that observed in wild-type individuals (Supplementary Fig. 1a). Serum leptin, GH and IGF-1 levels were normal in Super-PTEN mice, therefore indicating that the effect of PTEN on body size is unlikely to be mediated indirectly through effects on hormone production (Supplementary Fig. 1b).
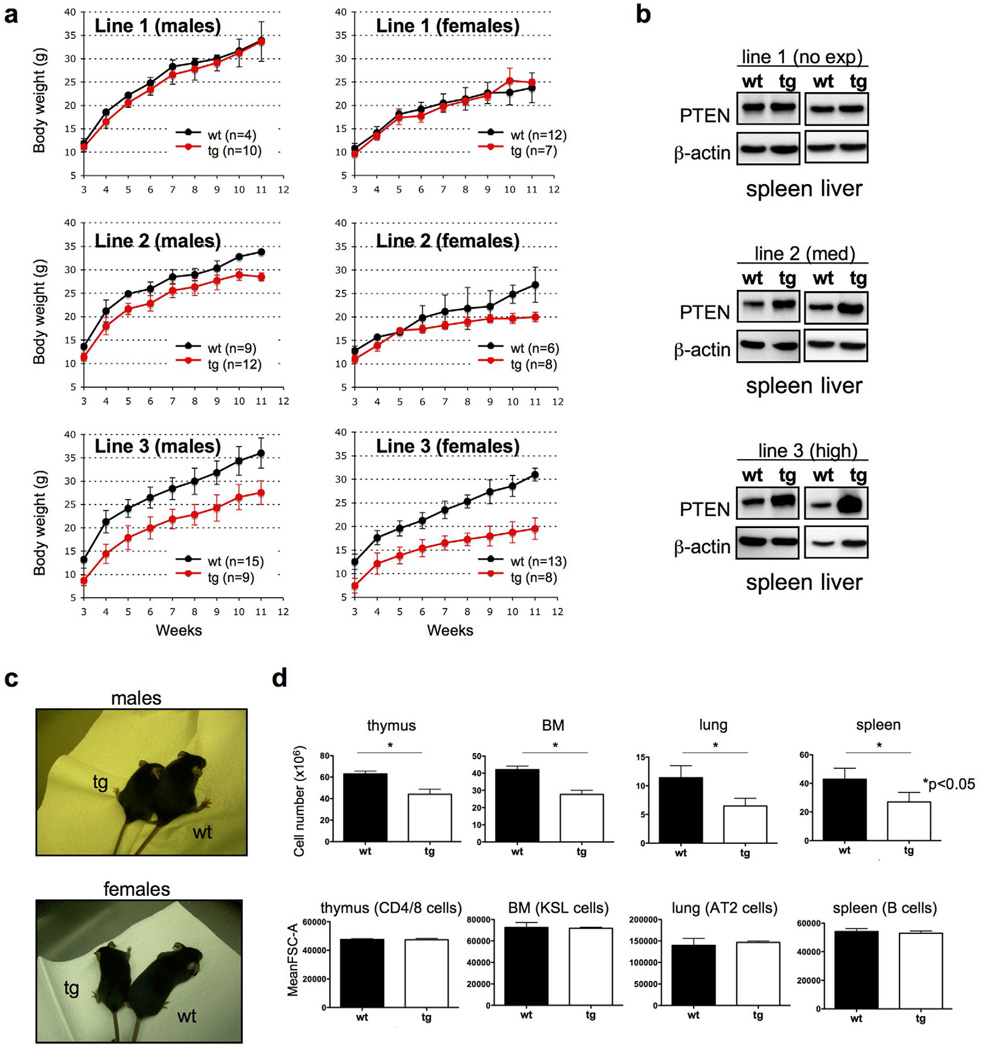
Interestingly, the effect on body weight and size (Fig. 2a, c) was more severe as PTEN levels increased, revealing that PTEN controls body size in a dose-dependent manner in a mammalian organism (Fig. 2a, b).
Figure 2. A Super-PTEN state is compatible with life but results in decreases in organ size due to reduced cell numbers.
a, Growth curves indicate that PTEN overexpression results in reduced body mass. Note that the effect is more severe as PTEN dose increases. b, Western blot showing PTEN levels in different tissues (spleen, liver) from different PTEN transgenic lines (line 1: 1.2-fold; line 2: 2-fold; line 3: 3.5-fold above the endogenous PTEN level). c, Effect of PTEN overexpression on body size (line 3). d, Cell number and cell size in organs isolated from Super-PTEN and wild-type mice (line 3, n=3 per genotype). FSC=Forward Scatter. CD4/8 cells: CD4+/Cd8+, KSL cells: Lin−/c-Kit+/Sca-1+, AT2 cells: Sca1−/CD45−/PECAM−/Autoflhi, B cells: B220mid+/IgMmid+. Error bars in a and d denote s.d. See also Figure S1.
A reduction in organ size can be the result of a decrease in cell size, cell number, or both. Unexpectedly, flow-cytometry analysis on dissociated cells from tissues (transgenic mice from line 3) revealed a normal cell size (as measured by forward scatter), whereas the total number of cells was decreased (Fig. 2d). Thus, PTEN elevation results in a reduced body size in mammals by controlling cell number, but not cell size.
PTEN elevation results in reduced cell proliferation, decreased c-Myc levels, and confers cancer resistance
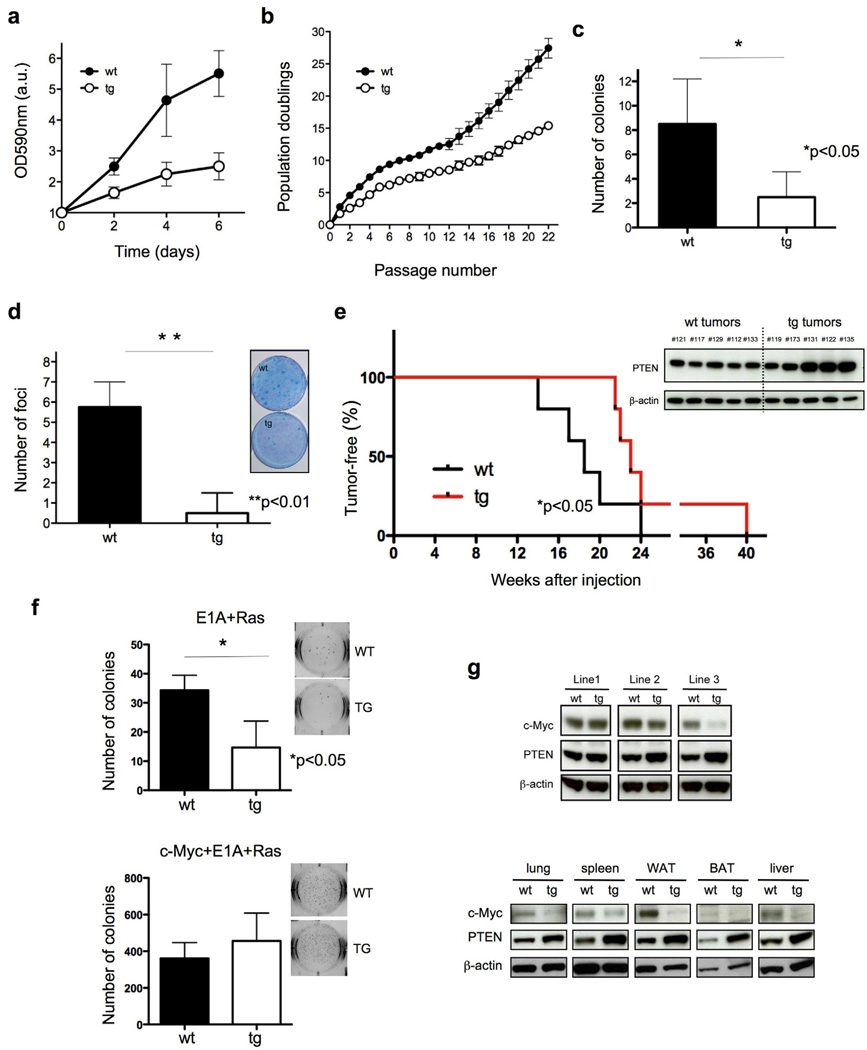
PTEN regulates a variety of biological processes to ensure correct cell homeostasis, and alterations of these functions contribute to cancer initiation and progression (Salmena et al., 2008). In line with this notion and with our previous results, Super-PTEN mouse embryonic fibroblasts (MEFs) showed a significantly slower growth rate than their wild-type counterparts (Fig. 3a and 3b: growth curve and serial 3T3 cultivation). The ability of primary MEFs to form colonies when seeded at low density is a reliable way to measure their proliferative potential. Indeed, Super-PTEN MEFs display a lower plating efficiency than those derived from wild-type embryos (Fig. 3c).
Figure 3. PTEN overexpression results in reduced growth rate in culture, resistance to oncogenic cellular transformation, decreased c-Myc levels, and confers cancer resistance in vivo.
a, Growth curves of wt and tg MEFs (n=3 per genotype). b, serial 3T3 cultivation of primary wt and tg MEFs (n=3). The figure shows the accumulated population doublings (PDL) at each passage. c, Colony-formation efficiency of wt and tg MEFs (n=4). d, Transformation susceptibility of wt and tg MEFs (n=4). The picture shows the numbers of neoplastic foci formed after transfection with a combination of oncogenic Ras and E1A. e, Susceptibility to 3MC-induced fibrosarcomas. Mice of the indicated genotypes were injected intramuscularly with 3MC in one of the rear legs and the latency for the development of fibrosarcomas was scored (line 3, n=5 per genotype). Super-PTEN mice developed tumors with a significantly longer latency than wild-type mice, as determined by Gehan-Breslow-Wilcoxon test (*p<0.05). Western blot shows PTEN expression levels in tumors derived from wild-type and Super-PTEN mice. f, Soft-agar assay in primary MEFs transformed by E1A+Ras (top) or cMyc+E1A+Ras (bottom). The graph shows the number of colonies per well (six-well plates, triplicate samples). A representative picture from the assay is showed. g, Immunoblotting of protein lysates from wt and tg MEFs and tissues. Total cell extracts were probed with antibodies towards c-Myc, PTEN and β-actin. Error bars in a, b, c, d and f denote s.d. See also Figure S2.
We next examined the impact of PTEN overexpression on oncogene-mediated cellular transformation. Employing classical focus-formation assays with E1A and Ras oncogenes in Super-PTEN and wild-type MEFs, we found the number of foci of morphologically transformed cells in Super-PTEN cells to be decreased (Fig. 3d). To validate this result in vivo, we evaluated the susceptibility of Super-PTEN mice to develop tumors upon chemical carcinogenesis (induction of fibrosarcomas by injection of 3MC: 3-methyl-cholantrene). As shown in Fig. 3e, wild-type mice started developing tumors at week 14 after 3MC injection, whereas Super-PTEN mice developed tumors with a significantly longer latency, starting at week 21. It has been observed that 3MC treatment induces the methylation and corresponding loss of expression of tumor suppressor genes in rats (Liu et al., 2010). However, PTEN expression is maintained in 3MC-induced tumors from both wild-type and Super-PTEN mice (Fig. 3e; Western Blot), suggesting that the tumor resistance phenotype of Super-PTEN mice is due to an enhanced PTEN-dependent tumor suppressive role rather than a reduced chance for Pten inactivation by 3MC. This data demonstrates that it is possible to confer resistance to oncogenic transformation both in vitro and in vivo by increasing PTEN levels.
The reduced organ weight, size, and cellularity observed in Super-PTEN mice were reminiscent of the phenotype observed in c-myc hypomorphic mice (Trumpp et al., 2001). c-Myc plays a key role in direct reprogramming of somatic cells into induced pluripotent stem (iPS) cells (Banito and Gil, 2010). iPS reprogramming is achieved by co-expressing pluripotency factors and oncogenes, including c-Myc, and can provide a readout of transformability and oncogenic potential (Banito and Gil, 2010), while it has been recently reported that tumor suppressors act as a barrier to reprogramming (Banito et al., 2009; Hong et al., 2009; Kawamura et al., 2009; Li et al., 2009; Marion et al., 2009; Utikal et al., 2009). Overexpression of PTEN in cells resulted in a reduced number of iPS colonies upon expression of reprogramming factors Oct4, Sox2 and Klf4 (Supplementary Fig. 2a), whereas, importantly, the negative effect of PTEN expression in iPS formation was overcome by c-Myc transduction. In order to determine the involvement of c-Myc in the resistance of Super-PTEN cells to oncogenic transformation, we performed soft-agar assays to evaluate anchorage-independent growth in cells infected with E1A-Ras alone or in the presence of c-Myc (cMyc+E1A-Ras). As shown in Fig. 3f (top), Super-PTEN cells form fewer colonies in soft-agar when compared to wild-type cells. Importantly, this resistance to transformation in Super-PTEN cells is completely rescued by the addition of c-Myc (Fig. 3f; bottom). On the basis of these observations we hypothesized that PTEN opposes c-Myc function or it suppresses its expression. Indeed, c-Myc expression was profoundly reduced in both Super-PTEN MEFs as well as in vivo, in various tissues (Fig. 3g).
Super-Pten mice exhibit increased energy expenditure
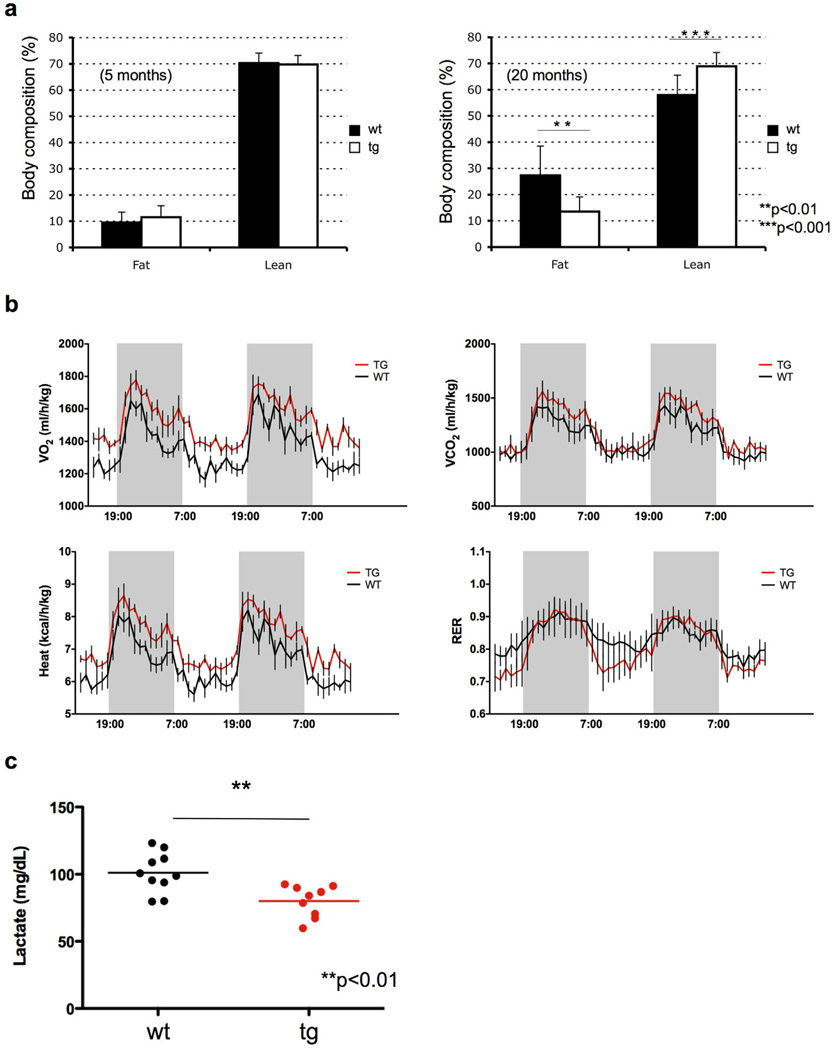
We next examined the effect of a Super-PTEN state on body fat accumulation and energy metabolism. Given the inhibitory role of PTEN on signaling pathways critical for growth factors and glucose uptake (Leevers et al., 1999), we anticipated that Super-PTEN mice would exhibit reduced metabolic activity. Surprisingly, Super-PTEN mice exhibited reduced body fat accumulation compared to wild-type counterparts, as determined by EchoMRI (Fig. 4a), suggesting that PTEN elevation impacts on nutrient adaptation and utilization. Furthermore, indirect calorimetry analysis revealed that Super-PTEN mice presented higher energy expenditure than wild-type mice (Fig. 4b and Supplementary Fig. 3a) although locomotor activity was not significantly affected (Supplementary Fig. 3b). Food intake was similar or even slightly higher in Super-PTEN mice (Supplementary Fig. 3c), indicating that the difference in energy expenditure observed between wild-type and Super-PTEN mice is not due to changes in food intake and rather due to an elevated metabolic state.
Figure 4. Super-PTEN mice show increased energy expenditure.
a, Percentage of fat and lean mass was determined in young (left panel: wt n=7; tg n=7) and old mice (right panel: wt n=12; tg n=10) by EchoMRI (line 3). b, Indirect calorimetry of Super-PTEN (red line) and wild-type mice (black line). Oxygen consumption (VO2), carbon dioxide release (VCO2), respiratory exchange ratio (RER; VCO2/VO2) and energy expenditure per kg of body weight were determined in Super-PTEN and wild-type mice in metabolic chambers (line 3, n=4 per genotype). c, Serum lactate levels in wt (n=10) and tg (n=9) mice (line 3). Error bars in a denote s.d.; error bars in b denote s.e.m. See also Figure S3.
FAO (fatty acid oxidation) represents a crucial process in energy metabolism and fat storage (Ruderman and Flier, 2001). Fatty acids can either be used for lipid synthesis and protein modification, or they can be degraded through mitochondrial β-oxidation, which produces substrates that maintain ATP generation through oxidative phosphorylation. In order to determine a potential contribution of altered FAO to the metabolic state of Super-PTEN mice and cells, we initially measured the expression levels of key enzymes in fatty acid metabolism (SCD1, CPT1-α, PPAR-α, PPAR-δ, PDK4, MCAD, Acox1). However, we could not find significant differences in the expression of these genes, with the exception of CPT1-α (Supplementary Fig. 3d), which is slightly increased in Super-PTEN cells. Interestingly, PI3K signalling reportedly suppresses CPT1α expression (Deberardinis et al., 2006). We could not find significant differences in the rate of FAO between wild-type and Super-PTEN cells, either primary MEFs (Supplementary Fig. 3e) or hepatocytes (Supplementary Fig. 3f). We next examined the use of glucose as a precursor for lipid synthesis and found that Super-PTEN cells show a significant reduction in the contribution of (6-14C)-glucose to lipid synthesis when compared to wild-type cells (Supplementary Fig. 3g). This decreased rate of lipogenesis is consistent with the reduction in body fat accumulation observed in Super-PTEN mice. We next decided to evaluate serum lactate levels as an indicator of glycolytic activity in Super-PTEN mice. As shown in Fig. 4c, we observed a reduction in lactate levels in Super-PTEN mice compared to wild-type littermates. Pyruvate generated from glucose catabolism can be reduced to lactic acid (anaerobic glycolysis) or further metabolised by the mitochondria (oxidative phosphorylation). This data suggested that the increase in energy expenditure observed in Super-PTEN mice could be reflective of increased mitochondrial oxidative phosphorylation and a concomitant reduction in anaerobic glycolysis.
PTEN elevation shifts cellular energy metabolism toward mitochondrial respiration
In order to determine the contribution of mitochondrial respiration to cellular bioenergetics we measured oxygen consumption in Super-PTEN and wild-type MEFs. The difference in energy expenditure was supported in vitro by the increase in mitochondrial oxygen consumption observed in primary Super-PTEN MEFs (Fig. 5a), which was accompanied in turn with increased mitochondrial ATP production (Fig. 5b) and the generation of reactive oxygen species (Supplementary Fig. 4a). It has previously been shown that hypoxia-inducible factor 1 (HIF-1) stimulates glycolytic energy production and negatively regulates mitochondrial biogenesis and O2 consumption (Denko, 2008). Since activation of the PI3K-Akt pathway leads to increased HIF1α levels and activity (Zundel et al., 2000), we compared the levels of HIF-1α in Super-PTEN versus wild-type MEFs. However, we found no substantial differences in HIF-1α protein levels (Supplementary Fig. 4b) or HIF-1α target genes (Supplementary Fig. 4c) between Super-PTEN and wild-type MEFs.
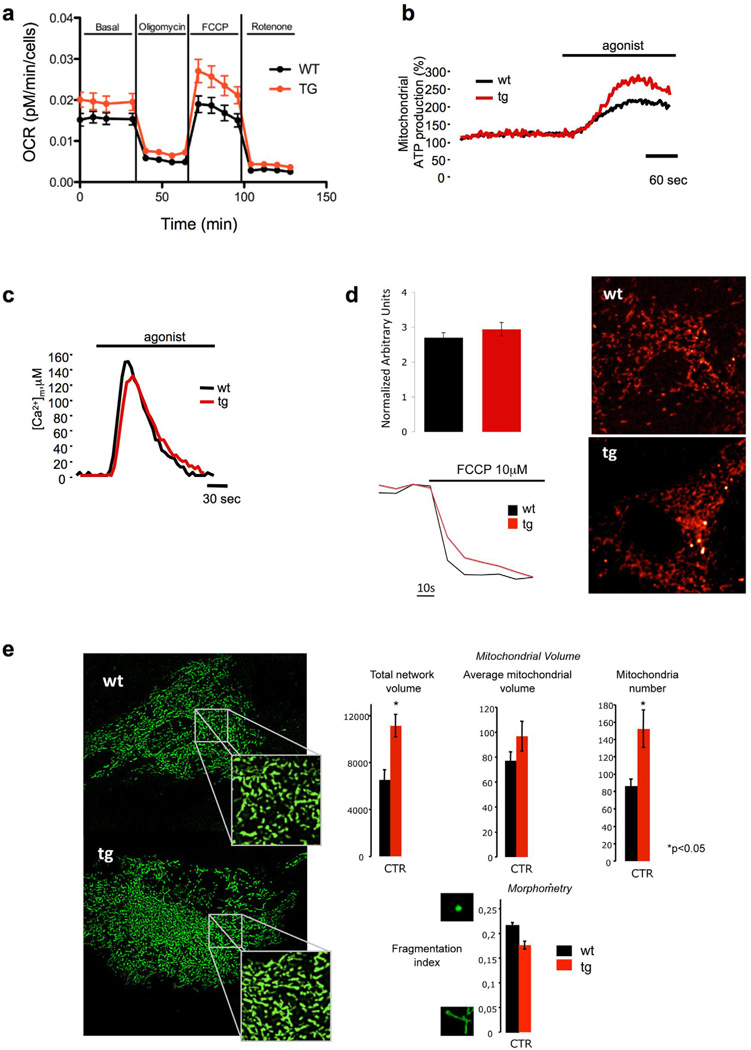
Figure 5. PTEN overexpression results in increased mitochondrial oxygen consumption, mitochondrial ATP production and mitochondrial number.
a, OCR (oxygen consumption rate) was measured (Seahorse XF24 analyzer) in primary wt and tg MEFs (n=3 per genotype) under basal conditions and after addition of oligomycin, FCCP and Rotenone. b, Mitochondrial ATP production in wt and tg MEFs after agonist stimulation as described in the methods section. Where indicated, cells were treated with 100 µM ATP. Data are expressed as a percentage of the initial value. wt: 162 ± 8%; tg: 251 ± 18%. n=15 from three independent experiments and p<0.05 (mean ± s.e.m). c, Mitochondrial Ca2+ homeostasis measurements after agonist stimulation as described in the methods section. Where indicated, cells were treated with 100 µM ATP. wt: [Ca2+]m peak 155 ± 8 µM. tg: [Ca2+]m peak 131 ± 5 µM. n=12 from three independent experiments (mean ± s.e.m). d, Analysis of mitochondrial membrane potential (ΔΨm) in wt and tg MEFs. Cells where loaded with TMRM as described in the methods section. Where indicated cells were treated with FCCP to collapse completely the ΔΨm. The traces are representative of single cell responses (wt n=28; tg n= 33). e, Analysis of total and single mitochondrial volume and mitochondrial numbers as described in the methods section (wt n=43; tg n= 40 from three independent experiments and p<0.05). Mitochondrial morphology in wt and tg MEFs as revealed by mitochondrial targeted GFP visualization. Mitochondrial fragmentation index was calculated as described in the methods section (wt n=43; tg n= 40 from three independent experiments). Ordinates for the graphs of Network volume, Average mitochondrial volume and Mitochondrial number are Voxel/Cell, Voxel/Object and N° Object/Cell respectively. Error bars in a denote s.d.; error bars in d, e denote s.e.m. See also Figure S4.
The increased mitochondrial ATP production could not be ascribed to a more efficient mitochondrial Ca2+ uptake (Griffiths and Rutter, 2009) as this parameter was if anything slightly lower in Super-PTEN MEFs (Fig. 5c). Moreover, Super-PTEN and wild-type MEFs showed similar mitochondrial membrane potential (Fig. 5d). Strikingly however, Super-PTEN MEFs exhibited greater mitochondrial biogenesis. Indeed, total network mitochondrial volume was higher in Super-PTEN MEFs due to an increase in mitochondrial number while average mitochondrial volume and morphology were unaffected (Fig. 5e).
Microarray analysis revealed that this was accompanied in Super-PTEN MEFs by a significant PGC1α gene enrichment signature (Supplementary Fig. 4d), in agreement with the fact that Akt is known to suppress PGC1α function (Li et al., 2007). PGC1α is a key regulator of energy metabolism that promotes mitochondrial oxidative phosphorylation and mitochondrial biogenesis (Puigserver and Spiegelman, 2003; Wu et al., 1999).
To functionally assess differences in mitochondrial activity, we next forced cells to rely on oxidative phosphorylation alone for energy production by substituting glucose for galactose in the growth media (Marroquin et al., 2007). In line with an increased mitochondrial function, Super-PTEN cells showed less of a reduction in growth in galactose versus glucose compared to wild-type cells (Supplementary Fig. 4e). Thus, through the generation of the Super-PTEN mouse we have identified a role for PTEN in energy homeostasis and mitochondrial biogenesis and function.
The Super-PTEN state is accompanied by repressed PI3K-Akt signaling and reduced glucose uptake in spite of high energy utilization
We then aimed to define the biochemical features of Super-PTEN mice that are responsible for this unexpected metabolic phenotype. PTEN is the main negative regulator of the PI3K pathway, a highly oncogenic and metabolic node (Engelman et al., 2006). Dephosphorylation of PIP3 by PTEN impairs Akt activation and thereby opposes the PI3K-Akt signaling pathway. Consistently, cells overexpressing PTEN show reduced levels of the substrate PIP3 (Supplementary Fig. 5a), reduced PI3K activity (as measured by phosphorylation of the downstream component Akt) (Supplementary Fig. 5b) and impaired glucose uptake (Supplementary Fig. 5c). The analysis of glucose and lactate metabolites in the extracellular media revealed that Super-PTEN cells consume less glucose and extrude less lactate into the media than wild-type cells (Fig. 6a). These data demonstrate that a Super-PTEN state is coherently accompanied by repressed PI3K-Akt signalling and reduced glucose uptake. While the prediction is that this would result in low energy utilization, it was surprisingly not the case in Super-PTEN cells and mice (see above).
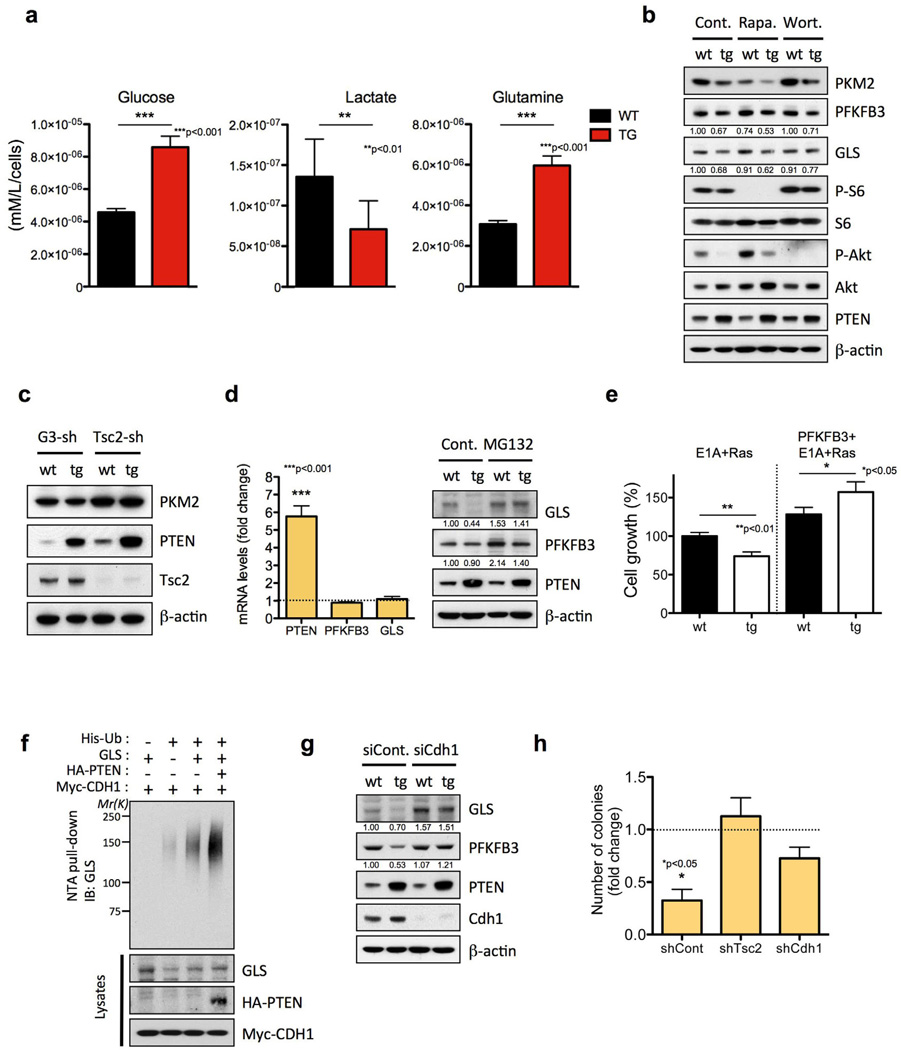
Figure 6. PTEN elevation induces a tumor suppressive metabolic state by regulating PI3K-dependent and independent pathways.
a, Glucose, lactate and glutamine levels in culture media were measured in wt and tg cells (n=2) and normalized to cell number. b, Effects of pharmacological inhibition of mTORC1 and PI3K/Akt pathway on glycolytic and glutaminolytic enzymes. Cell lysates from wt and tg MEFs treated with DMSO (Cont.), 20 nM Rapamycin (Rapa.) for 24 h, or 100 nM Wortmannin (Wort.) for 8 h were subjected to immunoblotting with antibodies against PKM2, PFKFB3, GLS, p-S6, S6, p-Akt, Akt, PTEN and β-actin. c, Effects of Tsc2 depletion-mediated mTORC1 activation on PKM2 protein level. Cell lysates from GL3-shRNA (control shRNA against the luciferase gene) and Tsc2-shRNA infected wt and tg MEFs were subjected to immunoblotting with antibodies against PKM2, Tsc2, PTEN and ²-actin. d, Fold change ratio (tg versus wt) in PTEN, FKFB3 and GLS mRNA levels determined by qRT-PCR of total RNA from wt and tg MEFs (left). Proteasome-mediated degradation of GLS and PFKFB3. Cell lysates from wt and tg MEFs treated with 10 µM of the proteasome inhibitor MG132 for 4 h were subjected to immunoblotting with antibodies against GLS, PFKFB3, PTEN and β-actin (right). e, Impact of PFKFB3 overexpression in cell growth (48 hours after plating) in primary MEFs transformed by E1A+Ras. Percentage of growth relative to wt MEFs+E1A+Ras is shown. f, PTEN enhances APC/C-Cdh1-mediated ubiquitination of GLS. PTEN-deficient PC-3 cells were co-transfected with GLS, His-ubiquitin (Ub), CDH1 and PTEN, and treated with MG132 (10 µM) for 4 hr before harvesting. His-Ub-conjugated GLS was purified from cell lysates using Ni2+-NTA spin column under denaturing conditions. g, Cdh1-silencing recovers the level of GLS and PFKFB3 in tg MEFs. Cell lysates from wt and tg MEFs transfected with siRNAs for Renilla luciferase (siCont.) or Cdh1 were subjected to immunoblotting with antibodies against GLS, PFKFB3, PTEN, Cdh1 and ²-actin. h, Number of colonies (fold change ratio: tg versus wt MEFs+Ras) formed in soft-agar assay after Tsc2 or Cdh1 knockdown in immortalized MEFs transformed by oncogenic Ras. The graph shows the number of colonies per well (six-well plates, triplicate samples). Error bars in a, d, e and h denote s.d. See also Figure S5.
PTEN regulates PKM2 levels through mTORC1
One key characteristic of cancer cells is the way glucose is utilized, switching from energy efficient oxidative phosphorylation to inefficient lactate production (Warburg effect) (Warburg, 1956), allowing for the accumulation of glycolysis intermediates and their redirection to biosynthetic pathways. Our data shows that Super-PTEN cells uptake less glucose, and yet they redirect a greater fraction of glycolytic products into mitochondrial oxidative phosphorylation. These features could be regarded as an anti-Warburg status, which would correlate with decreased proliferation and a cancer-resistant state. One of the central components in the metabolic switch is the enzyme responsible for converting phosphoenolpyruvate into pyruvate, pyruvate kinase (PK). The switch from PKM1 expression to PKM2 in cells results in a lower rate of pyruvate kinase activity, allowing the accumulation of glycolytic precursors for the production of redox power and biomass (Vander Heiden et al., 2009; Vander Heiden et al., 2010). We therefore studied the status of PKM2 in Super-PTEN cells. Interestingly, Super-PTEN cells had reduced levels of PKM2 (Fig. 6b) and decreased PK activity (Supplementary Fig. 5d), suggesting that altered activity of this Warburg effect regulatory node may contribute to the observed metabolic changes in Super-PTEN cells.
As the PKM1/M2 isoforms are generated through alternative splicing of two mutually exclusive exons (Christofk et al., 2008; Clower et al., 2010; David et al., 2010), we next compared splicing patterns of the Pkm gene in wild-type and Super-PTEN MEFs. However, Pkm2 was the predominant mRNA isoform in MEFs regardless of genotype (Supplementary Fig. 5e). This result was unanticipated given that PKM2 splicing is under control of c-Myc (David et al., 2010) and that we find Super-PTEN cells to express lower levels of c-Myc (Fig. 3g). Interestingly, in the iPS reprogramming experiments, addition of c-Myc rescues PKM2 expression in Super-PTEN cells (Supplementary Fig. 2b). However, splicing pattern analysis (Supplementary Fig. 2c) suggests that the c-Myc dependent rescue of PKM2 levels in Super-PTEN cells under iPS reprogramming conditions is mediated through a mechanism distinct to a switch in splicing patterns (see below).
In an effort to account for changes to PKM2 expression in Super-PTEN cells, we therefore investigated the potential involvement of other PTEN-regulated cellular pathways in the control of PKM2 expression. Rapamycin treatment was found to reduce PKM2 levels in MEFs, suggesting that decreased activity of mTORC1, a downstream target of the PI3K/Akt pathway (Zoncu et al., 2011), could potentially account for the reduced levels of PKM2 in Super-PTEN cells (Fig. 6b). To test this hypothesis we analyzed PKM2 expression in wild-type and Super-PTEN MEFs upon knockdown of Tsc2, a negative regulator of mTORC1 (Tee and Blenis, 2005). Importantly, hyperactivation of the mTORC1 pathway by Tsc2 knockdown rescued PKM2 levels in Super-PTEN MEFs and further increased PKM2 levels in wild-type cells (Fig. 6c), confirming that reduced mTORC1 activity in Super-PTEN cells accounts for the decreased expression of PKM2. Interestingly, Tsc2 knockdown also increased c-Myc expression in Super-PTEN cells although levels still remained substantially lower than those of wild-type cells (Supplementary Fig. 5f). These data are consistent with the described role of mTORC1 in regulating c-myc translation (West et al., 1998). However, the inability of Tsc2 knockdown to completely rescue c-Myc expression in Super-PTEN cells indicates that additional pathways downstream PTEN known to regulate c-Myc levels are co-ordinately in action (Gregory et al., 2003; Sears et al., 2000). Conversely, the ability of c-Myc to rescue PKM2 expression in Super-PTEN cells during iPS reprogramming may be related to enhanced mTORC1 signaling at multiple levels (Jones et al., 1996; Ravitz et al., 2007).
PTEN controls both glycolysis and glutaminolysis through regulation of PFKFB3 and GLS stability
The regulation of metabolism and growth must be tightly coupled to guarantee the efficient use of energy and anabolic substrates throughout the cell cycle. PFKFB3 (6-phosphofructo-2-kinase/fructose-2,6-biphosphatase isoform 3), potently stimulates glycolysis by catalyzing the formation of fructose 2,6-bisphosphate, an allosteric activator of 6-phosphofructo-1-kinase (PFK-1), a rate-limiting enzyme and essential control point in glycolysis. PFKFB3 has been shown to be abundantly expressed in human tumors (Atsumi et al., 2002) and required for the high glycolytic rate and anchorage-independent growth of Ras-transformed cells (Telang et al., 2006). As shown in Fig. 6b, PFKFB3 levels are downregulated in Super-PTEN cells.
Cancer cells depend on a high rate of glucose uptake and metabolism to maintain their viability despite being maintained in an oxygen-replete environment (Warburg effect). However, another remarkable metabolic feature of tumor cells is glutamine addiction. Interestingly, Super-PTEN cells show an impaired glutamine uptake (Fig. 6a) and reduced levels of glutaminase (GLS), the first enzyme in the glutaminolysis pathway (Fig. 6b). Therefore, PTEN elevation negatively impacts both glycolysis and glutaminolysis through the regulation of PFKFB3 and GLS.
Both GLS and PFKFB3 are substrates targeted for degradation by the E3 ubiquitin ligase anaphase-promoting complex/cyclosome-Cdh1 (APC/C-Cdh1) (Colombo et al., 2010; Najafov and Alessi, 2010), while PTEN has recently been demonstrated to promote APC/C-Cdh1 activity (Song et al., 2011). This function of PTEN is dependent on its ability to directly promote APC/C-Cdh1 complex assembly and independent of its ability to inhibit the PI3K signalling pathway through dephosphorylation of PIP3. In agreement with this, both PFKFB3 and GLS levels were unaffected by treatment with the PI3K-inhibitor Wortmannin (Fig. 6b). We found that the reduction in PFKFB3 and GLS levels was rescued by MG132 treatment, and no significant changes in mRNA levels were found in cells overexpressing PTEN compared to wild-type cells, indicating a proteosome-dependent downregulation of PFKFB3 and GLS (Fig. 6d). Super-PTEN cells are resistant to transformation and show a reduced growth rate in response to the oncogenic combination E1A+Ras. Importantly, the addition of PFKFB3 is able to completely rescue the deficient growth of transformed Super-PTEN cells (Fig. 6e). In vivo ubiquitination assay confirmed GLS as a substrate for APC/C-Cdh1, and increased GLS ubiquitination in the presence of PTEN (Fig. 6f). PFKFB3 and GLS levels were rescued by depletion of Cdh1 (Fig. 6g), indicating that PTEN negatively regulates PFKFB3 and GLS through the APC/Cdh1 complex.
Reduced PKM2, PFKFB3 and GLS protein levels characterize the tumor suppressive metabolic state of Super-PTEN cells. As discussed above, PKM2 levels are rescued by Tsc2 knockdown (Fig. 6c), while PFKFB3 and GLS levels are normalized by Cdh1 inactivation (Fig. 6g). As previously described (Li et al., 2008; Zhang et al., 2003), Tsc2 or Cdh1 knockdown resulted in a marked reduction in growth rate of primary MEFs (Supplementary Fig. 5g), although this effect was more pronounced with wild-type than Super-PTEN cells in the case of Tsc2 inactivation. To assess whether Tsc2 or Cdh1 inactivation could rescue the resistance to oncogenic transformation of Super-PTEN cells, we studied anchorage-independent growth of immortalized wild-type and Super-PTEN MEFs after introduction of oncogenic Ras. Strikingly, Tsc2 inactivation renders Super-PTEN cells as susceptible to oncogenic transformation as wild-type cells, whereas Cdh1 inactivation leads to a partial rescue in colony-forming ability in soft agar (Fig 6h).
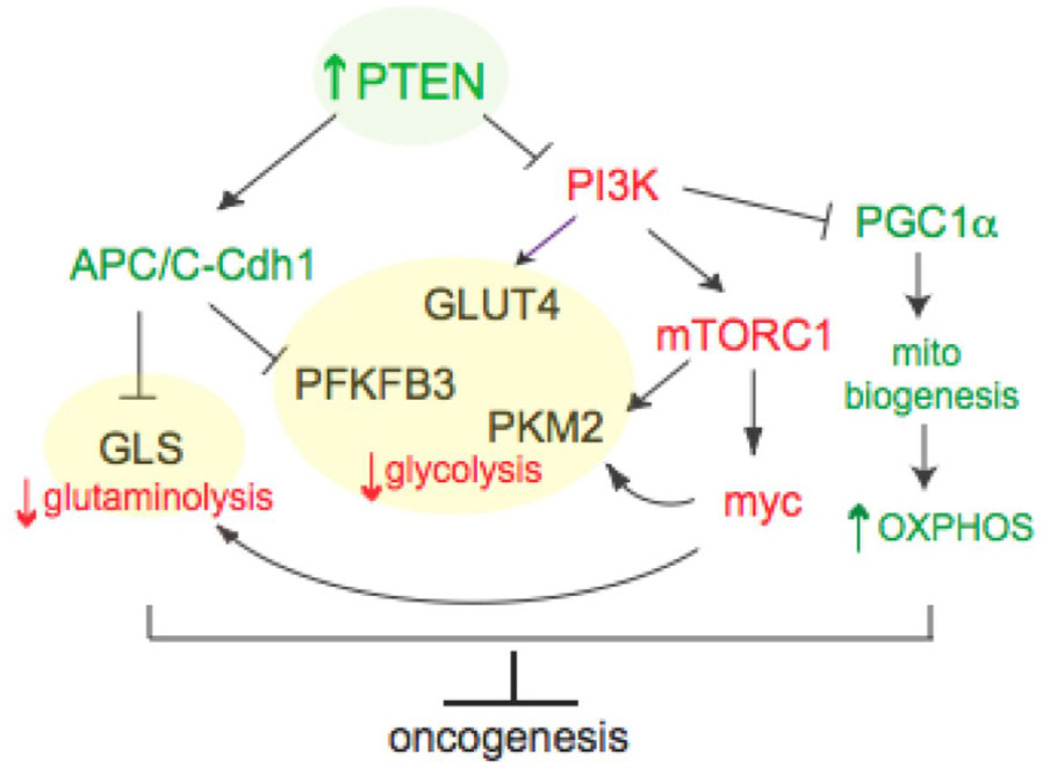
On the basis of these findings, we propose a model (Fig. 7) by which PTEN elevation induces a tumor suppressive metabolic state by regulating PI3K-dependent and independent pathways, and negatively impacts two of the most noticeable metabolic features of tumor cells: glutaminolysis and Warburg effect.
Figure 7. PTEN induces a tumor suppressive metabolic state by regulating PI3K-dependent and independent pathways.
Model by which PTEN elevation negatively impacts two of the most noticeable metabolic features of tumor cells: glutaminolysis and Warburg effect.
DISCUSSION
Our study reveals the profound impact of PTEN elevation in organismal homeostasis and cancer development. Studies in Drosophila melanogaster have highlighted roles of the Akt pathway in positively controlling cell number and cell size (Bohni et al., 1999; Gao et al., 2000; Goberdhan et al., 1999; Huang et al., 1999; Scanga et al., 2000; Verdu et al., 1999; Weinkove et al., 1999). However, our data shows that the reduction in organism and tissue size in Super-PTEN is due solely to a reduction in cell number but not cell size. Interestingly, cell growth phenotypes of some Drosophila mutations are not always paralleled by their mammalian counterparts. For instance, reduction of c-Myc expression in mice results in an overall decrease in body size due to a reduction in cell number (hypoplasia) without detectable changes in cell size (Trumpp et al., 2001), whereas Drosophila dmyc mutants are smaller as a result of reduced cell size (hypotrophy) (Johnston et al., 1999). Our Super-PTEN model displays an obvious reduction of c-Myc expression as well, yet reduced cell number but not cell size is the outcome at the organismal level. Nevertheless, the c-Myc down-regulation is a distinctive component of the Super-PTEN phenotype. Accordingly, c-Myc over-expression rescues the defective three-factor iPS reprogramming of Super-PTEN MEFs. Further, c-Myc controls genes regulating glucose metabolism and glutaminolysis, including PKM2 and GLS, which are decreased in Super-PTEN cells (David et al., 2010; Gao et al., 2009).
Tumor cells exhibit an altered metabolism that allows them to sustain higher proliferative rates and resist cell death signals (DeBerardinis et al., 2008; King and Gottlieb, 2009; Tennant et al., 2009). Microarray analysis revealed that genes of the glycolysis pathway are overexpressed in the majority of clinically relevant cancers (Altenberg and Greulich, 2004). Among these genes is pyruvate kinase, which regulates the rate-limiting final step of glycolysis. Recent work demonstrated that expression of the type II isoform of the pyruvate-kinase-M gene (PK-M2) is a critical determinant of the metabolic phenotype of cancer cells, and confers a selective proliferative advantage to tumor cells in vivo (Christofk et al., 2008). Our data shows that Super-PTEN cells have reduced levels of PKM2, and that this key regulator of glycolytic flux PKM2 is under the control of the mTORC1 pathway. Another key regulator of glycolysis is PFKFB3, which potently stimulates glycolysis by catalyzing the formation of fructose 2,6-bisphosphate, the allosteric activator of PFK1 (Hue and Rider, 1987), a rate-limiting enzyme of glycolysis. It has been previously reported that PFKFB3 is constitutively expressed by neoplastic cells and serves as an essential downstream metabolic mediator of oncogenic Ras (Telang et al., 2006). Importantly, we demonstrate that PTEN elevation triggers PFKFB3 degradation through the APC/Cdh1 complex. Super-PTEN cells show reduced protein levels of PFKFB3, with no changes at the mRNA level. Beyond the PTEN-dependent inhibition of glycolysis, we find that Super-PTEN cells have reduced levels of GLS, the first enzyme in glutaminolysis. Once again, this key tumor suppressive metabolic switch that opposes glutaminolysis is triggered by PTEN elevation through GLS degradation by the APC/Cdh1 complex. Interestingly, GLS has been reported to have increased activity in several tumor types, and is upregulated in c-Myc-transformed cells (Gao et al., 2009; Wise et al., 2008), highlighting it as a potential chemoterapeutic target.
In summary, Super-PTEN mice exhibit an unexpected cancer-resistant and very unique metabolic state, which is the outcome of the ability of PTEN to regulate metabolism at multiple levels both from the cytosol and from the nucleus. On the one hand, these mice exhibit increased oxygen consumption and energy expenditure. On the other hand, Super-PTEN mice and cells are less prone to transformation and cancer development. Thus, the features observed in Super-PTEN mice and cells resemble an “anti-Warburg state”, in which less glucose is up-taken but it is more efficiently directed to the mitochondrial tricarboxylic acid cycle. Moreover, PTEN elevations induce mitochondrial biogenesis and thus increased mitochondrial ATP production. Thus PTEN coordinates mitochondrial function and dynamics to the regulation of cell metabolism through PI3K-dependent and PI3K-independent mechanisms. It is therefore tempting to speculate that the unique metabolic state resulting from PTEN elevation contributes to the increased cancer resistance observed in these mice and cells, by opposing cancer associated metabolic reprogramming (Fig. 7). Our data unexpectedly identify PTEN as a key node for the control of obesity and tumorigenesis. Hence its elevation represents an attractive therapeutic approach, as this would act to prevent cancer development and also increase energy expenditure thereby opposing fat accumulation and obesity.
EXPERIMENTAL PROCEDURES
BAC transgenesis
For transgenesis, a large genomic insert (218.50 Kb) containing the entire Pten locus and cloned into the BAC vector pBACe3.6 was isolated from a mouse BAC genomic library (BAC RP23-215F15 clone; RPCI library, C57BL/6J). After digestion with AscI, linearized BAC DNA was used for microinjection into the pronuclei of fertilized oocytes, derived from intercrosses between (C57BL/6 X CBA)F1 mice. Subsequently, embryos were transferred into the oviducts of recipient pseudopregnant CD1 females. The resulting offspring were analyzed for the presence of the transgene using PCRs that detected the junction between the SP6 and T7 vector flanking elements and the genomic insert.
Cell culture
Isolation, culture, and analysis of mouse embryo fibroblasts (MEFs) was performed as previously described (Garcia-Cao et al., 2002). Generation of mouse iPS cells was performed as described (Li et al., 2009; Marion et al., 2009). A detailed description of the various assays involving MEFs and generation of iPS cells is provided in Extended Experimental Procedures.
Immunohistochemistry and immunoblot analysis
For immunohistochemistry, mouse tissues were fixed in 4% paraformaldehyde overnight, subsequently washed twice with PBS, and transferred into 70% ethanol. Tissues were embedded in paraffin, sectioned, and stained with anti-PTEN antibody (M3627: DAKO). Immunoblot analysis was carried out according to standard protocols. Antibodies used are listed in Extended Experimental Procedures.
shRNAs
Wild-type and Super-Pten MEFs were infected with lentiviruses expressing shRNA against mouse Cdh1 (clone ID TRCN0000027712, Open Biosystems), rat Tsc2 or corresponding control shRNA against luciferase (Di Nardo et al., 2009). To prepare lentiviral particles, 3 × 106 293T cells were plated per 10-cm culture dish and then shRNAs transfected with Lipofectamine 2000 (Invitrogen). For infection, MEFs were plated at a density of 6 × 105 cells per 10-cm culture dish and infected by virus from 293T cells 48 h after transfection. After reseeding, MEFs were used for the different assays.
In vivo ubiquitination Assay
Pten-deficient PC-3 cells were transfected with a combination of pCMV6-XL4-GLS (SC114750: OriGene), HA-PTEN, Myc-CDH1, and His-ubiquitin and His-Ub-conjugated GLS were purified from cell lysates using Ni2+-NTA spin column (Qiagen) under denaturing conditions. The extent of ubiquitination was then analyzed by immunoblotting.
qRT-PCR
RNA was isolated with the RNeasy Protect kit (Qiagen) and included a DNase digestion step using the RNase-free DNase kit (Qiagen). cDNA was obtained with Transcriptor (Roche). TaqMan probes were obtained from Applied Biosystems Inc. Amplifications were run in a 7900 Real-Time PCR System (Applied Biosystems Inc.). Each value was adjusted using β-glucuronidase levels as a reference.
Chemical carcinogenesis
For 3MC-induced carcinogenesis, four-month-old mice received a single intramuscular injection into one of their rear legs of a 100 µl solution containing 3-methyl-cholanthrene (Sigma), dissolved at a final concentration of 10 µg µl−1 in sesame oil (Sigma) as previously described (Garcia-Cao et al., 2002; Wexler and Rosenberg, 1979).
Metabolic studies
Metabolic performance was studied by using an automated combined indirect calorimetric system (TSE systems). Fat and lean masses were determined by EchoMRI (Echo Medical Systems). For detailed methodology please see Extended Experimental Procedures.
Measurement of oxygen consumption
For oxygen consumption, 2 × 104 cells were plated and, 24 hr later, oxygen consumption rate (OCR) was measured with the Seahorse XF24 instrument (Seahorse Bioscience) under basal conditions and after addition of oligomycin (1 µM), FCCP (2.5 µM) and Rotenone (1 µM). All the chemicals were purchased from Sigma.
Measurement of glucose, lactate and glutamine
Glucose, lactate and glutamine levels in culture media were measured using the BioProfile FLEX analyzer (NOVA biomedical) and normalized to cell number. Fresh media was added to a 6-well plate of cells and analyzed after 24h (triplicate samples).
Measurement of ROS production
Reactive oxygen species production was measured by flow cytometric assessment of DCF (2',7'-dichlorodihydrofluorescein) fluorescence. The details of this assay are described in Extended Experimental Procedures.
Microarray analysis
Gene expression was assessed in wt and tg MEFs (n=3 per genotype) with GeneChip HT MG-430 PM 24-Array Plate (Affymetrix, #901257). Data was processed using the Affymetrix Gene Chip Operating System.
Mitochondrial assays
The details of luciferase measurements, aequorin measurements, membrane potential measurements and microscopic analysis of mitochondrial morphology and number are described in Extended Experimental Procedures.
Phosphoinositide analysis
Phosphatidylinositides were separated by anion-exchange high-performance liquid chromatography (Beckman), detected by a flow scintillation analyzer (Perkin-Elmer), and quantified with ProFSA software (Perkin-Elmer) as described (Serunian et al., 1991). Please see Extended Experimental Procedures for details. The [32P]-PIP3 peak counts were normalized against the total phosphoinositide counts.
Radioactive RT-PCR assay
Splicing patterns of the Pkm gene in wt and tg cells were analyzed as previously described (Clower et al., 2010). For primer sequences and detailed methodology please see Extended Experimental Procedures.
Measurement of glucose metabolism
Cellular glucose metabolism rates were measured by following the conversion of 5-3H-glucose to 3H2O as previously described (Vander Heiden et al., 2001). The details of this assay are described in Extended Experimental Procedures.
Measurement of pyruvate kinase activity
Pyruvate kinase activity was measured by a continuous assay coupled to lactate dehydrogenase (LDH). The details of this assay are included in Extended Experimental Procedures.
Hepatocyte Isolation
Primary hepatocytes were isolated using a two-step perfusion protocol, based on a previous method (Lin et al., 2004). Please refer to Extended Experimental Procedures for details.
Fatty acid oxidation
Fatty acid oxidation was determined by measuring 3H2O produced during cellular oxidation of [3H] palmitate (Finck et al., 2006; Gerhart-Hines et al., 2007). Please refer to Extended Experimental Procedures for details.
Lipid synthesis assay
Lipid synthesis was assayed by measuring the amount of 14C incorporated into lipids after 2 hours incubation with [6-14C]glucose (Hatzivassiliou et al., 2005). For a detailed description of this assay please see Extended Experimental Procedures.
Statistical analysis
In vitro and in vivo data were analyzed with an unpaired t test (GraphPad Prism, GraphPad Software Inc.). Values of P < 0.05 were considered statistically significant (*P < 0.05; ** P <0.01; *** P < 0.001). Correlation analysis was performed with SigmaPlot 5.0 software (SPSS Inc.).
Supplementary Material
ACKNOWLEDGEMENTS
We thank M. Bhasin for help with microarray analysis, C. Clower for assistance with PKM splicing assays, W. J. Haveman for mouse genotyping and Kaitlyn Webster for IHC analysis. I.G.C. was supported by fellowships from the Human Frontier Science Program (HFSP) and the Spanish Ministry of Education and Science (MEC). This work was supported by NIH grants R01 CA-82328-09 to P.P.P., P01-CA089021 and R01-GM41890 to L.C.C., and Italian Association for Cancer Research (AIRC) to P.P..
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
The authors declare no competing financial interests.
REFERENCES
- Alimonti A, Carracedo A, Clohessy JG, Trotman LC, Nardella C, Egia A, Salmena L, Sampieri K, Haveman WJ, Brogi E, et al. Subtle variations in Pten dose determine cancer susceptibility. Nat Genet. 2010;42:454–458. doi: 10.1038/ng.556. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Altenberg B, Greulich KO. Genes of glycolysis are ubiquitously overexpressed in 24 cancer classes. Genomics. 2004;84:1014–1020. doi: 10.1016/j.ygeno.2004.08.010. [DOI] [PubMed] [Google Scholar]
- Atsumi T, Chesney J, Metz C, Leng L, Donnelly S, Makita Z, Mitchell R, Bucala R. High expression of inducible 6-phosphofructo-2-kinase/fructose-2,6-bisphosphatase (iPFK-2; PFKFB3) in human cancers. Cancer Res. 2002;62:5881–5887. [PubMed] [Google Scholar]
- Banito A, Rashid ST, Acosta JC, Li S, Pereira CF, Geti I, Pinho S, Silva JC, Azuara V, Walsh M, et al. Senescence impairs successful reprogramming to pluripotent stem cells. Genes Dev. 2009;23:2134–2139. doi: 10.1101/gad.1811609. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Banito A, Gil J. Induced pluripotent stem cells and senescence: learning the biology to improve the technology. EMBO Rep. 2010;11:353–359. doi: 10.1038/embor.2010.47. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bohni R, Riesgo-Escovar J, Oldham S, Brogiolo W, Stocker H, Andruss BF, Beckingham K, Hafen E. Autonomous control of cell and organ size by CHICO, a Drosophila homolog of vertebrate IRS1-4. Cell. 1999;97:865–875. doi: 10.1016/s0092-8674(00)80799-0. [DOI] [PubMed] [Google Scholar]
- Bonneau D, Longy M. Mutations of the human PTEN gene. Hum Mutat. 2000;16:109–122. doi: 10.1002/1098-1004(200008)16:2<109::AID-HUMU3>3.0.CO;2-0. [DOI] [PubMed] [Google Scholar]
- Cantley LC, Neel BG. New insights into tumor suppression: PTEN suppresses tumor formation by restraining the phosphoinositide 3-kinase/AKT pathway. Proc Natl Acad Sci U S A. 1999;96:4240–4245. doi: 10.1073/pnas.96.8.4240. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Christofk HR, Vander Heiden MG, Harris MH, Ramanathan A, Gerszten RE, Wei R, Fleming MD, Schreiber SL, Cantley LC. The M2 splice isoform of pyruvate kinase is important for cancer metabolism and tumour growth. Nature. 2008;452:230–233. doi: 10.1038/nature06734. [DOI] [PubMed] [Google Scholar]
- Clower CV, Chatterjee D, Wang Z, Cantley LC, Vander Heiden MG, Krainer AR. The alternative splicing repressors hnRNP A1/A2 and PTB influence pyruvate kinase isoform expression and cell metabolism. Proc Natl Acad Sci U S A. 2010;107:1894–1899. doi: 10.1073/pnas.0914845107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Colombo SL, Palacios-Callender M, Frakich N, De Leon J, Schmitt CA, Boorn L, Davis N, Moncada S. Anaphase-promoting complex/cyclosome-Cdh1 coordinates glycolysis and glutaminolysis with transition to S phase in human T lymphocytes. Proc. 2010;107:18868–18873. doi: 10.1073/pnas.1012362107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- David CJ, Chen M, Assanah M, Canoll P, Manley JL. HnRNP proteins controlled by c-Myc deregulate pyruvate kinase mRNA splicing in cancer. Nature. 2010;463:364–368. doi: 10.1038/nature08697. [DOI] [PMC free article] [PubMed] [Google Scholar]
- DeBerardinis RJ, Lum JJ, Thompson CB. Phosphatidylinositol 3-kinase-dependent modulation of carnitine palmitoyltransferase 1A expression regulates lipid metabolism during hematopoietic cell growth. J Biol Chem. 2006;281:37372–37380. doi: 10.1074/jbc.M608372200. [DOI] [PubMed] [Google Scholar]
- DeBerardinis RJ, Lum JJ, Hatzivassiliou G, Thompson CB. The biology of cancer: metabolic reprogramming fuels cell growth and proliferation. Cell Metab. 2008;7:11–20. doi: 10.1016/j.cmet.2007.10.002. [DOI] [PubMed] [Google Scholar]
- Denko NC. Hypoxia, HIF1 and glucose metabolism in the solid tumour. Nat Rev Cancer. 2008;8:705–713. doi: 10.1038/nrc2468. [DOI] [PubMed] [Google Scholar]
- Di Cristofano A, Pesce B, Cordon-Cardo C, Pandolfi PP. Pten is essential for embryonic development and tumour suppression. Nat Genet. 1998;19:348–355. doi: 10.1038/1235. [DOI] [PubMed] [Google Scholar]
- Di Nardo A, Kramvis I, Cho N, Sadowski A, Meikle L, Kwiatkowski DJ, Sahin M. Tuberous sclerosis complex activity is required to control neuronal stress responses in an mTOR-dependent manner. J Neurosci. 2009;29:5926–5937. doi: 10.1523/JNEUROSCI.0778-09.2009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eng C. PTEN: one gene, many syndromes. Hum Mutat. 2003;22:183–198. doi: 10.1002/humu.10257. [DOI] [PubMed] [Google Scholar]
- Engelman JA, Luo J, Cantley LC. The evolution of phosphatidylinositol 3-kinases as regulators of growth and metabolism. Nat Rev Genet. 2006;7:606–619. doi: 10.1038/nrg1879. [DOI] [PubMed] [Google Scholar]
- Finck BN, Gropler MC, Chen Z, Leone TC, Croce MA, Harris TE, Lawrence JC, Jr, Kelly DP. Lipin 1 is an inducible amplifier of the hepatic PGC-1alpha/PPARalpha regulatory pathway. Cell Metab. 2006;4:199–210. doi: 10.1016/j.cmet.2006.08.005. [DOI] [PubMed] [Google Scholar]
- Gao P, Tchernyshyov I, Chang TC, Lee YS, Kita K, Ochi T, Zeller KI, De Marzo AM, Van Eyk JE, Mendell JT, et al. c-Myc suppression of miR-23a/b enhances mitochondrial glutaminase expression and glutamine metabolism. Nature. 2009;458:762–765. doi: 10.1038/nature07823. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gao X, Neufeld TP, Pan D. Drosophila PTEN regulates cell growth and proliferation through PI3K-dependent and -independent pathways. Dev Biol. 2000;221:404–418. doi: 10.1006/dbio.2000.9680. [DOI] [PubMed] [Google Scholar]
- Garcia-Cao I, Garcia-Cao M, Martin-Caballero J, Criado LM, Klatt P, Flores JM, Weill JC, Blasco MA, Serrano M. "Super p53" mice exhibit enhanced DNA damage response, are tumor resistant and age normally. EMBO J. 2002;21:6225–6235. doi: 10.1093/emboj/cdf595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gerhart-Hines Z, Rodgers JT, Bare O, Lerin C, Kim SH, Mostoslavsky R, Alt FW, Wu Z, Puigserver P. Metabolic control of muscle mitochondrial function and fatty acid oxidation through SIRT1/PGC-1alpha. EMBO J. 2007;26:1913–1923. doi: 10.1038/sj.emboj.7601633. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goberdhan DC, Paricio N, Goodman EC, Mlodzik M, Wilson C. Drosophila tumor suppressor PTEN controls cell size and number by antagonizing the Chico/PI3-kinase signaling pathway. Genes Dev. 1999;13:3244–3258. doi: 10.1101/gad.13.24.3244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Graham FL, van der Eb AJ. A new technique for the assay of infectivity of human adenovirus 5 DNA. Virology. 1973;52:456–467. doi: 10.1016/0042-6822(73)90341-3. [DOI] [PubMed] [Google Scholar]
- Gregory MA, Qi Y, Hann SR. Phosphorylation by glycogen synthase kinase-3 controls c-myc proteolysis and subnuclear localization. J Biol Chem. 2003;278:51606–51612. doi: 10.1074/jbc.M310722200. [DOI] [PubMed] [Google Scholar]
- Griffiths EJ, Rutter GA. Mitochondrial calcium as a key regulator of mitochondrial ATP production in mammalian cells. Biochim Biophys Acta. 2009;1787:1324–1333. doi: 10.1016/j.bbabio.2009.01.019. Epub 2009 Feb 1323. [DOI] [PubMed] [Google Scholar]
- Hatzivassiliou G, Zhao F, Bauer DE, Andreadis C, Shaw AN, Dhanak D, Hingorani SR, Tuveson DA, Thompson CB. ATP citrate lyase inhibition can suppress tumor cell growth. Cancer Cell. 2005;8:311–321. doi: 10.1016/j.ccr.2005.09.008. [DOI] [PubMed] [Google Scholar]
- Hong H, Takahashi K, Ichisaka T, Aoi T, Kanagawa O, Nakagawa M, Okita K, Yamanaka S. Suppression of induced pluripotent stem cell generation by the p53-p21 pathway. Nature. 2009;460:1132–1135. doi: 10.1038/nature08235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang H, Potter CJ, Tao W, Li DM, Brogiolo W, Hafen E, Sun H, Xu T. PTEN affects cell size, cell proliferation and apoptosis during Drosophila eye development. Development. 1999;126:5365–5372. doi: 10.1242/dev.126.23.5365. [DOI] [PubMed] [Google Scholar]
- Hue L, Rider MH. Role of fructose 2,6-bisphosphate in the control of glycolysis in mammalian tissues. Biochem J. 1987;245:313–324. doi: 10.1042/bj2450313. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johnston LA, Prober DA, Edgar BA, Eisenman RN, Gallant P. Drosophila myc regulates cellular growth during development. Cell. 1999;98:779–790. doi: 10.1016/s0092-8674(00)81512-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jones RM, Branda J, Johnston KA, Polymenis M, Gadd M, Rustgi A, Callanan L, Schmidt EV. An essential E box in the promoter of the gene encoding the mRNA cap-binding protein (eukaryotic initiation factor 4E) is a target for activation by c-myc. Mol Cell Biol. 1996;16:4754–4764. doi: 10.1128/mcb.16.9.4754. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kawamura T, Suzuki J, Wang YV, Menendez S, Morera LB, Raya A, Wahl GM, Belmonte JC. Linking the p53 tumour suppressor pathway to somatic cell reprogramming. Nature. 2009;460:1140–1144. doi: 10.1038/nature08311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- King A, Gottlieb E. Glucose metabolism and programmed cell death: an evolutionary and mechanistic perspective. Curr Opin Cell Biol. 2009;21:885–893. doi: 10.1016/j.ceb.2009.09.009. [DOI] [PubMed] [Google Scholar]
- Leevers SJ, Vanhaesebroeck B, Waterfield MD. Signalling through phosphoinositide 3-kinases: the lipids take centre stage. Curr Opin Cell Biol. 1999;11:219–225. doi: 10.1016/s0955-0674(99)80029-5. [DOI] [PubMed] [Google Scholar]
- Li H, Collado M, Villasante A, Strati K, Ortega S, Canamero M, Blasco MA, Serrano M. The Ink4/Arf locus is a barrier for iPS cell reprogramming. Nature. 2009;460:1136–1139. doi: 10.1038/nature08290. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li M, Shin YH, Hou L, Huang X, Wei Z, Klann E, Zhang P. The adaptor protein of the anaphase promoting complex Cdh1 is essential in maintaining replicative lifespan and in learning and memory. Nat Cell Biol. 2008;10:1083–1089. doi: 10.1038/ncb1768. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li X, Monks B, Ge Q. Akt/PKB regulates hepatic metabolism by directly inhibiting PGC-1alpha transcription coactivator. Nature. 2007;447:1012–1016. doi: 10.1038/nature05861. [DOI] [PubMed] [Google Scholar]
- Lin J, Wu PH, Tarr PT, Lindenberg KS, St-Pierre J, Zhang CY, Mootha VK, Jager S, Vianna CR, Reznick RM, et al. Defects in adaptive energy metabolism with CNS-linked hyperactivity in PGC-1alpha null mice. Cell. 2004;119:121–135. doi: 10.1016/j.cell.2004.09.013. [DOI] [PubMed] [Google Scholar]
- Liu WB, Ao L, Zhou ZY, Cui ZH, Zhou YH, Yuan XY, Xiang YL, Cao J, Liu JY. CpG island hypermethylation of multiple tumor suppressor genes associated with loss of their protein expression during rat lung carcinogenesis induced by 3-methylcholanthrene and diethylnitrosamine. Biochem Biophys Res Commun. 2010;402:507–514. doi: 10.1016/j.bbrc.2010.10.061. [DOI] [PubMed] [Google Scholar]
- Maehama T, Dixon JE. The tumor suppressor, PTEN/MMAC1, dephosphorylates the lipid second messenger, phosphatidylinositol 3,4,5-trisphosphate. J Biol Chem. 1998;273:13375–13378. doi: 10.1074/jbc.273.22.13375. [DOI] [PubMed] [Google Scholar]
- Marion RM, Strati K, Li H, Murga M, Blanco R, Ortega S, Fernandez-Capetillo O, Serrano M, Blasco MA. A p53-mediated DNA damage response limits reprogramming to ensure iPS cell genomic integrity. Nature. 2009;460:1149–1153. doi: 10.1038/nature08287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marroquin LD, Hynes J, Dykens JA, Jamieson JD, Will Y. Circumventing the Crabtree effect: replacing media glucose with galactose increases susceptibility of HepG2 cells to mitochondrial toxicants. Toxicol Sci. 2007;97:539–547. doi: 10.1093/toxsci/kfm052. [DOI] [PubMed] [Google Scholar]
- Najafov A, Alessi DR. Uncoupling the Warburg effect from cancer. Proc. 2010;107:19135–19136. doi: 10.1073/pnas.1014047107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Podsypanina K, Ellenson LH, Nemes A, Gu J, Tamura M, Yamada KM, Cordon-Cardo C, Catoretti G, Fisher PE, Parsons R. Mutation of Pten/Mmac1 in mice causes neoplasia in multiple organ systems. Proc Natl Acad Sci U S A. 1999;96:1563–1568. doi: 10.1073/pnas.96.4.1563. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ravitz MJ, Chen L, Lynch M, Schmidt EV. c-myc Repression of TSC2 contributes to control of translation initiation and Myc-induced transformation. Cancer Res. 2007;67:11209–11217. doi: 10.1158/0008-5472.CAN-06-4351. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Salmena L, Carracedo A, Pandolfi PP. Tenets of PTEN tumor suppression. Cell. 2008;133:403–414. doi: 10.1016/j.cell.2008.04.013. [DOI] [PubMed] [Google Scholar]
- Scanga SE, Ruel L, Binari RC, Snow B, Stambolic V, Bouchard D, Peters M, Calvieri B, Mak TW, Woodgett JR, et al. The conserved PI3'K/PTEN/Akt signaling pathway regulates both cell size and survival in Drosophila. Oncogene. 2000;19:3971–3977. doi: 10.1038/sj.onc.1203739. [DOI] [PubMed] [Google Scholar]
- Sears R, Nuckolls F, Haura E, Taya Y, Tamai K, Nevins JR. Multiple Ras-dependent phosphorylation pathways regulate Myc protein stability. Genes Dev. 2000;14:2501–2514. doi: 10.1101/gad.836800. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Serunian LA, Auger KR, Cantley LC. Identification and quantification of polyphosphoinositides produced in response to platelet-derived growth factor stimulation. Methods Enzymol. 1991;198:78–87. doi: 10.1016/0076-6879(91)98010-4. [DOI] [PubMed] [Google Scholar]
- Simpson L, Parsons R. PTEN: life as a tumor suppressor. Exp Cell Res. 2001;264:29–41. doi: 10.1006/excr.2000.5130. [DOI] [PubMed] [Google Scholar]
- Song MS, Carracedo A, Salmena L, Song SJ, Egia A, Malumbres M, Pandolfi PP. Nuclear PTEN regulates the APC-CDH1 tumor-suppressive complex in a phosphatase-independent manner. Cell. 2011;144:187–199. doi: 10.1016/j.cell.2010.12.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Suzuki A, de la Pompa JL, Stambolic V, Elia AJ, Sasaki T, del Barco Barrantes I, Ho A, Wakeham A, Itie A, Khoo W, et al. High cancer susceptibility and embryonic lethality associated with mutation of the PTEN tumor suppressor gene in mice. Curr Biol. 1998;8:1169–1178. doi: 10.1016/s0960-9822(07)00488-5. [DOI] [PubMed] [Google Scholar]
- Tee AR, Blenis J. mTOR, translational control and human disease. Semin Cell Dev Biol. 2005;16:29–37. doi: 10.1016/j.semcdb.2004.11.005. [DOI] [PubMed] [Google Scholar]
- Telang S, Yalcin A, Clem AL, Bucala R, Lane AN, Eaton JW, Chesney J. Ras transformation requires metabolic control by 6-phosphofructo-2-kinase. Oncogene. 2006;25:7225–7234. doi: 10.1038/sj.onc.1209709. [DOI] [PubMed] [Google Scholar]
- Tennant DA, Duran RV, Boulahbel H, Gottlieb E. Metabolic transformation in cancer. Carcinogenesis. 2009;30:1269–1280. doi: 10.1093/carcin/bgp070. [DOI] [PubMed] [Google Scholar]
- Tennant DA, Duran RV, Gottlieb E. Targeting metabolic transformation for cancer therapy. Nat Rev Cancer. 2010;10:267–277. doi: 10.1038/nrc2817. [DOI] [PubMed] [Google Scholar]
- Todaro GJ, Green H. Quantitative studies of the growth of mouse embryo cells in culture and their development into established lines. J Cell Biol. 1963;17:299–313. doi: 10.1083/jcb.17.2.299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tong X, Zhao F, Thompson CB. The molecular determinants of de novo nucleotide biosynthesis in cancer cells. Curr Opin Genet Dev. 2009;19:32–37. doi: 10.1016/j.gde.2009.01.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Trotman LC, Niki M, Dotan ZA, Koutcher JA, Di Cristofano A, Xiao A, Khoo AS, Roy-Burman P, Greenberg NM, Van Dyke T, et al. Pten dose dictates cancer progression in the prostate. PLoS Biol. 2003;1:E59. doi: 10.1371/journal.pbio.0000059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Trumpp A, Refaeli Y, Oskarsson T, Gasser S, Murphy M, Martin GR, Bishop JM. c-Myc regulates mammalian body size by controlling cell number but not cell size. Nature. 2001;414:768–773. doi: 10.1038/414768a. [DOI] [PubMed] [Google Scholar]
- Utikal J, Polo JM, Stadtfeld M, Maherali N, Kulalert W, Walsh RM, Khalil A, Rheinwald JG, Hochedlinger K. Immortalization eliminates a roadblock during cellular reprogramming into iPS cells. Nature. 2009;460:1145–1148. doi: 10.1038/nature08285. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vander Heiden MG, Plas DR, Rathmell JC, Fox CJ, Harris MH, Thompson CB. Growth factors can influence cell growth and survival through effects on glucose metabolism. Mol Cell Biol. 2001;21:5899–5912. doi: 10.1128/MCB.21.17.5899-5912.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vander Heiden MG, Cantley LC, Thompson CB. Understanding the Warburg effect: the metabolic requirements of cell proliferation. Science. 2009;324:1029–1033. doi: 10.1126/science.1160809. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vander Heiden MG, Locasale JW, Swanson KD, Sharfi H, Heffron GJ, Amador-Noguez D, Christofk HR, Wagner G, Rabinowitz JD, Asara JM, et al. Evidence for an alternative glycolytic pathway in rapidly proliferating cells. Science. 2010;329:1492–1499. doi: 10.1126/science.1188015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Verdu J, Buratovich MA, Wilder EL, Birnbaum MJ. Cell-autonomous regulation of cell and organ growth in Drosophila by Akt/PKB. Nat Cell Biol. 1999;1:500–506. doi: 10.1038/70293. [DOI] [PubMed] [Google Scholar]
- Warburg O. On the origin of cancer cells. Science. 1956;123:309–314. doi: 10.1126/science.123.3191.309. [DOI] [PubMed] [Google Scholar]
- Weinkove D, Neufeld TP, Twardzik T, Waterfield MD, Leevers SJ. Regulation of imaginal disc cell size, cell number and organ size by Drosophila class I(A) phosphoinositide 3-kinase and its adaptor. Curr Biol. 1999;9:1019–1029. doi: 10.1016/s0960-9822(99)80450-3. [DOI] [PubMed] [Google Scholar]
- West MJ, Stoneley M, Willis AE. Translational induction of the c-myc oncogene via activation of the FRAP/TOR signalling pathway. Oncogene. 1998;17:769–780. doi: 10.1038/sj.onc.1201990. [DOI] [PubMed] [Google Scholar]
- Wexler H, Rosenberg SA. Pulmonary metastases from autochthonous 3-methylcholanthrene-induced murine tumors. J Natl Cancer Inst. 1979;63:1393–1395. [PubMed] [Google Scholar]
- Wise DR, DeBerardinis RJ, Mancuso A, Sayed N, Zhang XY, Pfeiffer HK, Nissim I, Daikhin E, Yudkoff M, McMahon SB, et al. Myc regulates a transcriptional program that stimulates mitochondrial glutaminolysis and leads to glutamine addiction. Proc Natl Acad Sci U S A. 2008;105:18782–18787. doi: 10.1073/pnas.0810199105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zoncu R, Efeyan A, Sabatini DM. mTOR: from growth signal integration to cancer, diabetes and ageing. Nat. 2011;12:21–35. doi: 10.1038/nrm3025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zundel W, Schindler C, Haas-Kogan D, Koong A, Kaper F, Chen E, Gottschalk AR, Ryan HE, Johnson RS, Jefferson AB, et al. Loss of PTEN facilitates HIF-1-mediated gene expression. Genes Dev. 2000;14:391–396. [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.