Abstract
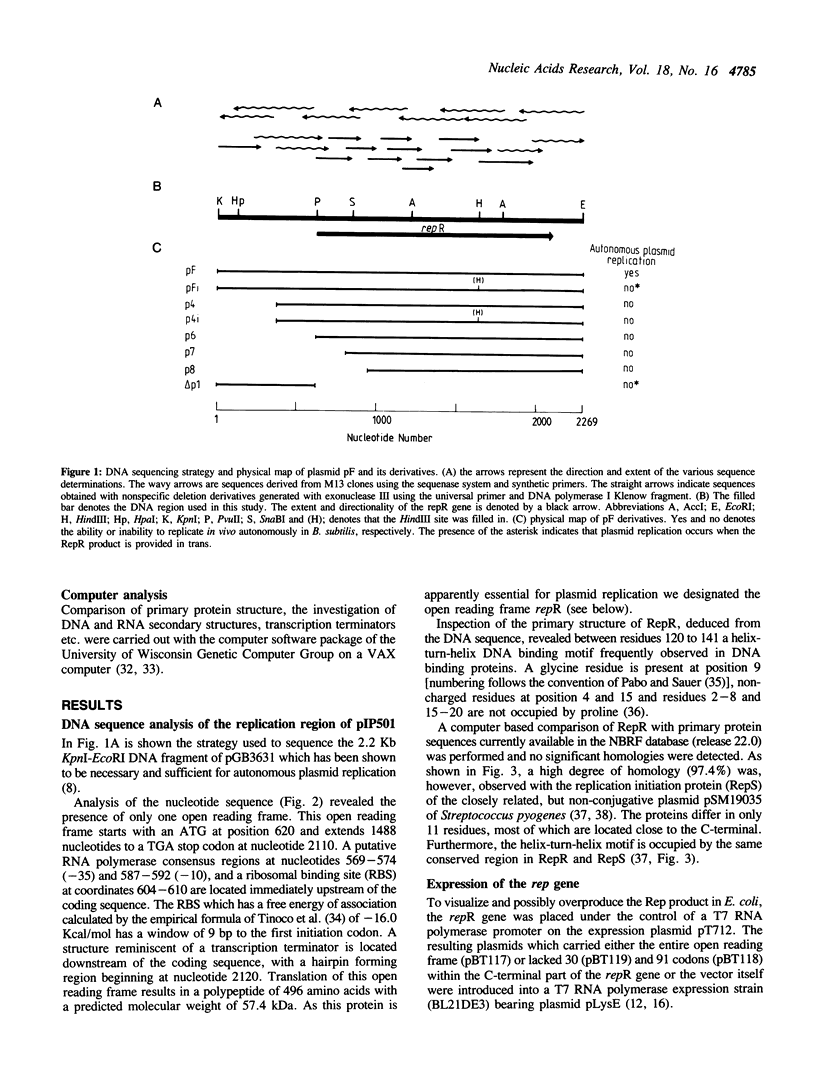
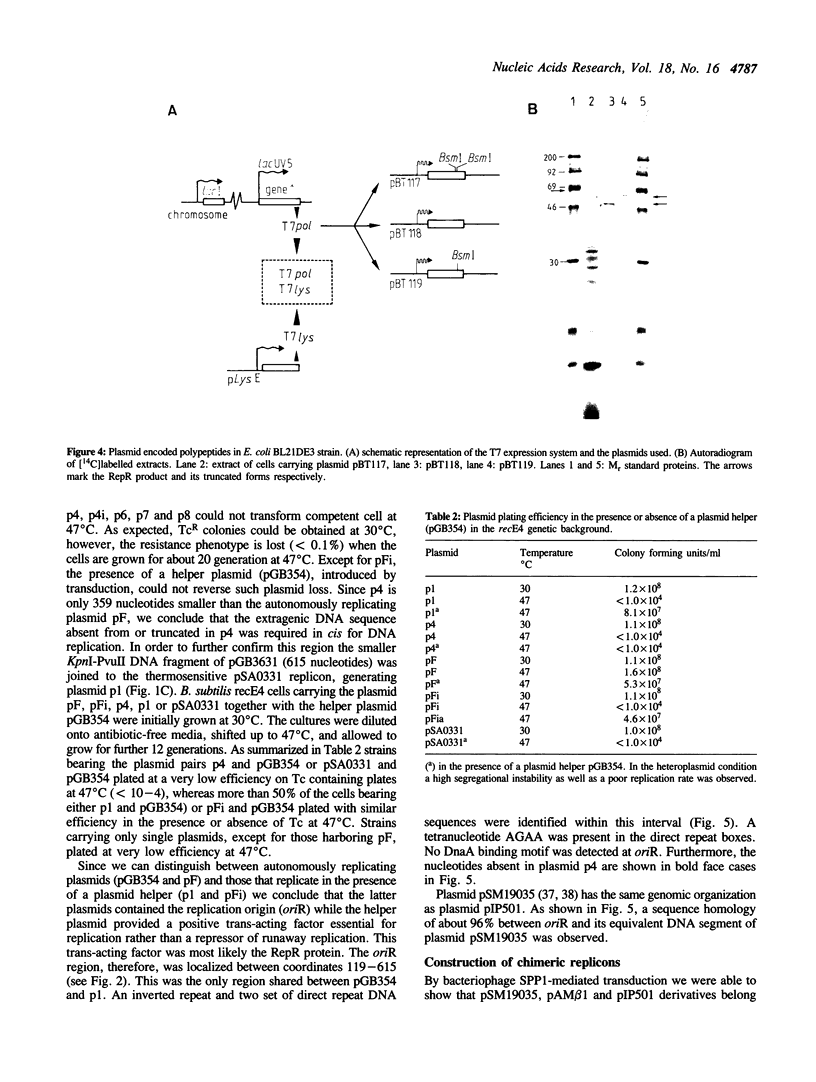
The large conjugative plasmid pIP501 was originally isolated from Streptococcus agalactiae. To study the molecular basis of pIP501 replication we determined the nucleotide sequence of a 2.2 kb DNA segment which is essential and sufficient for autonomous replication of pIP501 derived plasmids, in Bacillus subtilis cells. This region can be divided into two functionally discrete segments: a 496 bp region (oriR) that acts as an origin of replication, and a 1488 bp segment coding for an essential replication protein (RepR). The RepR protein, which has a molecular mass of 57.4 kDa, could complement in trans a thermosensitive replicon bearing the pIP501 origin. Chimeric Rep proteins and replicons were obtained by domain swapping between rep genes of closely related streptococcal plasmids belonging to the inc18 group (pIP501, pAM beta 1 and pSM19035). The chimeras were functional in B. subtilis.
Full text
PDF





Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Alonso J. C. DNA replication of plasmids from gram-positive bacteria in Bacillus subtilis. Plasmid pUB110 as a model system. Microbiologia. 1989 Jun;5(1):5–12. [PubMed] [Google Scholar]
- Alonso J. C., Lüder G., Trautner T. A. Requirements for the formation of plasmid-transducing particles of Bacillus subtilis bacteriophage SPP1. EMBO J. 1986 Dec 20;5(13):3723–3728. doi: 10.1002/j.1460-2075.1986.tb04706.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alonso J. C., Tailor R. H. Initiation of plasmid pC194 replication and its control in Bacillus subtilis. Mol Gen Genet. 1987 Dec;210(3):476–484. doi: 10.1007/BF00327200. [DOI] [PubMed] [Google Scholar]
- Alonso J. C., Trautner T. A. A gene controlling segregation of the Bacillus subtilis plasmid pC194. Mol Gen Genet. 1985;198(3):427–431. doi: 10.1007/BF00332934. [DOI] [PubMed] [Google Scholar]
- Alonso J. C., Viret J. F., Tailor R. H. Plasmid maintenance in Bacillus subtilis recombination-deficient mutants. Mol Gen Genet. 1987 Jun;208(1-2):349–352. doi: 10.1007/BF00330464. [DOI] [PubMed] [Google Scholar]
- Behnke D., Gerlach D. Cloning and expression in Escherichia coli, Bacillus subtilis, and Streptococcus sanguis of a gene for staphylokinase--a bacterial plasminogen activator. Mol Gen Genet. 1987 Dec;210(3):528–534. doi: 10.1007/BF00327208. [DOI] [PubMed] [Google Scholar]
- Behnke D., Gilmore M. S., Ferretti J. J. Plasmid pGB301, a new multiple resistance streptococcal cloning vehicle and its use in cloning of a gentamicin/kanamycin resistance determinant. Mol Gen Genet. 1981;182(3):414–421. doi: 10.1007/BF00293929. [DOI] [PubMed] [Google Scholar]
- Behnke D., Gilmore M. S. Location of antibiotic resistance determinants, copy control, and replication functions on the double-selective streptococcal cloning vector pGB301. Mol Gen Genet. 1981;184(1):115–120. doi: 10.1007/BF00271206. [DOI] [PubMed] [Google Scholar]
- Behnke D., Klaus S. Double or triple sets of replication functions as inverted and direct repeats on in vitro reconstructed streptococcal MLS resistance plasmids. Z Allg Mikrobiol. 1983;23(9):539–547. doi: 10.1002/jobm.3630230902. [DOI] [PubMed] [Google Scholar]
- Birnboim H. C., Doly J. A rapid alkaline extraction procedure for screening recombinant plasmid DNA. Nucleic Acids Res. 1979 Nov 24;7(6):1513–1523. doi: 10.1093/nar/7.6.1513. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brendel V., Trifonov E. N. A computer algorithm for testing potential prokaryotic terminators. Nucleic Acids Res. 1984 May 25;12(10):4411–4427. doi: 10.1093/nar/12.10.4411. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brennan R. G., Matthews B. W. The helix-turn-helix DNA binding motif. J Biol Chem. 1989 Feb 5;264(4):1903–1906. [PubMed] [Google Scholar]
- Bron S., Luxen E. Segregational instability of pUB110-derived recombinant plasmids in Bacillus subtilis. Plasmid. 1985 Nov;14(3):235–244. doi: 10.1016/0147-619x(85)90007-1. [DOI] [PubMed] [Google Scholar]
- Clewell D. B. Plasmids, drug resistance, and gene transfer in the genus Streptococcus. Microbiol Rev. 1981 Sep;45(3):409–436. doi: 10.1128/mr.45.3.409-436.1981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deichelbohrer I., Alonso J. C., Lüder G., Trautner T. A. Plasmid transduction by Bacillus subtilis bacteriophage SPP1: effects of DNA homology between plasmid and bacteriophage. J Bacteriol. 1985 Jun;162(3):1238–1243. doi: 10.1128/jb.162.3.1238-1243.1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Devereux J., Haeberli P., Smithies O. A comprehensive set of sequence analysis programs for the VAX. Nucleic Acids Res. 1984 Jan 11;12(1 Pt 1):387–395. doi: 10.1093/nar/12.1part1.387. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gruss A., Ehrlich S. D. The family of highly interrelated single-stranded deoxyribonucleic acid plasmids. Microbiol Rev. 1989 Jun;53(2):231–241. doi: 10.1128/mr.53.2.231-241.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Henikoff S. Unidirectional digestion with exonuclease III creates targeted breakpoints for DNA sequencing. Gene. 1984 Jun;28(3):351–359. doi: 10.1016/0378-1119(84)90153-7. [DOI] [PubMed] [Google Scholar]
- Horodniceanu T., Bouanchaud D. H., Bieth G., Chabbert Y. A. R plasmids in Streptococcus agalactiae (group B). Antimicrob Agents Chemother. 1976 Nov;10(5):795–801. doi: 10.1128/aac.10.5.795. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Iordănescu S. Recombinant plasmid obtained from two different, compatible staphylococcal plasmids. J Bacteriol. 1975 Nov;124(2):597–601. doi: 10.1128/jb.124.2.597-601.1975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Iordănescu S., Surdeanu M. Complementation of a plasmid replication defect by autonomous incompatible plasmids in Staphylococcus aureus. Plasmid. 1980 Jul;4(1):1–7. doi: 10.1016/0147-619x(80)90078-5. [DOI] [PubMed] [Google Scholar]
- Krah E. R., 3rd, Macrina F. L. Genetic analysis of the conjugal transfer determinants encoded by the streptococcal broad-host-range plasmid pIP501. J Bacteriol. 1989 Nov;171(11):6005–6012. doi: 10.1128/jb.171.11.6005-6012.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Laemmli U. K. Cleavage of structural proteins during the assembly of the head of bacteriophage T4. Nature. 1970 Aug 15;227(5259):680–685. doi: 10.1038/227680a0. [DOI] [PubMed] [Google Scholar]
- Maciag I. E., Viret J. F., Alonso J. C. Replication and incompatibility properties of plasmid pUB110 in Bacillus subtilis. Mol Gen Genet. 1988 May;212(2):232–240. doi: 10.1007/BF00334690. [DOI] [PubMed] [Google Scholar]
- Moffatt B. A., Studier F. W. T7 lysozyme inhibits transcription by T7 RNA polymerase. Cell. 1987 Apr 24;49(2):221–227. doi: 10.1016/0092-8674(87)90563-0. [DOI] [PubMed] [Google Scholar]
- Novick R. P. Staphylococcal plasmids and their replication. Annu Rev Microbiol. 1989;43:537–565. doi: 10.1146/annurev.mi.43.100189.002541. [DOI] [PubMed] [Google Scholar]
- Pabo C. O., Sauer R. T. Protein-DNA recognition. Annu Rev Biochem. 1984;53:293–321. doi: 10.1146/annurev.bi.53.070184.001453. [DOI] [PubMed] [Google Scholar]
- Projan S. J., Novick R. Comparative analysis of five related Staphylococcal plasmids. Plasmid. 1988 May;19(3):203–221. doi: 10.1016/0147-619x(88)90039-x. [DOI] [PubMed] [Google Scholar]
- Rabinovich P. M., Haykinson MYa, Arutyunova L. S., Yomantas YuV, Stepanov A. I. The structure and source of plasmid DNA determine the cloning properties of vectors for Bacillus subtilis. Basic Life Sci. 1985;30:635–656. doi: 10.1007/978-1-4613-2447-8_44. [DOI] [PubMed] [Google Scholar]
- Rottländer E., Trautner T. A. Genetic and transfection studies with B, subtilis phage SP 50. I. Phage mutants with restricted growth on B. subtilis strain 168. Mol Gen Genet. 1970;108(1):47–60. doi: 10.1007/BF00343184. [DOI] [PubMed] [Google Scholar]
- Sanger F., Nicklen S., Coulson A. R. DNA sequencing with chain-terminating inhibitors. Proc Natl Acad Sci U S A. 1977 Dec;74(12):5463–5467. doi: 10.1073/pnas.74.12.5463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shields D. C., Sharp P. M. Synonymous codon usage in Bacillus subtilis reflects both translational selection and mutational biases. Nucleic Acids Res. 1987 Oct 12;15(19):8023–8040. doi: 10.1093/nar/15.19.8023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Simon D., Chopin A. Construction of a vector plasmid family and its use for molecular cloning in Streptococcus lactis. Biochimie. 1988 Apr;70(4):559–566. doi: 10.1016/0300-9084(88)90093-4. [DOI] [PubMed] [Google Scholar]
- Studier F. W., Moffatt B. A. Use of bacteriophage T7 RNA polymerase to direct selective high-level expression of cloned genes. J Mol Biol. 1986 May 5;189(1):113–130. doi: 10.1016/0022-2836(86)90385-2. [DOI] [PubMed] [Google Scholar]
- Swinfield T. J., Oultram J. D., Thompson D. E., Brehm J. K., Minton N. P. Physical characterisation of the replication region of the Streptococcus faecalis plasmid pAM beta 1. Gene. 1990 Mar 1;87(1):79–90. [PubMed] [Google Scholar]
- Tinoco I., Jr, Borer P. N., Dengler B., Levin M. D., Uhlenbeck O. C., Crothers D. M., Bralla J. Improved estimation of secondary structure in ribonucleic acids. Nat New Biol. 1973 Nov 14;246(150):40–41. doi: 10.1038/newbio246040a0. [DOI] [PubMed] [Google Scholar]
- Vieira J., Messing J. Production of single-stranded plasmid DNA. Methods Enzymol. 1987;153:3–11. doi: 10.1016/0076-6879(87)53044-0. [DOI] [PubMed] [Google Scholar]
- Yanisch-Perron C., Vieira J., Messing J. Improved M13 phage cloning vectors and host strains: nucleotide sequences of the M13mp18 and pUC19 vectors. Gene. 1985;33(1):103–119. doi: 10.1016/0378-1119(85)90120-9. [DOI] [PubMed] [Google Scholar]



