Abstract
A single retroviral protein, Gag, is sufficient for virus particle assembly. While Gag is capable of specifically packaging the genomic RNA into the particle, this RNA species is unnecessary for particle assembly in vivo. In vitro, nucleic acids profoundly enhance the efficiency of assembly by recombinant Gag proteins, apparently by acting as “scaffolding” in the particle. To address the participation of RNA in retrovirus assembly in vivo, we analyzed murine leukemia virus particles that lack genomic RNA because of a deletion in the packaging signal of the viral RNA. We found that these particles contain cellular mRNA in place of genomic RNA. This result was particularly evident when Gag was expressed by using a Semliki Forest virus-derived vector: under these conditions, the Semliki Forest virus vector-directed mRNA became very abundant in the cells and was readily identified in the retroviral virus-like particles. Furthermore, we found that the retroviral cores were disrupted by treatment with RNase. Taken together, the data strongly suggest that RNA is a structural element in retrovirus particles.
Keywords: murine leukemia virus, packaging cell lines, retrovirus assembly, RNA packaging, Semliki Forest virus gene expression system
In all virus particles, the nucleic acid genome is enclosed in a particle consisting largely of virus-coded protein subunits. Therefore, assembly of the complete particle always involves both protein–protein and nucleic acid–protein interactions. In the case of some viruses, protein–protein interactions are evidently sufficient for assembly of the outer shell, because empty capsids can be formed in the absence of nucleic acid (1–4). In other viruses, the nucleic acid is essential for assembly of the shell, acting as a nucleation point or as scaffolding for the capsid proteins (5–9)
It is not known which of these two broad categories retroviruses fall into. Expression of a single virus-coded protein, termed Gag, is sufficient for the efficient assembly and release of retrovirus-like particles from cells of higher eukaryotes (10–12). A typical newly released particle of a conventional retrovirus contains ≈1,500 Gag molecules, arranged radially in a roughly spherical array with their N termini facing outward and their C termini inward (12–15). In the interior of the particle is the genomic RNA of the virus (a pair of identical, sense-strand RNA molecules joined in a dimeric form), along with ≈30 molecules of tRNA and other small cellular RNAs (16), and the virus-coded enzymes—i.e., protease (PR), reverse transcriptase (RT), and integrase. The Gag molecules are associated with a lipid bilayer that is derived from the plasma membrane of the virus-producing cell, and molecules of the virus-coded envelope protein project through the bilayer.
After the particle is released from the cell, the Gag protein is cleaved by PR into a series of cleavage products. These cleavage events are termed “maturation” of the particle, and the products always include at least three proteins, i.e., matrix (MA), capsid (CA), and nucleocapsid (NC) (12). Maturation results in a reorganization of the internal structure of the virion and is required for the infectivity of the particle. NC is complexed with RNA in the interior of the mature particle.
Selection of the genomic RNA for packaging in the nascent particle is not understood in detail, but it involves recognition by the Gag protein of a cis-acting “packaging signal,” termed Ψ, in the 5′ leader region of the RNA (for a review see ref. 16). Deletion of this signal or depletion of this RNA species severely impairs viral RNA packaging in the particle but does not interfere with particle assembly (17, 18). As far as is known, it is the NC domain of Gag that interacts with the RNA and selects the genome for packaging into retroviral particles. Mutations in this domain frequently prevent the specific packaging of the RNA (reviewed in ref. 16). In addition, deletions in the NC domain may impair assembly or lead to the production of aberrant particles that have a low buoyant density (12, 19–21). Taken together, these studies suggest that the NC domain of Gag plays an important role in particle formation.
The fact that genomic RNA is completely unnecessary for particle formation might imply that assembly is driven purely by protein–protein interactions between Gag molecules. On the other hand, in vitro studies using fragments of recombinant Gag proteins containing the NC domain suggest a role for RNA in assembly, because nucleic acids dramatically facilitate assembly in these experiments (22–26). One hypothesis consistent with all these observations is that the small cellular RNAs, principally tRNAs, that are present in retrovirus particles are used in assembly in vivo, as suggested earlier (14, 17). Another possibility is that particles lacking genomic RNA use other, cell-derived RNAs as scaffolding.
We have now explored the possibility that in mammalian cells Gag–RNA interactions are essential in the formation and maintenance of the structure of the retrovirus particle. One prediction of this hypothesis is that all particles always contain a roughly constant amount of RNA: thus, when genomic RNA is not incorporated into virus-like particles (VLPs) in vivo, it will be replaced by other, cell-derived RNA molecules. A second prediction is that the internal core of a retrovirus particle might be disrupted by exposure to RNase. As shown below, our results confirm both predictions, lending strong support to the idea that RNA has a structural role in retrovirus particles.
Materials and Methods
Plasmid Constructs.
Wild-type Moloney murine leukemia virus (MuLV) and the PR− active-site mutant D32L proviral clones used in this study are in pCGcos3neo, a vector derived from pSV2Neo (27, 28). The Ψ− clone is a chimera in which the 5′ region of the genome, up to the XhoI site at nucleotide 1560, is from pPAM3 and contains a deletion from nucleotide 215 to 563 of the MuLV genome, eliminating the packaging signal Ψ (29), whereas the remainder is from our wild-type clone. The Semliki Forest virus (SFV) vector constructs pSFVC-Pr65gag and pSFV1 have been described previously (30) and were a kind gift of Henrik Garoff.
Mammalian Cell Transfection and Virus Production.
293T human embryonic kidney cells and BHK 21 baby hamster kidney cells were used in this study.
Culture fluids containing virus particles were harvested at 24, 48, and 72 h after transfection of 293T cells and clarified by filtration (0.45-μm pore diameter; Nalge), as described previously (31). Ten micrograms of plasmid DNA was used to transfect 106 cells per 10-cm culture dish.
RNA transcripts of pSFVC-Pr65gag or pSFV1 plasmids linearized by SpeI were synthesized in vitro and introduced into BHK cells by electroporation (32). The cells were then plated on 10-cm tissue culture dishes and incubated at 37°C. The supernatant containing the VLPs was harvested at 24 h after electroporation and clarified by centrifugation at low speed (1,000 × g).
Immunoblotting and RT Assays.
Relative amounts of virus were measured by immunoblotting with rabbit antiserum against MuLV p30CA, using the ECL reagent (Amersham Pharmacia Life Sciences). The quantification of the ECL signal on the membrane (from multiple dilutions of each sample) was performed either by scanning the x-ray film or directly by measuring the chemiluminescence with a phosphorimager (Bio-Rad), using the quantification program Quantity One (Bio-Rad). Wild-type and Ψ− virus were compared with respect to p30CA, whereas PR−, Ψ−PR−, and SFV-derived VLPs were compared with respect to Pr65Gag levels. In some experiments, virus production was also determined by assaying RT activity in the MuLV particles (31). The RT results were always in close agreement with the immunoblotting measurements.
Virus Preparation and RNA Isolation.
Virions and VLPs were purified from filtered culture supernatants by pelleting through a cushion of 20% sucrose/TNE (100 mM NaCl/10 mM Tris⋅HCl, pH 7.4/1 mM EDTA), at 25,000 rpm for 45 min at 4°C in a Beckman SW28 rotor. The virus pellets were resuspended for 1 h at 4°C in TNE and either stored at −20°C for protein quantification or disrupted by addition of 1 vol of a 2× concentrated lysis buffer containing 100 mM Tris⋅HCl (pH 7.4), 20 mM EDTA, 2% SDS, 200 mM NaCl, and 200 μg of proteinase K per ml, followed by an incubation of 30 min at 37°C. The RNAs were then extracted with phenol/chloroform and precipitated with 3 vol of 100% ethanol/0.3 M sodium acetate, pH 5.2/0.02% linear acrylamide. “Mock” preparations were obtained from 293T cells transfected with the empty pCGcos3neo vector, or from BHK cells electroporated with pSFV1 vector RNA, and treated exactly as in the viral preparations. The volume of mock supernatant assayed was always the same as the wild-type sample.
Total cellular RNAs were extracted by the Trizol procedure (GIBCO/BRL), dissolved in 20 μl of RNase-free water, and quantified by absorbance at 260 nm.
RNA 3′-End-Labeling.
RNA extracted from viral particles or BHK cells was dissolved in 10 μl of RNase-free water and mixed with 17 μl of a buffer containing 10% DMSO (Sigma), 1× T4 RNA ligase buffer (Roche), 4 units of recombinant RNasin (Promega), and 10 μg/ml BSA. The RNA molecules were labeled at their 3′ ends by incubation of the reaction mixture with 1 μl of [32P]pCp at 3,000 Ci/mmol (ICN; 1 Ci = 37 GBq) and 2 μl of T4 RNA ligase at 10 units/μl (Roche), overnight at 10°C. The labeled RNA molecules were then purified with a Sephadex G-50 spin column (Roche), extracted with phenol/chloroform, and precipitated with ethanol after addition of linear polyacrylamide.
The radiolabeled RNA samples were pelleted from the ethanol, dissolved in 10 μl of RNase-free water, and analyzed on a denaturing 1% agarose gel containing formaldehyde, in Mops 1× running buffer (33). The gel was fixed for 20 min in 10% acetic acid/20% methanol, and then dried for 3 h at 65°C before visualization by autoradiography (Biomax Amersham Pharmacia film and screen). Radioactivity in RNA bands was quantified with the Storm 860 PhosphorImager (Molecular Dynamics).
Poly(A)+ RNA Purification.
Poly(A)+ RNAs were purified with the poly(A)+ RNA isolation kit (Roche). Mock preparations always gave values ≤2% of the experimental values.
RNA Quantification by Ribogreen Reagent.
Amounts of total viral RNA and poly(A)+ RNA were measured by using the Ribogreen quantification kit (Molecular Probes). RNA samples were first treated by RNase-free DNase RQ1 (Promega) in 10 μl of a DNase buffer containing 10 mM Tris⋅HCl (pH 7.4), 100 mM NaCl, 5 mM MgCl2, for 1 h at 37°C, and the RNA was then purified with a Sephadex G-50 spin column (Roche). The purified RNA samples in 10 mM Tris⋅HCl, pH 7.4/1 mM EDTA (TE buffer) were then mixed with 1 vol of the Ribogreen reagent that had been diluted 2000-fold in TE buffer. Excitation was at 480 nm, and the emission (520 nm) was measured on a TD700 fluorometer (Turner Designs, Sunnyvale, CA).
Extensive control experiments showed that the intensity of fluorescence is linearly proportional to the RNA concentration over the concentration range used here and that equal amounts of different RNAs purified by sucrose gradients [tRNA, viral genomic RNA, and cellular poly(A)+ RNA extracted from 293T cells] gave equal fluorescence (data not shown).
RT-PCR Assay.
RNA was isolated from pellets of wild-type and Ψ− mutant virions (containing the same number of particles as determined by immunoblotting for CA), and serial dilutions of the RNA were analyzed for human “housekeeping” gene mRNAs by RT-PCR, as described (34), using PCR primers for human β-actin (giving an 838-bp product) and glyceraldehyde-3-phosphate dehydrogenase (G3PDH) (giving a 983-bp product) from CLONTECH. Levels of RNA species in various samples were estimated by determining the dilution at which the PCR product was no longer visible by ethidium bromide fluorescence.
RNA Analysis by Northern Blotting.
RNAs were analyzed by denaturing Northern blotting as described (33). RNA samples were heated for 10 min at 65°C in denaturing RNA loading dye before electrophoresis on a 0.9% agarose denaturing gel. The 32P-labeled MuLV cDNA probe was generated from the entire Moloney MuLV proviral clone that had been digested by XbaI, using a random-primer cDNA kit (Roche).
Retrovirus Core Isolation and RNase A Treatment.
PR− and Ψ−PR− virions, isolated as described above, were resuspended in 50 mM Tris⋅HCl, pH 7.4/100 mM NaCl (TN buffer) and incubated at room temperature for 30 min, in the presence of 1% Nonidet P-40 or 1% SDS or in the absence of detergent, in a final volume of 20 μl. Afterward, the samples were incubated at room temperature for 2 h with or without RNase A (40 μg per sample) in TN buffer in a final volume of 30 μl. The samples were then layered onto 10 μl of 20% sucrose/TNE cushions containing the same detergents present during incubation, and centrifuged in a microcentrifuge tube at 18°C for 1 h at 13,000 rpm in a Jouan AB2.14 rotor (BR4i centrifuge). The Gag proteins present in the supernatant and pellet were fractionated by SDS/PAGE and analyzed by immunoblotting with a rabbit antiserum to MuLV CA, as described above.
Results
Ψ− MuLV Particles Contain a Roughly Constant Amount of RNA.
If RNA molecules are involved in forming and maintaining the structure of retroviruses, then all particles should contain nearly the same amount of RNA. We tested this hypothesis by comparing the total RNA contents of wild-type and Ψ− particles of Moloney MuLV. MuLV Gag is synthesized as a 65-kDa precursor polyprotein (Pr65Gag), which is cleaved during maturation into the four mature proteins—i.e., matrix, p12, CA, and NC (12). The Ψ− particles are formed from wild-type viral proteins, but do not package their genomic RNA efficiently because of a deletion of the cis-acting packaging signal (Ψ) in the viral RNA (18). In all of the experiments described in this paper, RNAs from equal amounts of Ψ+ (wild-type) and Ψ− (or Ψ+PR− and Ψ−PR−) virions are compared. Relative particle concentrations were determined by immunoblotting with anti-CA serum or by RT assays. RNA was extracted from Ψ+ and Ψ− particles and the RNA concentrations were measured by using the Ribogreen assay, in which a fluorescent dye can measure as little as 1 ng/ml RNA (see Materials and Methods). We found that the RNA content of the Ψ− particles was ≈57% of that of the wild-type particles (Table 1). To establish that this RNA was actually packaged within virus particles, and not just present in contaminating cell debris, we measured the RNA level in mock virus preparations (see Materials and Methods). This level was always ≤7% of that in viral pellets (Table 1). This question is considered further below. Because viral genomic RNA represents the majority of the RNA in a wild-type particle (16), other RNAs from the cell have evidently replaced a significant fraction of this RNA in the Ψ− particles.
Table 1.
Relative RNA content of virus particles and Gag VLPs
| MuLV virions | Genomic RNA, % | Total RNA content, % | Poly(A)+ mRNA, % |
|---|---|---|---|
| Wild-type (*) | 100 | 100 | 100 |
| Ψ− (*) | <0.5 | 57.7 ± 8 | 34.4 ± 6 |
| PR− | 100 | 100 | 100 |
| Ψ−PR− | <0.5 | 65.3 ± 2 | 27.1 ± 3 |
| SFV-encoded Gag VLPs | — | 107 ± 11 | ND |
Results are expressed relative to RNA content of wild-type (*) or PR− virions, determined by Ribogreen assay. Each sample contains the same amount of p30CA (wild-type and Ψ−) or Pr65Gag (PR−, Ψ−PR−, and Gag VLPs), or the same RT activity. Each value is expressed as a percentage of wild-type or PR− RNA content and represents the mean (±SD) of values obtained from at least three independent experiments. Each experimental value shown has been reduced by subtraction of the RNA content of the mock preparation—i.e., supernatant harvested from 293T cells transfected with pGCcos3neo vector (equal to 6% ± 1%) or from BHK cells electroporated with pSFV1 vector (equal to 13% ± 0.7%), and treated and assayed in parallel with the virion samples. ND, not determined.
We also performed analogous experiments on PR− virions. While Pr65Gag processing is required for infectivity, MuLV with a defect in PR still assembles into immature particles containing Pr65Gag and the viral RNA. As shown in Table 1, Ψ−PR− MuLV particles contained ≈65% as much RNA as did PR− particles: thus, just as with mature particles, cellular RNAs are present in immature MuLV particles in place of genomic RNA.
Analysis of End-Labeled RNA Molecules of MuLV Particles Lacking Genomic RNA.
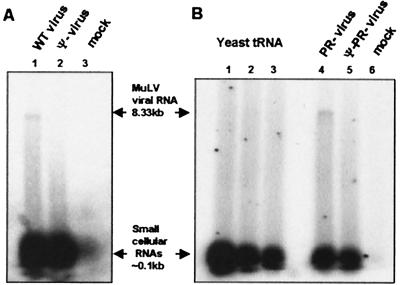
The Ribogreen measurements suggested that Ψ− particles contain cellular RNA in place of genomic RNA. We attempted to detect and identify such “compensating” cellular RNA by end-labeling it in vitro after extraction from the virions, and then analyzing it by electrophoresis on denaturing gels (Fig. 1). We observed that wild-type MuLV particles contain a faint band near the top of the gel, representing the 8.3-kb genomic RNA (lane 1). There is a very faint smear down the lane, and then a very broad, intense band of low-molecular-weight RNA, with size of 100 nucleotides or less. When the radioactivity is counted and the sizes of these end-labeled RNAs taken into account, the amount of low-molecular-weight RNA is found to be 20–25% of the amount of genomic RNA in these preparations. This ratio is in excellent agreement with previous biochemical descriptions of retroviruses (reviewed in ref. 16).
Figure 1.
Analysis of wild-type and mutant virion RNA content by RNA end-labeling. RNA extracted from virions was end-labeled with T4 RNA ligase and [32P]pCp, and was analyzed on a denaturing 1% agarose gel. (A) RNAs extracted from the same number of wild-type (lane 1) and Ψ− (lane 2) virion particles or from a mock preparation (lane 3). (B) RNAs extracted from the same number of PR− (lane 4) and Ψ−PR− (lane 5) virion particles or from a mock preparation (lane 6). Lanes 1, 2, and 3 of B represent 2, 1, and 0.5 pmol of yeast tRNA, respectively, which were labeled by the same technique as the other RNA samples.
As expected, labeling the RNAs extracted from Ψ− particles shows no visible band of genomic RNA (Fig. 1A, lane 2). However, there is a faint smear down the length of the lane, and an intense band of low-molecular-weight RNA at the bottom of the gel. Remarkably, neither the smear down the lane nor the band of small RNAs is more intense than in the wild-type control in lane 1; thus, the identity of the RNA replacing genomic RNA in Ψ− particles is not evident from these results. Identical results were also obtained in the comparison of PR− with Ψ−PR− particles (Fig. 1B, lanes 4 and 5).
Control experiments showed that the labeling of an RNA species is proportional to the amount of this species in the sample (shown for yeast tRNA in Fig. 1B, lanes 1–3). Thus, phosphorimager analysis (not shown) of the low-molecular-weight RNA bands of Fig. 1 showed that the Ψ− particles contained 78–86% as much small RNA as the wild-type particles. Thus, the level of these RNAs is not altered significantly by the absence of genomic RNA in Ψ− MuLV particles.
We also analyzed end-labeled RNA from a mock virus preparation. As shown (Fig. 1A, lane 3; Fig. 1B, lane 6), these samples contained only ≈7–10% as much low-molecular-weight RNA as the wild-type pellet. We also found that the small RNAs in viral preparations were in structures with a buoyant density of ≈1.14 g/ml, the density of retrovirus particles (data not shown). Still more evidence that the RNAs in viral pellets are actually packaged within virus particles was obtained by exposing them to RNase: as shown in Fig. 5 and its accompanying text, which are published as supplemental data on the PNAS web site, www.pnas.org, they were completely resistant to digestion unless the membrane of the virus was disrupted with detergent.
Ψ− MuLV Particles Package Cellular Messenger RNAs.
While the results in Table 1 clearly indicated that other RNAs take the place of genomic RNA in Ψ− MuLV particles, the end-labeling analysis (Fig. 1) failed to identify these RNAs as a discrete band. The only RNA detectable in the Ψ− particles, other than small cellular RNAs, was the faint, polydisperse smear extending over a very broad molecular-weight range (Fig. 1A, lane 2; Fig. 1B, lane 5).
It seemed possible that this smear represents a polydisperse population of cellular mRNA molecules, and that these are the RNAs compensating for genomic RNA in Ψ− particles. We measured the amount of poly(A)+ RNA in the Ψ− particles by using the Ribogreen assay, and we found that the mass of poly(A)+ RNA in the Ψ− RNA preparations was 27–34% of that in wild-type preparations (Table 1).
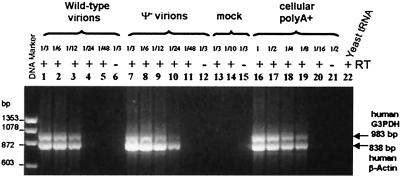
We also used semiquantitative RT-PCR assays to analyze Ψ− mutant virions, in comparison to wild-type virions, for the presence of abundant cellular mRNAs—i.e., human β-actin and G3PDH mRNAs (Fig. 2). We observed that the dilution end-point for detection of these mRNAs was 1/24 in Ψ− RNAs (Fig. 2, lane 10) and 1/12 in wild-type RNAs (Fig. 2, lane 3). Thus, Ψ− particles contain 2- to 4-fold more β-actin and G3PDH mRNAs than do wild-type particles. The ratio of these two species to each other in the viral preparations also appears similar to that in mRNA of the virus-producing cells (Fig. 2, lanes 16–20), suggesting that the particles may contain a random sampling of cellular mRNAs.
Figure 2.
RT-PCR analysis of human β-actin and G3PDH mRNAs packaged in wild-type and Ψ− mutant particles. Lanes 1–6, 2-fold serial dilutions of total RNA extracted from wild-type virus; lanes 7–12, 2-fold dilutions of RNA from Ψ− virions; lanes 13–15, RNA from dilutions of mock supernatant; lanes 16–21, 2-fold dilutions starting from 200 ng of purified cellular poly(A)+ RNA from 293T cells; lane 22, yeast tRNA (50 ng). Starting material represented the same amount of wild-type and Ψ− virions. As controls for DNA contamination, lanes 6, 12, 15, and 21 do not contain any RT.
The results presented above are all consistent with the hypothesis that Ψ− particles package cellular mRNA in place of genomic RNA. However, because it is so difficult to identify and accurately quantify polydisperse mRNAs, we attempted to solve this problem by analyzing Ψ− particles produced from cells in which the cellular mRNA population was practically monodisperse, rather than polydisperse. We achieved this goal by expressing the MuLV Gag polyprotein from an SFV-derived vector and examining the RNA in the resulting Gag particles.
RNA Content of SFV-Derived MuLV Gag VLPs.
SFV is an alphavirus. SFV-derived vectors, like SFV itself, encode proteins capable of extremely efficient RNA-directed RNA synthesis. They produce two major species of RNA: a full-length (≈12–13 kb) genomic RNA and a subgenomic RNA. The latter RNA encodes viral structural proteins, and in alphavirus vectors the gene to be expressed replaces the coding region for these proteins (for a review, see ref. 35). SFV-derived vectors expressing the gag gene of Moloney MuLV have been described (30, 36). These vectors contain no retroviral sequences other than the gag coding region (Fig. 3A).
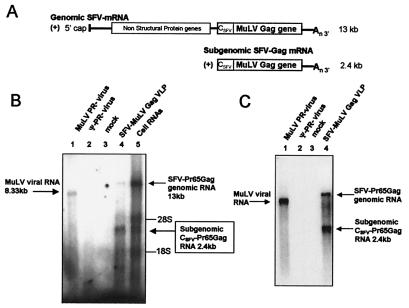
Figure 3.
RNA content of SFV-derived MuLV Gag VLPs. (A) Schematic representation of the two (+) strand RNAs produced by the SFV vector expressing MuLV Gag: the SFV genomic and subgenomic mRNAs. CSFV represents the sequence of SFV CA, which is removed from Gag cotranslationally (30). (B) RNA end-labeling analysis of VLPs and cell lysates. RNAs were analyzed as in Fig. 1. Lanes 1 and 2, RNAs were obtained from the same amount of PR− (lane 1) and Ψ−PR− (lane 2) virions; lane 3, “VLP” preparations from BHK 21 cells electroporated with pSFV1 RNA (“mock”); lane 4, VLPs produced by BHK21 cells electroporated with pSFVC-Pr65gag RNA (Gag VLPs); and lane 5, cellular RNA from the same electroporated culture as in lane 4. Lanes 1, 2, and 4 contain the same number of particles. (C) Identification of Pr65Gag mRNA by Northern blotting using a full-length proviral MuLV probe. RNA from the same amount of PR− (lane 1) and Ψ−PR− (lane 2) virions, and Gag VLPs (lane 4) were analyzed. Lane 3 is a mock preparation as in B. MuLV genomic RNA (8.33 kb), the SFV-Pr65Gag genomic RNA (13 kb), and the SFV subgenomic Pr65Gag mRNA (2.4 kb) are indicated, as are 28S (≈5 kb) and 18S (≈1.5 kb) ribosomal RNAs.
The RNA content of MuLV Gag VLPs produced from the SFV c-Gag vector was analyzed by RNA end-labeling and compared with the RNA content of immature PR− virions (Fig. 3, Table 1). The SFV c-Gag-derived VLPs contained two major discrete species of high-molecular-weight RNA (Fig. 3B, lane 4). Their molecular masses were ≈13 kb and 2.4 kb. These were evidently packaged within the VLPs, since a mock preparation did not contain them (lane 3), and since they were protected from RNase digestion by a detergent-sensitive membrane (see supplemental data). We could also detect some 28S and 18S rRNA and heterodisperse RNAs smaller than 2.4 kb that seemed to be associated with the VLPs (lane 4). The molecular weights of the two prominent bands in the SFV c-Gag-derived VLPs correspond to those of the two RNA species—i.e., full-length and subgenomic—expressed from the SFV c-Gag vector (Fig. 3A). RNAs of these sizes were present at extremely high levels in the cells producing the VLPs (Fig. 3B, lane 5).
The identity of the 2.4- and 13-kb RNAs in the VLPs as SFV-derived RNAs was confirmed by Northern blotting with an MuLV-specific DNA probe (Fig. 3C, lane 4). The probe also reacted with the 8.3-kb genomic RNA in PR− virions (lane 1), whereas no MuLV-specific band was detectable in the Ψ−PR− particles (lane 2), as expected.
We also measured the amount of total RNA in the SFV-encoded Gag VLPs by the Ribogreen assay. These particles contain as much RNA as PR− virions did (Table 1). These results are fully consistent with the qualitative data in Fig. 3. Taken together, the findings show that the SFV-derived mRNAs totally compensate for the absence of retroviral RNA in the VLPs, despite the fact that these mRNA molecules do not contain any retroviral Ψ sequences.
MuLV Virion Cores Are Disrupted by RNase.
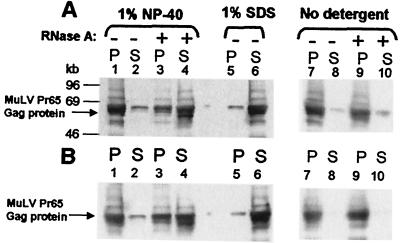
We also tested the possibility that the RNAs in MuLV particles are involved in maintenance of the structure of the particles, as suggested by in vitro results with retroviral Gag proteins (22, 24). We have approached this question by testing the effect of RNase digestion on the stability of immature virion cores [this cannot be studied in mature cores, because they are unstable in detergent (37)]. We found that immature MuLV cores were stable in 1% Nonidet P-40: nearly all of the Gag proteins remained in the pellet after centrifugation (Fig. 4, lanes 1 and 2). However, when the cores were exposed to RNase, a large fraction of the MuLV Gag proteins were no longer pelletable (lanes 3 and 4), indicating that they had been held together by RNA. This effect of RNase required removal of the viral membrane with Nonidet P-40 (lanes 9 and 10). In some experiments (as shown in lanes 3 and 4), only 2/3, rather than nearly all, of the Pr65Gag was released by RNase: either the RNA is not completely accessible to RNase, or some Gag–Gag interactions in the core are not RNase sensitive. Identical results were obtained with PR− (Fig. 4A) and Ψ−PR− (Fig. 4B) virions, suggesting that the RNAs in Ψ−PR− virions play the same role as the viral RNA in PR− virions. The disruption of retroviral cores by RNase is direct evidence that the Gag molecules are held together in the core by their interaction with RNA, and that contacts with RNA, as well as Gag–Gag interactions, are essential for the virion structure.
Figure 4.
Effects of RNase A on virion core stability. PR− (A) and Ψ−PR− (B) virions were incubated with or without detergent as indicated. The samples were then incubated in the presence or absence of RNase A before fractionation by centrifugation. The Gag proteins present in the supernatant (S) and pellet (P) were fractionated by SDS/PAGE and analyzed by immunoblotting with rabbit anti-MuLV CA antiserum. Lanes 1–4, virion cores in 1% Nonidet P-40; lanes 7–10, no detergent; lanes 5 and 6, virions lysed in 1% SDS; lanes 3, 4, 9, and 10, + RNase A.
Discussion
The results presented in this work can be briefly summarized as follows. First, Ψ− MuLV particles, which do not package the viral genome efficiently, contain mRNA molecules from the cell in place of the viral RNA. Second, treatment of cores from immature MuLV particles with RNase results in the release of the Gag molecules in soluble form. Taken together, these two findings strongly suggest that the formation and maintenance of the structure of the MuLV particle depend on Gag–RNA, as well as Gag–Gag, interactions.
All particles formed from MuLV Gag contain a roughly constant amount of RNA, suggesting that this amount of RNA could be the minimum required for assembly of an MuLV particle. We checked that the absence of viral RNA in Ψ− particles did not affect the size or morphology of the particles: electron microscopic examination showed that Ψ− and Ψ+ particles are all ≈100–110 nm in diameter, with similar morphologies (data not shown).
We attempted to identify the RNA packaged in Ψ− particles and found that the Ψ− particles contained some polydisperse RNA (Fig. 1) and a substantial amount of poly(A)+ RNA (Table 1). However, it was not clear at first that poly(A)+ RNAs were the “compensating” RNA, because wild-type particles also contain polydisperse RNA (Fig. 1). Northern analysis showed that at least some of the latter is degraded viral RNA (data not shown). We also assayed the viral preparations for specific abundant cellular mRNA species by RT-PCR. These experiments (Fig. 2) confirmed that Ψ− particles contain more cellular mRNA than Ψ+ particles. However, it is striking that wild-type particles contain approximately ¼–½ as much β-actin and G3PDH mRNA as Ψ− particles do (Fig. 2). If these two RNAs are reflective of bulk cellular mRNA, then these results suggest that the high-molecular-weight RNA in wild-type particles could be a mixture of ≈80% genomic RNA and ≈20% cellular mRNA. In other words, while the presence of Ψ in genomic RNA increases its incorporation in virions by >100-fold (data not shown), packaging is evidently still not completely specific. We obviously have no information on the fraction of wild-type particles containing cellular mRNA molecules, and cannot exclude the possibility that these molecules contribute to the assembly of Ψ+, as well as Ψ−, virus particles. Packaging of cellular mRNAs in wild-type retrovirus particles has previously been documented (38, 39), and cellular mRNAs have also been observed in avian Ψ− particles (40), especially those released by the SE21Q1b cell line, which produces particularly high levels of particles (41).
The presence of mRNA molecules in particles formed without genomic RNA became obvious when Gag was expressed in an SFV expression system. Under these conditions, the two SFV-derived RNA species were easily detectable in the VLPs assembled from MuLV Gag proteins (Fig. 3). These VLPs should be quite analogous to Ψ− MuLV, because they are formed from wild-type Gag in cells lacking Ψ+ MuLV RNA. It seems likely that the SFV-derived RNAs are the major RNA species in the particles simply because of their abundance in the cells (as Fig. 3 shows, they are roughly equimolar with ribosomal RNA). We observed that other RNAs, migrating more rapidly than the subgenomic SFV RNA, were also packaged inside the VLPs; the RNA population seems to reflect the cellular RNA distribution (Fig. 3). However, the ratio of 2.4-kb SFV-derived RNA to 13-kb SFV-derived RNA is higher in the VLP preparation (Fig. 3B, lane 4) than in the total cellular RNA (lane 5). This might imply a preferential packaging of the smaller RNA over the 13-kb SFV genomic RNA. It is possible that RNAs significantly larger than the MuLV genome, such as the SFV genome, are not packaged efficiently by MuLV Gag-derived VLPs, or that gag mRNAs are preferentially packaged in cis by Gag proteins.
It is remarkable that the level of low-molecular-weight RNA, such as tRNA, is unaffected by the loss of viral RNA in Ψ− particles (Fig. 1). Our results confirm that the small RNAs are taken into nascent virions independently of viral RNAs, as previously shown (42, 43). We do not know whether the small RNAs are structural elements in the particle.
We also found that immature MuLV cores, with or without viral genomic RNA, are disrupted by treatment with RNase (Fig. 4), showing clearly that the Gag molecules are held together in the core by their interaction with RNA molecules of cellular or viral origin, and that contacts with RNA are essential for the maintenance, as well as the formation, of the virion. Further experiments will be required to determine the physical state of the Gag molecules released by RNase treatment.
How does RNA contribute to retrovirus assembly? It is known that NC can bind nucleic acids with little specificity in vitro [at least at low ionic strength (44)], and that its basic residues are critical for this binding (reviewed in ref. 16). These residues also play a crucial role in both virion assembly and RNA packaging in vivo, and Gag proteins lacking the entire NC domain fail to assemble virus particles efficiently (refs. 19–21, 45, 46; reviewed in refs. 12 and 16). We propose that the RNAs attached to the NC domain of Gag in nascent retrovirus particles serve as a nucleation point or “scaffolding” for concentrating Gag proteins and facilitating Gag–Gag interactions. It should also be noted that retrovirus particles, unlike most “spherical” viruses, lack icosahedral symmetry (13, 14); the lack of absolute regularity in the arrangement of Gag molecules in these particles has previously led us to suggest that Gag–Gag interactions may not be sufficient to drive assembly, and that Gag–RNA interactions may be important in retrovirus structure (14).
Our results have some parallels in the assembly of other viruses. For example, spherical plant viruses, such as cowpea chlorotic mottle virus or brome mosaic virus, have an assembly process that involves interactions between viral capsid proteins and RNA, but this RNA need not be the genomic RNA (5). The assembly of alphaviruses, such as Sindbis virus, into core-like particles in vitro also requires the association of the nucleocapsid protein with nucleic acid (7, 9), as observed for retroviruses (22). Furthermore, when the proteins of flock house virus, a positive-strand RNA virus in the Nodaviridae, are expressed in the absence of genomic RNA, they assemble into virions containing cellular mRNAs (47). These results, and others in the same report, demonstrate the role of RNA in the structural integrity of this virus, and are strikingly comparable to our results with retroviruses.
In summary, our data show that retroviruses fall into the category of viruses whose assembly depends on RNA–protein interactions (as well as protein–protein interactions). It was previously known that Gag interacts with RNA in two distinct ways during virus assembly. First, Gag is able to specifically select the viral genomic RNA for encapsidation (for a review see ref. 16). Second, Gag, by virtue of its nucleic acid chaperone activity, anneals the primer tRNA to the viral RNA (31, 48, 49) and may also facilitate the dimerization of genomic RNA. We now demonstrate a third mode of Gag–RNA interaction: Gag is able to interact nonspecifically with RNA to construct a virion, using either Ψ+ or Ψ− RNA as a scaffold.
Supplementary Material
Acknowledgments
We thank Dr. Robert Gorelick and Ms. Tracy Gagliardi for the RT assays, Dr. José Casas-Finet for the introduction to fluorescence principles, Dr. Barbara Felber and Ms. Jennifer Bear for the use of the phosphorimager, Dr. Henrik Garoff for the gift of SFV-derived vectors, Drs. Stephen Hughes and Stephen Campbell for helpful discussions, and Robert Gorelick, Catherine Hibbert, and Judith Levin for comments on the manuscript.
Abbreviations
- SFV
Semliki Forest virus
- MuLV
murine leukemia virus
- PR
protease
- RT
reverse transcriptase
- CA
capsid
- NC
nucleocapsid
- Ψ
RNA encapsidation signal
- VLP
virus-like particle
- G3PDH
glyceraldehyde-3-phosphate dehydrogenase
Footnotes
This paper was submitted directly (Track II) to the PNAS office.
References
- 1.Salunke D M, Caspar D L, Garcea R L. Cell. 1986;46:895–904. doi: 10.1016/0092-8674(86)90071-1. [DOI] [PubMed] [Google Scholar]
- 2.Rose R C, Bonnez W, Reichman R C, Garcea R L. J Virol. 1993;67:1936–1944. doi: 10.1128/jvi.67.4.1936-1944.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Homa F L, Brown J C. Rev Med Virol. 1997;7:107–122. doi: 10.1002/(sici)1099-1654(199707)7:2<107::aid-rmv191>3.0.co;2-m. [DOI] [PubMed] [Google Scholar]
- 4.Zlotnick A, Johnson J M, Wingfield P W, Stahl S J, Endres D. Biochemistry. 1999;38:14644–14652. doi: 10.1021/bi991611a. [DOI] [PubMed] [Google Scholar]
- 5.Bancroft J B. Adv Virus Res. 1970;16:99–134. doi: 10.1016/s0065-3527(08)60022-6. [DOI] [PubMed] [Google Scholar]
- 6.Abouhaidar M, Bancroft J B. Virology. 1978;90:54–59. doi: 10.1016/0042-6822(78)90332-x. [DOI] [PubMed] [Google Scholar]
- 7.Wengler G, Boege U, Bischoff H, Wahn K. Virology. 1982;118:401–410. doi: 10.1016/0042-6822(82)90359-2. [DOI] [PubMed] [Google Scholar]
- 8.Sorger P K, Stockley P G, Harrison S C. J Mol Biol. 1986;191:639–658. doi: 10.1016/0022-2836(86)90451-1. [DOI] [PubMed] [Google Scholar]
- 9.Tellinghuisen T L, Hamburger A E, Fisher B R, Ostendorp R, Kuhn R J. J Virol. 1999;73:5309–5319. doi: 10.1128/jvi.73.7.5309-5319.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Shields A, Witte O N, Rothenberg E, Baltimore D. Cell. 1978;14:601–609. doi: 10.1016/0092-8674(78)90245-3. [DOI] [PubMed] [Google Scholar]
- 11.Gheysen D, Jacobs E, de Foresta F, Thiriart C, Francotte M, Thines D, De Wilde M. Cell. 1989;59:103–112. doi: 10.1016/0092-8674(89)90873-8. [DOI] [PubMed] [Google Scholar]
- 12.Swanstrom R, Wills J W. In: Retroviruses. Coffin J M, Hughes S H, Varmus H E, editors. Plainview, NY: Cold Spring Harbor Lab. Press; 1997. pp. 263–334. [Google Scholar]
- 13.Fuller S D, Wilk T, Gowen B E, Krausslich H G, Vogt V M. Curr Biol. 1997;7:729–738. doi: 10.1016/s0960-9822(06)00331-9. [DOI] [PubMed] [Google Scholar]
- 14.Yeager M, Wilson-Kubalek E M, Weiner S G, Brown P O, Rein A. Proc Natl Acad Sci USA. 1998;95:7299–7304. doi: 10.1073/pnas.95.13.7299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Vogt V M, Simon M N. J Virol. 1999;73:7050–7055. doi: 10.1128/jvi.73.8.7050-7055.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Berkowitz R, Fisher J, Goff S P. Curr Top Microbiol Immunol. 1996;214:177–218. doi: 10.1007/978-3-642-80145-7_6. [DOI] [PubMed] [Google Scholar]
- 17.Levin J G, Grimley P M, Ramseur J M, Berezesky I K. J Virol. 1974;14:152–161. doi: 10.1128/jvi.14.1.152-161.1974. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Mann R, Mulligan R C, Baltimore D. Cell. 1983;33:153–159. doi: 10.1016/0092-8674(83)90344-6. [DOI] [PubMed] [Google Scholar]
- 19.Zhang Y, Barklis E. J Virol. 1997;71:6765–6776. doi: 10.1128/jvi.71.9.6765-6776.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Zhang Y, Qian H, Love Z, Barklis E. J Virol. 1998;72:1782–1789. doi: 10.1128/jvi.72.3.1782-1789.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Bowzard J B, Bennett R P, Krishna N K, Ernst S M, Rein A, Wills J W. J Virol. 1998;72:9034–9044. doi: 10.1128/jvi.72.11.9034-9044.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Campbell S, Vogt V M. J Virol. 1995;69:6487–6497. doi: 10.1128/jvi.69.10.6487-6497.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Gross I, Hohenberg H, Krausslich H G. Eur J Biochem. 1997;249:592–600. doi: 10.1111/j.1432-1033.1997.t01-1-00592.x. [DOI] [PubMed] [Google Scholar]
- 24.Campbell S, Rein A. J Virol. 1999;73:2270–2279. doi: 10.1128/jvi.73.3.2270-2279.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Ganser B K, Li S, Klishko V Y, Finch J T, Sundquist W I. Science. 1999;283:80–83. doi: 10.1126/science.283.5398.80. [DOI] [PubMed] [Google Scholar]
- 26.Zuber G, McDermott J, Karanjia S, Zhao W, Schmid M F, Barklis E. J Virol. 2000;74:7431–7441. doi: 10.1128/jvi.74.16.7431-7441.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Southern P J, Berg P. J Mol Appl Genet. 1982;1:327–341. [PubMed] [Google Scholar]
- 28.Fu W, Rein A. J Virol. 1993;67:5443–5449. doi: 10.1128/jvi.67.9.5443-5449.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Miller A D, Buttimore C. Mol Cell Biol. 1986;6:2895–2902. doi: 10.1128/mcb.6.8.2895. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Li K J, Garoff H. Proc Natl Acad Sci USA. 1996;93:11658–11663. doi: 10.1073/pnas.93.21.11658. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Fu W, Ortiz-Conde B A, Gorelick R J, Hughes S H, Rein A. J Virol. 1997;71:6940–6946. doi: 10.1128/jvi.71.9.6940-6946.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Ekstrom M, Garoff H, Andersson H. Cell Biology: A Laboratory Handbook. Vol. 4. San Diego: Academic; 1998. pp. 218–224. [Google Scholar]
- 33.Sambrook J, Fritsch E F, Maniatis T. Molecular Cloning: A Laboratory Manual. 2nd Ed. Plainview, NY: Cold Spring Harbor Lab. Press; 1989. [Google Scholar]
- 34.Rein A, Harvin D P, Mirro J, Ernst S M, Gorelick R J. J Virol. 1994;68:6124–6129. doi: 10.1128/jvi.68.9.6124-6129.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Schlesinger S, Schlesinger M J. In: Virology. Fields B N, Knipe D M, Howley P M, editors. Philadelphia: Lippincott-Raven; 1996. pp. 825–841. [Google Scholar]
- 36.Suomalainen M, Hultenby K, Garoff H. J Cell Biol. 1996;135:1841–1852. doi: 10.1083/jcb.135.6.1841. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Yoshinaka Y, Luftig R B. Proc Natl Acad Sci USA. 1977;74:3446–3450. doi: 10.1073/pnas.74.8.3446. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Ikawa Y, Ross J, Leder P. Proc Natl Acad Sci USA. 1974;71:1154–1158. doi: 10.1073/pnas.71.4.1154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Adkins B, Hunter T. J Virol. 1981;39:471–480. doi: 10.1128/jvi.39.2.471-480.1981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Aronoff R, Linial M. J Virol. 1991;65:71–80. doi: 10.1128/jvi.65.1.71-80.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Anderson D J, Stone J, Lum R, Linial M L. J Virol. 1995;69:7319–7323. doi: 10.1128/jvi.69.11.7319-7323.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Levin J G, Seidman J G. J Virol. 1979;29:328–335. doi: 10.1128/jvi.29.1.328-335.1979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Peters G G, Hu J. J Virol. 1980;36:692–700. doi: 10.1128/jvi.36.3.692-700.1980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Fisher R J, Rein A, Fivash M, Urbaneja M A, Casas-Finet J R, Medaglia M, Henderson L E. J Virol. 1998;72:1902–1909. doi: 10.1128/jvi.72.3.1902-1909.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Poon D T, Wu J, Aldovini A. J Virol. 1996;70:6607–6616. doi: 10.1128/jvi.70.10.6607-6616.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Cimarelli A, Sandin S, Hoglund S, Luban J. J Virol. 2000;74:3046–3057. doi: 10.1128/jvi.74.7.3046-3057.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Bothner B, Schneemann A, Marshall D, Reddy V, Johnson J E, Siuzdak G. Nat Struct Biol. 1999;6:114–116. doi: 10.1038/5799. [DOI] [PubMed] [Google Scholar]
- 48.Cen S, Huang Y, Khorchid A, Darlix J L, Wainberg M A, Kleiman L. J Virol. 1999;73:4485–4488. doi: 10.1128/jvi.73.5.4485-4488.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Feng Y X, Campbell S, Harvin D, Ehresmann B, Ehresmann C, Rein A. J Virol. 1999;73:4251–4256. doi: 10.1128/jvi.73.5.4251-4256.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.