Abstract
The third intron from Physarum polycephalum (Pp LSU 3) is one of the closest known relatives to the well-studied Tetrahymena group I intron. Both introns are located at the same position in the 26S rRNA gene, and with the exception of an open reading frame in Pp LSU 3, are highly homologous. While Pp LSU 3 has been shown to self splice, little is known about its activity in vitro. We have examined the requirements for self splicing in greater detail. Despite its similarity to the Tetrahymena intron, Pp LSU 3 is 1500-fold less reactive, demonstrates a preference for high salt, and exhibits a low Km for GTP. Removal of the open reading frame results in a modest increase of activity. This system provides an opportunity to understand how sequence variations in two related introns alter the efficiency of autoexcision, and how this relates to adaptation of group I introns to their particular sequence context.
Full text
PDF
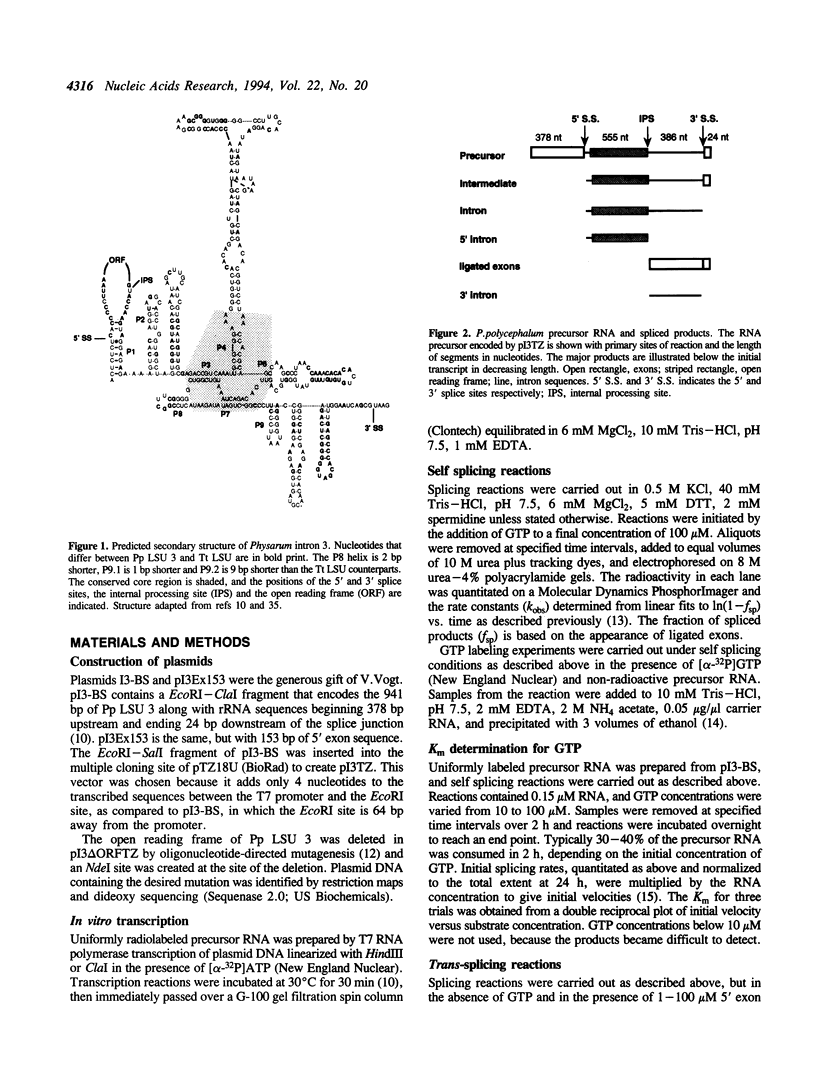
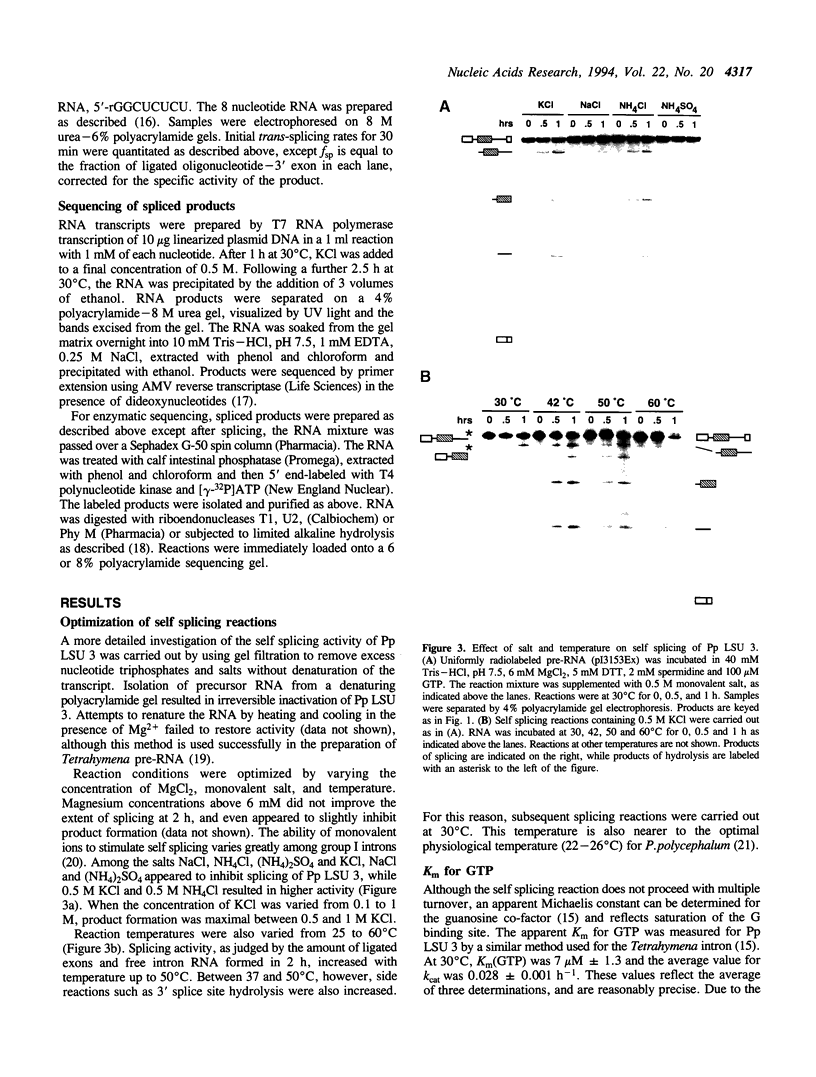


Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Bass B. L., Cech T. R. Ribozyme inhibitors: deoxyguanosine and dideoxyguanosine are competitive inhibitors of self-splicing of the Tetrahymena ribosomal ribonucleic acid precursor. Biochemistry. 1986 Aug 12;25(16):4473–4477. doi: 10.1021/bi00364a001. [DOI] [PubMed] [Google Scholar]
- Bass B. L., Cech T. R. Specific interaction between the self-splicing RNA of Tetrahymena and its guanosine substrate: implications for biological catalysis by RNA. 1984 Apr 26-May 2Nature. 308(5962):820–826. doi: 10.1038/308820a0. [DOI] [PubMed] [Google Scholar]
- Been M. D., Cech T. R. One binding site determines sequence specificity of Tetrahymena pre-rRNA self-splicing, trans-splicing, and RNA enzyme activity. Cell. 1986 Oct 24;47(2):207–216. doi: 10.1016/0092-8674(86)90443-5. [DOI] [PubMed] [Google Scholar]
- Belfort M. Bacteriophage introns: parasites within parasites? Trends Genet. 1989 Jul;5(7):209–213. doi: 10.1016/0168-9525(89)90083-8. [DOI] [PubMed] [Google Scholar]
- Cech T. R., Bass B. L. Biological catalysis by RNA. Annu Rev Biochem. 1986;55:599–629. doi: 10.1146/annurev.bi.55.070186.003123. [DOI] [PubMed] [Google Scholar]
- Cech T. R., Herschlag D., Piccirilli J. A., Pyle A. M. RNA catalysis by a group I ribozyme. Developing a model for transition state stabilization. J Biol Chem. 1992 Sep 5;267(25):17479–17482. [PubMed] [Google Scholar]
- Cech T. R. Self-splicing of group I introns. Annu Rev Biochem. 1990;59:543–568. doi: 10.1146/annurev.bi.59.070190.002551. [DOI] [PubMed] [Google Scholar]
- Donis-Keller H. Site specific enzymatic cleavage of RNA. Nucleic Acids Res. 1979 Sep 11;7(1):179–192. doi: 10.1093/nar/7.1.179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Doudna J. A., Szostak J. W. Miniribozymes, small derivatives of the sunY intron, are catalytically active. Mol Cell Biol. 1989 Dec;9(12):5480–5483. doi: 10.1128/mcb.9.12.5480. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dávila-Aponte J. A., Huss V. A., Sogin M. L., Cech T. R. A self-splicing group I intron in the nuclear pre-rRNA of the green alga, Ankistrodesmus stipitatus. Nucleic Acids Res. 1991 Aug 25;19(16):4429–4436. doi: 10.1093/nar/19.16.4429. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Emerick V. L., Woodson S. A. Self-splicing of the Tetrahymena pre-rRNA is decreased by misfolding during transcription. Biochemistry. 1993 Dec 21;32(50):14062–14067. doi: 10.1021/bi00213a040. [DOI] [PubMed] [Google Scholar]
- Gampel A., Cech T. R. Binding of the CBP2 protein to a yeast mitochondrial group I intron requires the catalytic core of the RNA. Genes Dev. 1991 Oct;5(10):1870–1880. doi: 10.1101/gad.5.10.1870. [DOI] [PubMed] [Google Scholar]
- Garriga G., Lambowitz A. M. RNA splicing in neurospora mitochondria: self-splicing of a mitochondrial intron in vitro. Cell. 1984 Dec;39(3 Pt 2):631–641. doi: 10.1016/0092-8674(84)90470-7. [DOI] [PubMed] [Google Scholar]
- Gutell R. R., Schnare M. N., Gray M. W. A compilation of large subunit (23S-like) ribosomal RNA sequences presented in a secondary structure format. Nucleic Acids Res. 1990 Apr 25;18 (Suppl):2319–2330. doi: 10.1093/nar/18.suppl.2319. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hicke B. J., Christian E. L., Yarus M. Stereoselective arginine binding is a phylogenetically conserved property of group I self-splicing RNAs. EMBO J. 1989 Dec 1;8(12):3843–3851. doi: 10.1002/j.1460-2075.1989.tb08562.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Inoue T., Sullivan F. X., Cech T. R. Intermolecular exon ligation of the rRNA precursor of Tetrahymena: oligonucleotides can function as 5' exons. Cell. 1985 Dec;43(2 Pt 1):431–437. doi: 10.1016/0092-8674(85)90173-4. [DOI] [PubMed] [Google Scholar]
- Kunkel T. A., Roberts J. D., Zakour R. A. Rapid and efficient site-specific mutagenesis without phenotypic selection. Methods Enzymol. 1987;154:367–382. doi: 10.1016/0076-6879(87)54085-x. [DOI] [PubMed] [Google Scholar]
- MITTERMAYER C., BRAUN R., RUSCH H. P. THE EFFECT OF ACTINOMYCIN D ON THE TIMING OF MITOSIS IN PHYSARUM POLYCEPHALUM. Exp Cell Res. 1965 Apr;38:33–41. doi: 10.1016/0014-4827(65)90424-6. [DOI] [PubMed] [Google Scholar]
- Murphy F. L., Cech T. R. An independently folding domain of RNA tertiary structure within the Tetrahymena ribozyme. Biochemistry. 1993 May 25;32(20):5291–5300. doi: 10.1021/bi00071a003. [DOI] [PubMed] [Google Scholar]
- Muscarella D. E., Ellison E. L., Ruoff B. M., Vogt V. M. Characterization of I-Ppo, an intron-encoded endonuclease that mediates homing of a group I intron in the ribosomal DNA of Physarum polycephalum. Mol Cell Biol. 1990 Jul;10(7):3386–3396. doi: 10.1128/mcb.10.7.3386. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Muscarella D. E., Vogt V. M. A mobile group I intron in the nuclear rDNA of Physarum polycephalum. Cell. 1989 Feb 10;56(3):443–454. doi: 10.1016/0092-8674(89)90247-x. [DOI] [PubMed] [Google Scholar]
- Otsuka T., Nomiyama H., Yoshida H., Kukita T., Kuhara S., Sakaki Y. Complete nucleotide sequence of the 26S rRNA gene of Physarum polycephalum: its significance in gene evolution. Proc Natl Acad Sci U S A. 1983 Jun;80(11):3163–3167. doi: 10.1073/pnas.80.11.3163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ruoff B., Johansen S., Vogt V. M. Characterization of the self-splicing products of a mobile intron from the nuclear rDNA of Physarum polycephalum. Nucleic Acids Res. 1992 Nov 25;20(22):5899–5906. doi: 10.1093/nar/20.22.5899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Walstrum S. A., Uhlenbeck O. C. The self-splicing RNA of Tetrahymena is trapped in a less active conformation by gel purification. Biochemistry. 1990 Nov 20;29(46):10573–10576. doi: 10.1021/bi00498a022. [DOI] [PubMed] [Google Scholar]
- Williamson C. L., Tierney W. M., Kerker B. J., Burke J. M. Site-directed mutagenesis of core sequence elements 9R', 9L, 9R, and 2 in self-splicing Tetrahymena pre-rRNA. J Biol Chem. 1987 Oct 25;262(30):14672–14682. [PubMed] [Google Scholar]
- Woodson S. A., Cech T. R. Alternative secondary structures in the 5' exon affect both forward and reverse self-splicing of the Tetrahymena intervening sequence RNA. Biochemistry. 1991 Feb 26;30(8):2042–2050. doi: 10.1021/bi00222a006. [DOI] [PubMed] [Google Scholar]
- Woodson S. A., Cech T. R. Reverse self-splicing of the tetrahymena group I intron: implication for the directionality of splicing and for intron transposition. Cell. 1989 Apr 21;57(2):335–345. doi: 10.1016/0092-8674(89)90971-9. [DOI] [PubMed] [Google Scholar]
- Woodson S. A., Emerick V. L. An alternative helix in the 26S rRNA promotes excision and integration of the Tetrahymena intervening sequence. Mol Cell Biol. 1993 Feb;13(2):1137–1145. doi: 10.1128/mcb.13.2.1137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Woodson S. A. Exon sequences distant from the splice junction are required for efficient self-splicing of the Tetrahymena IVS. Nucleic Acids Res. 1992 Aug 11;20(15):4027–4032. doi: 10.1093/nar/20.15.4027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu M. Q., Shub D. A. The catalytic core of the sunY intron of bacteriophage T4. Gene. 1989 Oct 15;82(1):77–82. doi: 10.1016/0378-1119(89)90032-2. [DOI] [PubMed] [Google Scholar]
- Zaug A. J., Grabowski P. J., Cech T. R. Autocatalytic cyclization of an excised intervening sequence RNA is a cleavage-ligation reaction. Nature. 1983 Feb 17;301(5901):578–583. doi: 10.1038/301578a0. [DOI] [PubMed] [Google Scholar]
- Zaug A. J., Grosshans C. A., Cech T. R. Sequence-specific endoribonuclease activity of the Tetrahymena ribozyme: enhanced cleavage of certain oligonucleotide substrates that form mismatched ribozyme-substrate complexes. Biochemistry. 1988 Dec 13;27(25):8924–8931. doi: 10.1021/bi00425a008. [DOI] [PubMed] [Google Scholar]
- Zaug A. J., Kent J. R., Cech T. R. A labile phosphodiester bond at the ligation junction in a circular intervening sequence RNA. Science. 1984 May 11;224(4649):574–578. doi: 10.1126/science.6200938. [DOI] [PubMed] [Google Scholar]
- Zaug A. J., McEvoy M. M., Cech T. R. Self-splicing of the group I intron from Anabaena pre-tRNA: requirement for base-pairing of the exons in the anticodon stem. Biochemistry. 1993 Aug 10;32(31):7946–7953. doi: 10.1021/bi00082a016. [DOI] [PubMed] [Google Scholar]