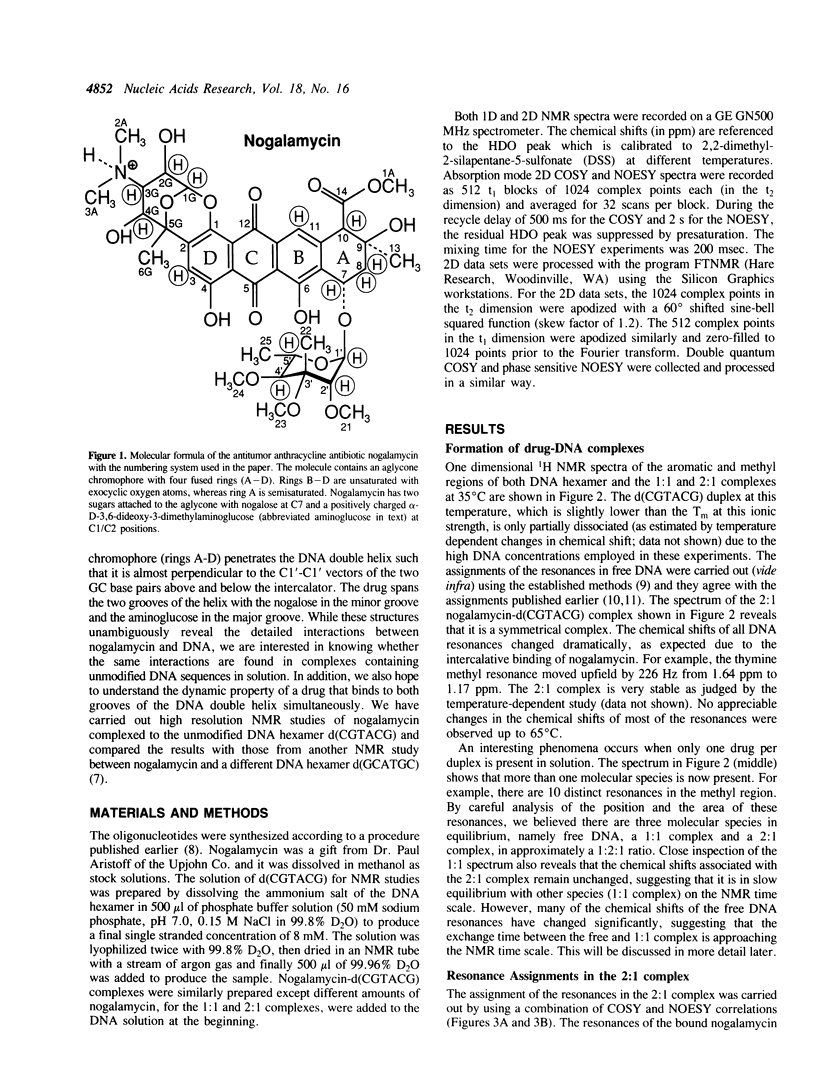
Abstract
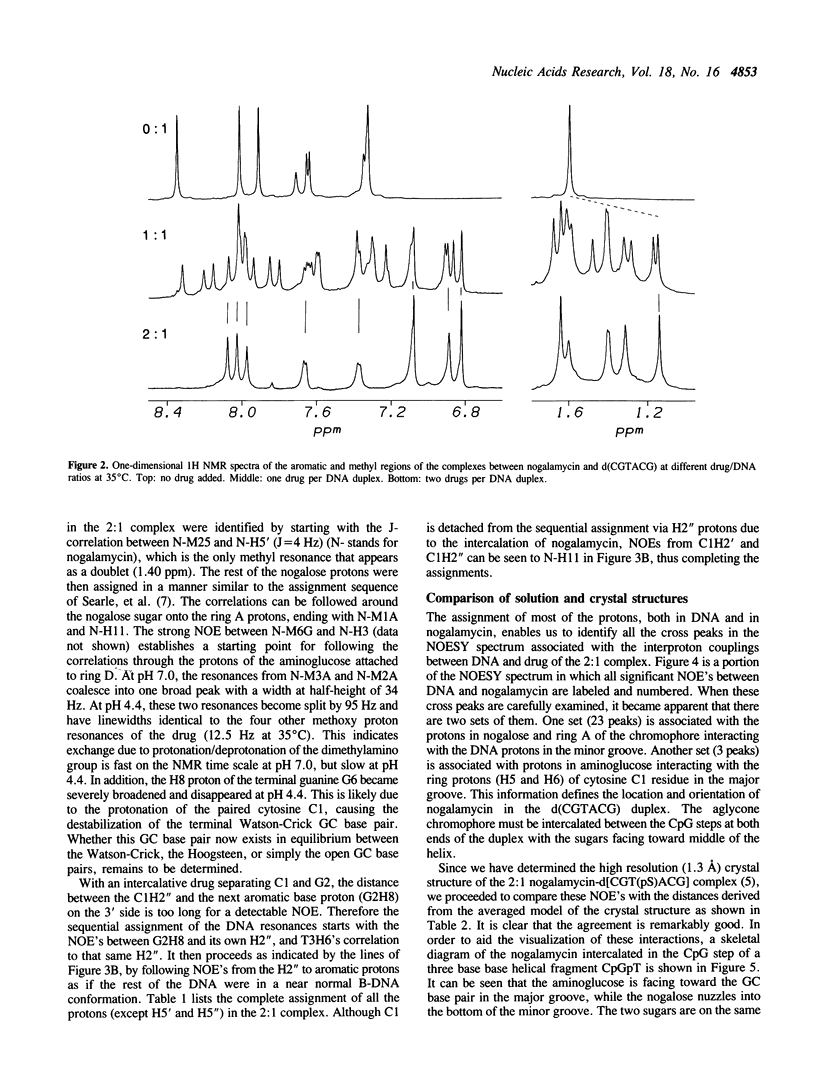
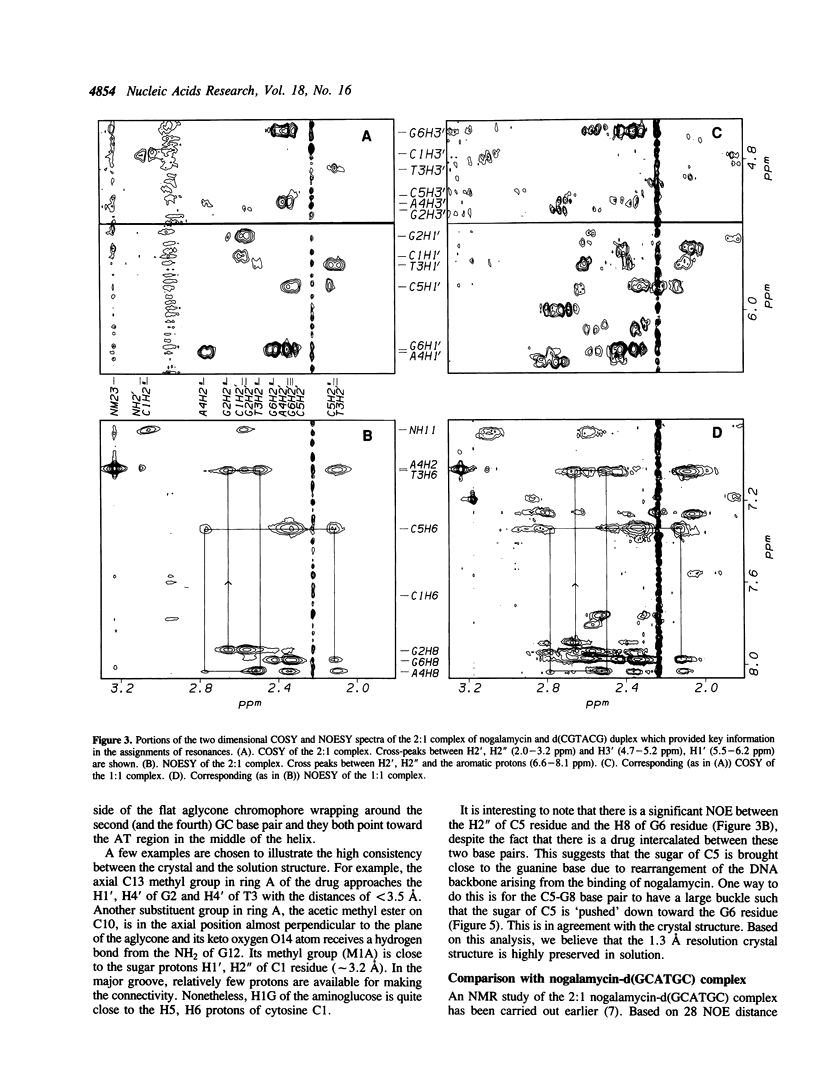
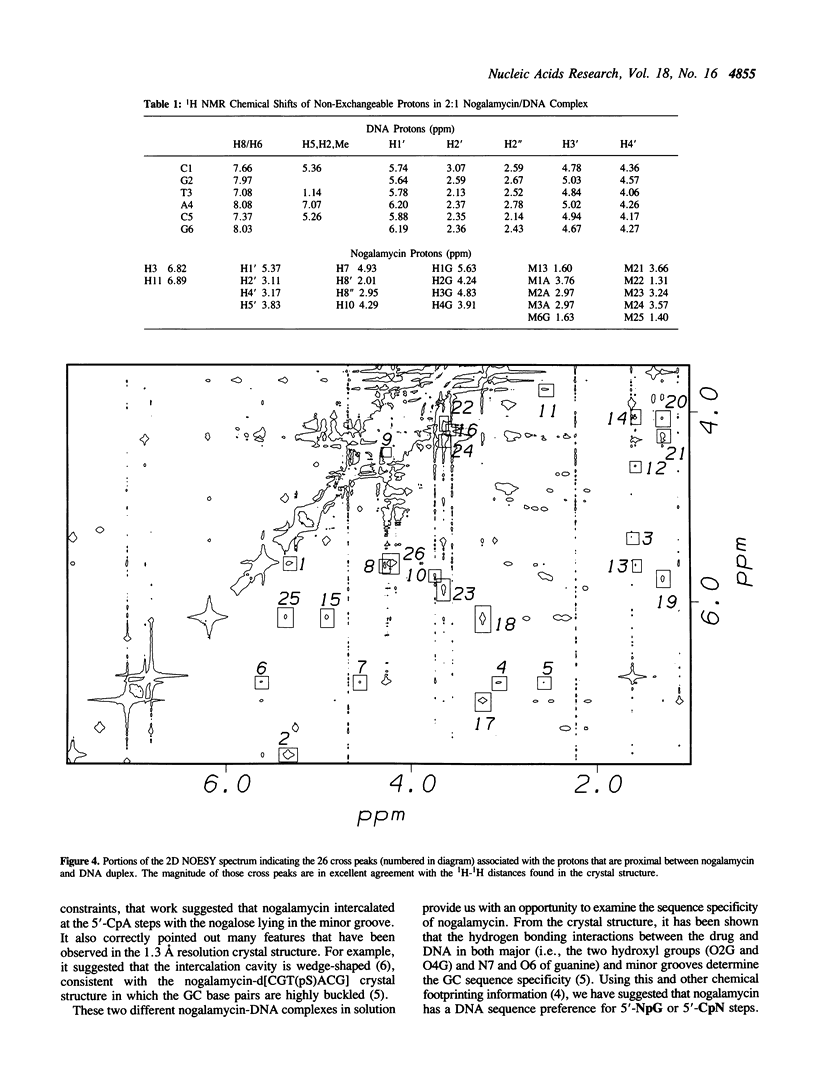
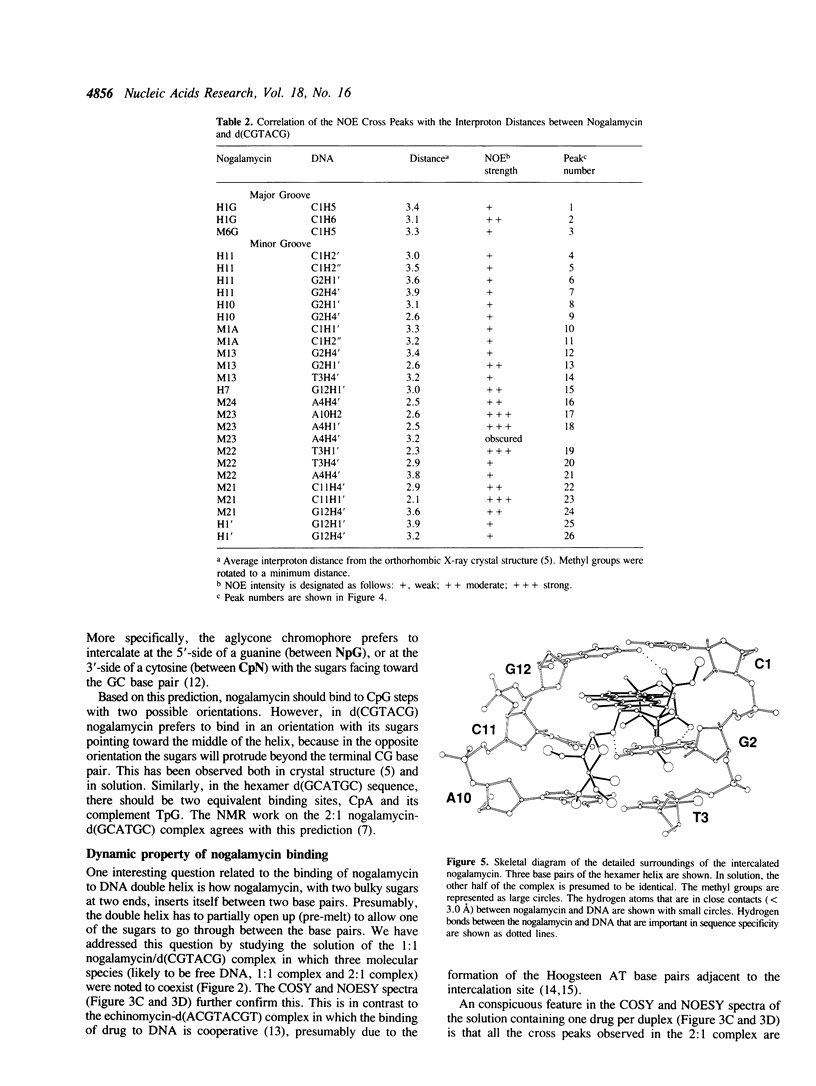
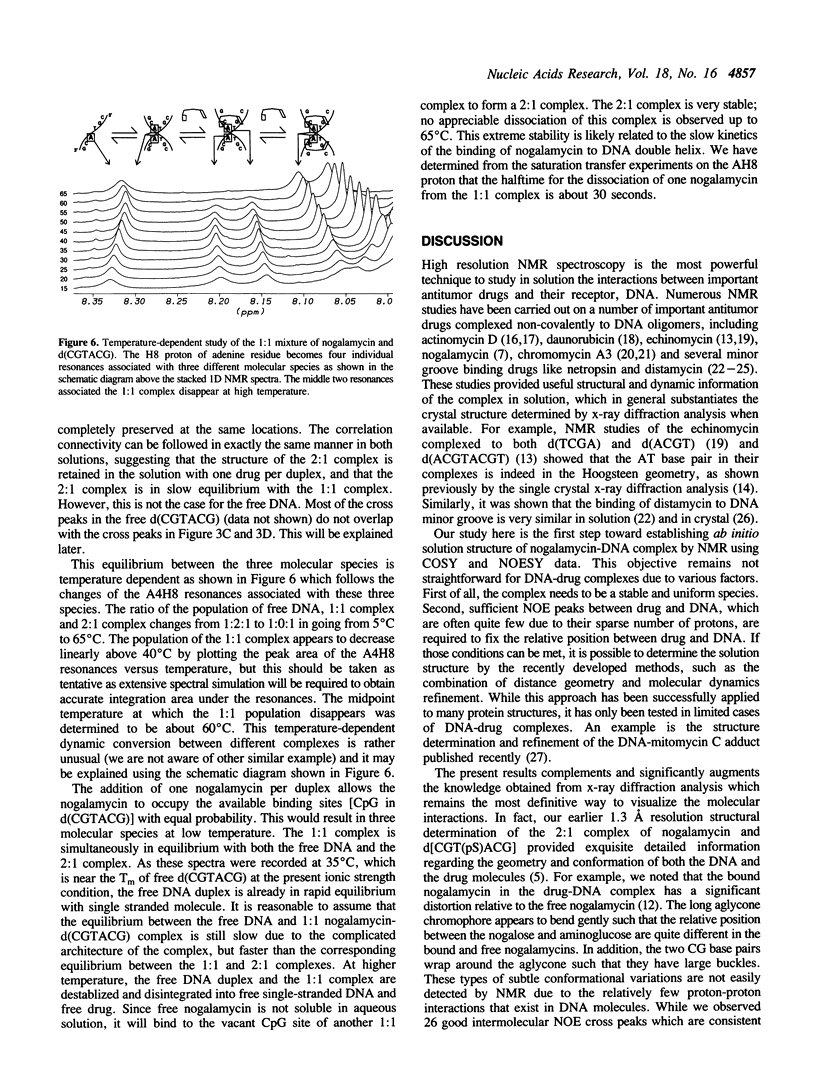
The interactions between a novel antitumor drug nogalamycin with the self-complementary DNA hexamer d(CGTACG) have been studied by 500 MHz two dimensional proton nuclear magnetic resonance spectroscopy. When two nogalamycins are mixed with the DNA hexamer duplex in a 2:1 ratio, a symmetrical complex is formed. All non-exchangeable proton resonances (except H5' & H5") of this complex have been assigned using 2D-COSY and 2D-NOESY methods at pH 7.0. The observed NOE cross peaks are fully consistent with the 1.3 A resolution x-ray crystal structure (Liaw et al., Biochemistry 28, 9913-9918, 1989) in which the elongated aglycone chromophore is intercalated between the CpG steps at both ends of the helix. The aglycone chromophore spans across the GC Watson-Crick base pairs with its nogalose lying in the minor groove and the aminoglucose lying in the major groove of the distorted B-DNA double helix. The binding conformation suggests that specific hydrogen bonds exist in the complex between the drug and guanine-cytosine bases in both grooves of the helix. When only one drug per DNA duplex is present in solution, there are three molecular species (free DNA, 1:1 complex and 2:1 complex) in slow exchange on the NMR time scale. This equilibrium is temperature dependent. At high temperature the free DNA hexamer duplex and the 1:1 complex are completely destabilized such that at 65 degrees C only free single-stranded DNA and the 2:1 complex co-exist. At 35 degrees C the equilibrium between free DNA and the 1:1 complex is relatively fast, while that between the 1:1 complex and the 2:1 complex is slow. This may be rationalized by the fact that the binding of nogalamycin to DNA requires that the base pairs in DNA open up transiently to allow the bulky sugars to go through. A separate study of the 2:1 complex at low pH showed that the terminal GC base pair is destabilized.
Full text
PDF

Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Adams W. J., McGovren J. P., Dalm E. A., Brewer J. E., Hosley J. D. Pharmacokinetics and systemic bioavailability of menogaril, an anthracycline antitumor agent, in the mouse, dog, and monkey. Cancer Res. 1989 Nov 15;49(22):6328–6336. [PubMed] [Google Scholar]
- Banville D. L., Keniry M. A., Kam M., Shafer R. H. NMR studies of the interaction of chromomycin A3 with small DNA duplexes. Binding to GC-containing sequences. Biochemistry. 1990 Jul 10;29(27):6521–6534. doi: 10.1021/bi00479a026. [DOI] [PubMed] [Google Scholar]
- Bhuyan B. K., Reusser F. Comparative biological activity of nogalamycin and its analogs. Cancer Res. 1970 Apr;30(4):984–989. [PubMed] [Google Scholar]
- Chaires J. B., Fox K. R., Herrera J. E., Britt M., Waring M. J. Site and sequence specificity of the daunomycin-DNA interaction. Biochemistry. 1987 Dec 15;26(25):8227–8236. doi: 10.1021/bi00399a031. [DOI] [PubMed] [Google Scholar]
- Fox K. R., Waring M. J. Nucleotide sequence binding preferences of nogalamycin investigated by DNase I footprinting. Biochemistry. 1986 Jul 29;25(15):4349–4356. doi: 10.1021/bi00363a026. [DOI] [PubMed] [Google Scholar]
- Gao X. L., Patel D. J. NMR studies of echinomycin bisintercalation complexes with d(A1-C2-G3-T4) and d(T1-C2-G3-A4) duplexes in aqueous solution: sequence-dependent formation of Hoogsteen A1.T4 and Watson--Crick T1.A4 base pairs flanking the bisintercalation site. Biochemistry. 1988 Mar 8;27(5):1744–1751. doi: 10.1021/bi00405a054. [DOI] [PubMed] [Google Scholar]
- Gao X. L., Patel D. J. Solution structure of the chromomycin-DNA complex. Biochemistry. 1989 Jan 24;28(2):751–762. doi: 10.1021/bi00428a051. [DOI] [PubMed] [Google Scholar]
- Gilbert D. E., van der Marel G. A., van Boom J. H., Feigon J. Unstable Hoogsteen base pairs adjacent to echinomycin binding sites within a DNA duplex. Proc Natl Acad Sci U S A. 1989 May;86(9):3006–3010. doi: 10.1073/pnas.86.9.3006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gronenborn A. M., Clore G. M., Kimber B. J. An investigation into the solution structures of two self-complementary DNA oligomers, 5'-d(C-G-T-A-C-G) and 5'-d(A-C-G-C-G-C-G-T), by means of nuclear-Overhauser-enhancement measurements. Biochem J. 1984 Aug 1;221(3):723–736. doi: 10.1042/bj2210723. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hare D. R., Wemmer D. E., Chou S. H., Drobny G., Reid B. R. Assignment of the non-exchangeable proton resonances of d(C-G-C-G-A-A-T-T-C-G-C-G) using two-dimensional nuclear magnetic resonance methods. J Mol Biol. 1983 Dec 15;171(3):319–336. doi: 10.1016/0022-2836(83)90096-7. [DOI] [PubMed] [Google Scholar]
- Jones R. L., Scott E. V., Zon G., Marzilli L. G., Wilson W. D. An NMR investigation of the binding of the anticancer drug actinomycin D to oligodeoxyribonucleotides with isolated 5'd(GC)3' binding sites. Biochemistry. 1988 Aug 9;27(16):6021–6026. doi: 10.1021/bi00416a029. [DOI] [PubMed] [Google Scholar]
- Lane A. N., Forster M. J. Determination of internal dynamics of deoxyriboses in the DNA hexamer d(CGTACG)2 by 1H NMR. Eur Biophys J. 1989;17(4):221–232. doi: 10.1007/BF00284729. [DOI] [PubMed] [Google Scholar]
- Leupin W., Chazin W. J., Hyberts S., Denny W. A., Wüthrich K. NMR studies of the complex between the decadeoxynucleotide d-(GCATTAATGC)2 and a minor-groove-binding drug. Biochemistry. 1986 Oct 7;25(20):5902–5910. doi: 10.1021/bi00368a010. [DOI] [PubMed] [Google Scholar]
- Liaw Y. C., Gao Y. G., Robinson H., van der Marel G. A., van Boom J. H., Wang A. H. Antitumor drug nogalamycin binds DNA in both grooves simultaneously: molecular structure of nogalamycin-DNA complex. Biochemistry. 1989 Dec 26;28(26):9913–9918. doi: 10.1021/bi00452a006. [DOI] [PubMed] [Google Scholar]
- Norman D., Live D., Sastry M., Lipman R., Hingerty B. E., Tomasz M., Broyde S., Patel D. J. NMR and computational characterization of mitomycin cross-linked to adjacent deoxyguanosines in the minor groove of the d(T-A-C-G-T-A).d(T-A-C-G-T-A) duplex. Biochemistry. 1990 Mar 20;29(11):2861–2875. doi: 10.1021/bi00463a032. [DOI] [PubMed] [Google Scholar]
- Patel D. J., Shapiro L. Molecular recognition in noncovalent antitumor agent-DNA complexes: NMR studies of the base and sequence dependent recognition of the DNA minor groove by netropsin. Biochimie. 1985 Jul-Aug;67(7-8):887–915. doi: 10.1016/s0300-9084(85)80181-4. [DOI] [PubMed] [Google Scholar]
- Pelton J. G., Wemmer D. E. Structural modeling of the distamycin A-d(CGCGAATTCGCG)2 complex using 2D NMR and molecular mechanics. Biochemistry. 1988 Oct 18;27(21):8088–8096. doi: 10.1021/bi00421a018. [DOI] [PubMed] [Google Scholar]
- Searle M. S., Hall J. G., Denny W. A., Wakelin L. P. NMR studies of the interaction of the antibiotic nogalamycin with the hexadeoxyribonucleotide duplex d(5'-GCATGC)2. Biochemistry. 1988 Jun 14;27(12):4340–4349. doi: 10.1021/bi00412a022. [DOI] [PubMed] [Google Scholar]
- Ughetto G., Wang A. H., Quigley G. J., van der Marel G. A., van Boom J. H., Rich A. A comparison of the structure of echinomycin and triostin A complexed to a DNA fragment. Nucleic Acids Res. 1985 Apr 11;13(7):2305–2323. doi: 10.1093/nar/13.7.2305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang A. H., Cottens S., Dervan P. B., Yesinowski J. P., van der Marel G. A., van Boom J. H. Interactions between a symmetrical minor groove binding compound and DNA oligonucleotides: 1H and 19F NMR studies. J Biomol Struct Dyn. 1989 Aug;7(1):101–117. doi: 10.1080/07391102.1989.10507754. [DOI] [PubMed] [Google Scholar]
- Wang A. H., Ughetto G., Quigley G. J., Hakoshima T., van der Marel G. A., van Boom J. H., Rich A. The molecular structure of a DNA-triostin A complex. Science. 1984 Sep 14;225(4667):1115–1121. doi: 10.1126/science.6474168. [DOI] [PubMed] [Google Scholar]
- Wang A. H., Ughetto G., Quigley G. J., Rich A. Interactions of quinoxaline antibiotic and DNA: the molecular structure of a triostin A-d(GCGTACGC) complex. J Biomol Struct Dyn. 1986 Dec;4(3):319–342. doi: 10.1080/07391102.1986.10506353. [DOI] [PubMed] [Google Scholar]
- Wiley P. F. Improved antitumor activity by modification of nogalamycin. J Nat Prod. 1979 Nov-Dec;42(6):569–582. doi: 10.1021/np50006a002. [DOI] [PubMed] [Google Scholar]
- Williams L. D., Egli M., Qi G., Bash P., van der Marel G. A., van Boom J. H., Rich A., Frederick C. A. Structure of nogalamycin bound to a DNA hexamer. Proc Natl Acad Sci U S A. 1990 Mar;87(6):2225–2229. doi: 10.1073/pnas.87.6.2225. [DOI] [PMC free article] [PubMed] [Google Scholar]


