Abstract
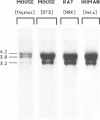
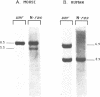
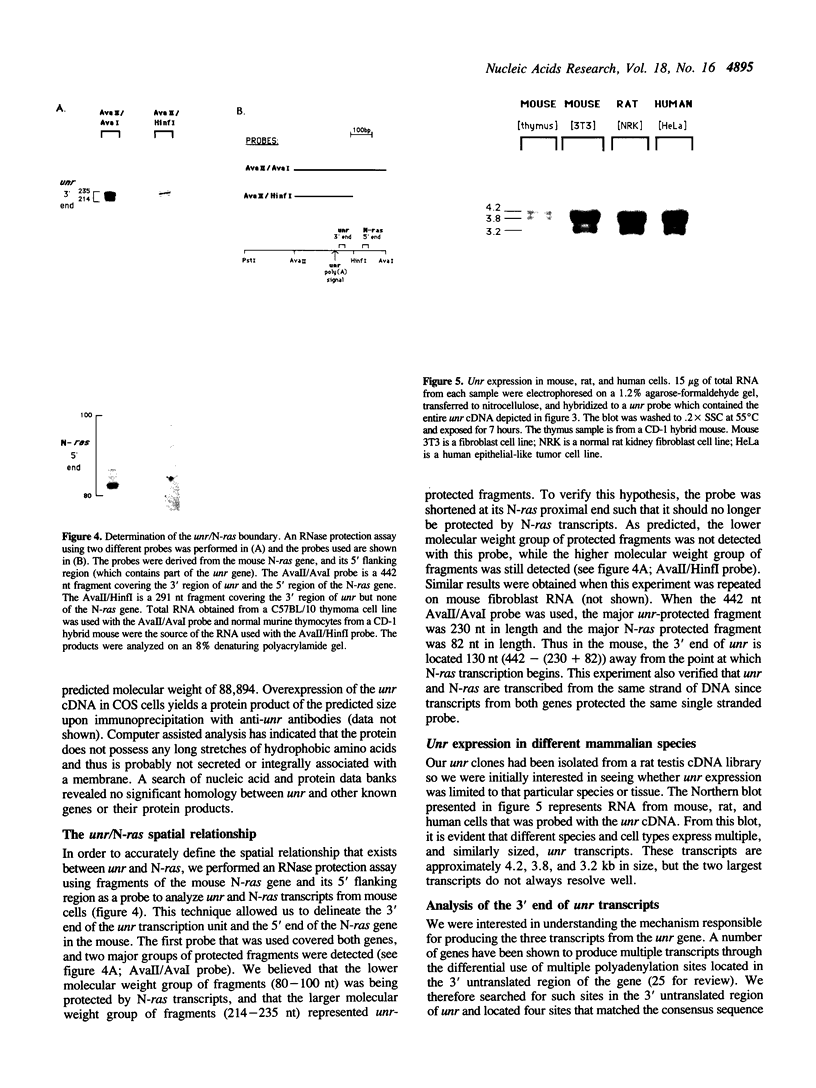
The mammalian N-ras gene is believed to play a role in cellular proliferation, differentiation, and transformation. While investigating N-ras, we isolated cDNA's that originate from a closely linked upstream gene. RNase protection assays reveal that this gene, unr, is transcribed in the same direction as N-ras and that its 3' end is located just 130 base pairs away from the point at which N-ras transcription begins. The close spatial relationship between the two genes is conserved in all species from which the N-ras gene has been isolated. An open reading frame, potentially encoding a 798 amino acid protein, is contained within the unr cDNA. Neither the primary protein structure nor the nucleic acid sequence of unr is homologous to any other known gene, including N-ras. Unr transcripts are detected in mouse, rat and human cells, and Southern analysis indicates that the unr locus found immediately upstream of the N-ras gene is transcriptionaly active in the mouse since only a single copy of unr is detected in this species. Unr produces multiple transcripts that differ in their 3' ends and are apparently created through the differential use of multiple polyadenylation sites located in the 3' untranslated region of the gene. Both unr and N-ras are expressed in all tissues examined. In the testis, both genes are developmentally regulated, with an increase in expression occurring upon testicular maturation. Thus the two genes may be coordinately regulated, at least in certain circumstances. Our findings suggest that a thorough analysis of the relationship that exists between the two genes could potentially provide insights into the regulation and/or function of N-ras.
Full text
PDF






Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Barbacid M. ras genes. Annu Rev Biochem. 1987;56:779–827. doi: 10.1146/annurev.bi.56.070187.004023. [DOI] [PubMed] [Google Scholar]
- Bray P. F., Barsh G., Rosa J. P., Luo X. Y., Magenis E., Shuman M. A. Physical linkage of the genes for platelet membrane glycoproteins IIb and IIIa. Proc Natl Acad Sci U S A. 1988 Nov;85(22):8683–8687. doi: 10.1073/pnas.85.22.8683. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brown R., Marshall C. J., Pennie S. G., Hall A. Mechanism of activation of an N-ras gene in the human fibrosarcoma cell line HT1080. EMBO J. 1984 Jun;3(6):1321–1326. doi: 10.1002/j.1460-2075.1984.tb01970.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chang E. H., Furth M. E., Scolnick E. M., Lowy D. R. Tumorigenic transformation of mammalian cells induced by a normal human gene homologous to the oncogene of Harvey murine sarcoma virus. Nature. 1982 Jun 10;297(5866):479–483. doi: 10.1038/297479a0. [DOI] [PubMed] [Google Scholar]
- Chang H. Y., Guerrero I., Lake R., Pellicer A., D'Eustachio P. Mouse N-ras genes: organization of the functional locus and of a truncated cDNA-like pseudogene. Oncogene Res. 1987 Jul;1(2):129–136. [PubMed] [Google Scholar]
- Chang S. C., Erwin A. E., Lee A. S. Glucose-regulated protein (GRP94 and GRP78) genes share common regulatory domains and are coordinately regulated by common trans-acting factors. Mol Cell Biol. 1989 May;9(5):2153–2162. doi: 10.1128/mcb.9.5.2153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chirgwin J. M., Przybyla A. E., MacDonald R. J., Rutter W. J. Isolation of biologically active ribonucleic acid from sources enriched in ribonuclease. Biochemistry. 1979 Nov 27;18(24):5294–5299. doi: 10.1021/bi00591a005. [DOI] [PubMed] [Google Scholar]
- Chomczynski P., Sacchi N. Single-step method of RNA isolation by acid guanidinium thiocyanate-phenol-chloroform extraction. Anal Biochem. 1987 Apr;162(1):156–159. doi: 10.1006/abio.1987.9999. [DOI] [PubMed] [Google Scholar]
- Doniger J., DiPaolo J. A. Coordinate N-ras mRNA up-regulation with mutational activation in tumorigenic guinea pig cells. Nucleic Acids Res. 1988 Feb 11;16(3):969–980. doi: 10.1093/nar/16.3.969. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Doniger J. Differential conservation of non-coding regions within human and guinea pig N-ras genes. Oncogene. 1987;1(3):331–334. [PubMed] [Google Scholar]
- Gilman A. G. G proteins and dual control of adenylate cyclase. Cell. 1984 Mar;36(3):577–579. doi: 10.1016/0092-8674(84)90336-2. [DOI] [PubMed] [Google Scholar]
- Grosveld F., van Assendelft G. B., Greaves D. R., Kollias G. Position-independent, high-level expression of the human beta-globin gene in transgenic mice. Cell. 1987 Dec 24;51(6):975–985. doi: 10.1016/0092-8674(87)90584-8. [DOI] [PubMed] [Google Scholar]
- Hall A., Brown R. Human N-ras: cDNA cloning and gene structure. Nucleic Acids Res. 1985 Jul 25;13(14):5255–5268. doi: 10.1093/nar/13.14.5255. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ishii S., Merlino G. T., Pastan I. Promoter region of the human Harvey ras proto-oncogene: similarity to the EGF receptor proto-oncogene promoter. Science. 1985 Dec 20;230(4732):1378–1381. doi: 10.1126/science.2999983. [DOI] [PubMed] [Google Scholar]
- Ishii S., Xu Y. H., Stratton R. H., Roe B. A., Merlino G. T., Pastan I. Characterization and sequence of the promoter region of the human epidermal growth factor receptor gene. Proc Natl Acad Sci U S A. 1985 Aug;82(15):4920–4924. doi: 10.1073/pnas.82.15.4920. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kozak M. The scanning model for translation: an update. J Cell Biol. 1989 Feb;108(2):229–241. doi: 10.1083/jcb.108.2.229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kyte J., Doolittle R. F. A simple method for displaying the hydropathic character of a protein. J Mol Biol. 1982 May 5;157(1):105–132. doi: 10.1016/0022-2836(82)90515-0. [DOI] [PubMed] [Google Scholar]
- Leff S. E., Rosenfeld M. G., Evans R. M. Complex transcriptional units: diversity in gene expression by alternative RNA processing. Annu Rev Biochem. 1986;55:1091–1117. doi: 10.1146/annurev.bi.55.070186.005303. [DOI] [PubMed] [Google Scholar]
- Leon J., Guerrero I., Pellicer A. Differential expression of the ras gene family in mice. Mol Cell Biol. 1987 Apr;7(4):1535–1540. doi: 10.1128/mcb.7.4.1535. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Manley J. L. Polyadenylation of mRNA precursors. Biochim Biophys Acta. 1988 May 6;950(1):1–12. doi: 10.1016/0167-4781(88)90067-x. [DOI] [PubMed] [Google Scholar]
- Marie J., Simon M. P., Lone Y. C., Cognet M., Kahn A. Tissue-specific heterogeneity of the 3'-untranslated region of L-type pyruvate kinase mRNAs. Eur J Biochem. 1986 Jul 1;158(1):33–41. doi: 10.1111/j.1432-1033.1986.tb09717.x. [DOI] [PubMed] [Google Scholar]
- McKay I. A., Marshall C. J., Calés C., Hall A. Transformation and stimulation of DNA synthesis in NIH-3T3 cells are a titratable function of normal p21N-ras expression. EMBO J. 1986 Oct;5(10):2617–2621. doi: 10.1002/j.1460-2075.1986.tb04542.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Melton D. A., Krieg P. A., Rebagliati M. R., Maniatis T., Zinn K., Green M. R. Efficient in vitro synthesis of biologically active RNA and RNA hybridization probes from plasmids containing a bacteriophage SP6 promoter. Nucleic Acids Res. 1984 Sep 25;12(18):7035–7056. doi: 10.1093/nar/12.18.7035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ponzetto C., Wolgemuth D. J. Haploid expression of a unique c-abl transcript in the mouse male germ line. Mol Cell Biol. 1985 Jul;5(7):1791–1794. doi: 10.1128/mcb.5.7.1791. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Povey S., Morton N. E., Sherman S. L. Report of the Committee on the Genetic Constitution of Chromosomes 1 and 2. Cytogenet Cell Genet. 1985;40(1-4):67–106. doi: 10.1159/000132170. [DOI] [PubMed] [Google Scholar]
- Povey S., Parrington J. M. Chromosome 1 in relation to human disease. J Med Genet. 1986 Apr;23(2):107–115. doi: 10.1136/jmg.23.2.107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Proudfoot N. J. How RNA polymerase II terminates transcription in higher eukaryotes. Trends Biochem Sci. 1989 Mar;14(3):105–110. doi: 10.1016/0968-0004(89)90132-1. [DOI] [PubMed] [Google Scholar]
- Proudfoot N. J. Transcriptional interference and termination between duplicated alpha-globin gene constructs suggests a novel mechanism for gene regulation. Nature. 1986 Aug 7;322(6079):562–565. doi: 10.1038/322562a0. [DOI] [PubMed] [Google Scholar]
- Reynolds G. A., Goldstein J. L., Brown M. S. Multiple mRNAs for 3-hydroxy-3-methylglutaryl coenzyme A reductase determined by multiple transcription initiation sites and intron splicing sites in the 5'-untranslated region. J Biol Chem. 1985 Aug 25;260(18):10369–10377. [PubMed] [Google Scholar]
- Sanger F., Nicklen S., Coulson A. R. DNA sequencing with chain-terminating inhibitors. Proc Natl Acad Sci U S A. 1977 Dec;74(12):5463–5467. doi: 10.1073/pnas.74.12.5463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Santos E., Nebreda A. R. Structural and functional properties of ras proteins. FASEB J. 1989 Aug;3(10):2151–2163. doi: 10.1096/fasebj.3.10.2666231. [DOI] [PubMed] [Google Scholar]
- Sorrentino V., McKinney M. D., Giorgi M., Geremia R., Fleissner E. Expression of cellular protooncogenes in the mouse male germ line: a distinctive 2.4-kilobase pim-1 transcript is expressed in haploid postmeiotic cells. Proc Natl Acad Sci U S A. 1988 Apr;85(7):2191–2195. doi: 10.1073/pnas.85.7.2191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Southern E. M. Detection of specific sequences among DNA fragments separated by gel electrophoresis. J Mol Biol. 1975 Nov 5;98(3):503–517. doi: 10.1016/s0022-2836(75)80083-0. [DOI] [PubMed] [Google Scholar]
- Sweet R. W., Yokoyama S., Kamata T., Feramisco J. R., Rosenberg M., Gross M. The product of ras is a GTPase and the T24 oncogenic mutant is deficient in this activity. Nature. 1984 Sep 20;311(5983):273–275. doi: 10.1038/311273a0. [DOI] [PubMed] [Google Scholar]
- Tollefsen S. E., Sadow J. L., Rotwein P. Coordinate expression of insulin-like growth factor II and its receptor during muscle differentiation. Proc Natl Acad Sci U S A. 1989 Mar;86(5):1543–1547. doi: 10.1073/pnas.86.5.1543. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Willingham M. C., Pastan I., Shih T. Y., Scolnick E. M. Localization of the src gene product of the Harvey strain of MSV to plasma membrane of transformed cells by electron microscopic immunocytochemistry. Cell. 1980 Apr;19(4):1005–1014. doi: 10.1016/0092-8674(80)90091-4. [DOI] [PubMed] [Google Scholar]
- Wu L. C., Morley B. J., Campbell R. D. Cell-specific expression of the human complement protein factor B gene: evidence for the role of two distinct 5'-flanking elements. Cell. 1987 Jan 30;48(2):331–342. doi: 10.1016/0092-8674(87)90436-3. [DOI] [PubMed] [Google Scholar]
- Yamamoto F., Perucho M. Characterization of the human c-K-ras gene promoter. Oncogene Res. 1988 Sep;3(2):125–130. [PubMed] [Google Scholar]
- von Heijne G. A new method for predicting signal sequence cleavage sites. Nucleic Acids Res. 1986 Jun 11;14(11):4683–4690. doi: 10.1093/nar/14.11.4683. [DOI] [PMC free article] [PubMed] [Google Scholar]