Abstract
The purpose of this review is to explore the feasibility of bioaerosol fingerprinting based on current understanding of cellular debris (with an emphasis on human-emitted particulates) in aerosols and arguments regarding sampling, sensitivity, separations and detection schemes. Target aerosol particles include cellular material and proteins emitted by humans, animals and plants and can be considered information- rich packets that carry biochemical information specific to the living organisms present in the collection settings. In this work we discuss sampling and analysis techniques that can be integrated with molecular (e.g. protein) detection protocols to properly assess the aerosolized cellular material of interest. Developing a detailed understanding of bioaerosol molecular profiles in various environments suggests exciting possibilities of bioaerosol analysis with applications ranging from military defense to medical diagnosis and wildlife identification.
Keywords: bioaerosols, skin debris, fingerprinting, identification
1. Introduction
Bioaerosols are diverse and complex dispersed particles that are either living or of biological origin. These include viruses, pollen, fungal spores, bacteria, and debris from vertebrates, including humans, and other biota (plants, insects, etc). These particles range from ~10 nm to 100 μm [1], and their existence has been recognized for well over a century [2]. Currently, the central topics of bioaerosol studies focus on health hazards, effects on the atmosphere, terrorism detection, and global climate.
Over the past decade, several studies focused on molecular and isotopic markers that can be used to track bioaerosol; specially for tracing particles released from soils and various agricultural environments [3–5]. Molecular marker studies have mainly focused on organic marker compounds such as saccharides, alkanes and steroids for the tracing of soil dust and plant bioaerosols [4, 5]. Beyond these studies, viewing animal and human bioaerosols as an information-rich marker of its source has not been seriously considered. Reasons for this absence are lack of sufficient (bio) analytical capabilities and poor understanding of the biochemical fingerprints that are likely present in this type of debris. Considering the body of evidence that does exist indicating abundant cellular material and proteins in the atmosphere [6–8], this is somewhat surprising—limited analytical capabilities notwithstanding. Even though there is imperfect knowledge of “dead” and fragmented biological fraction of particles in the atmosphere, the mere existence of this type of debris creates an opportunity for its use in many potential applications. Living organisms, including humans, constantly emit a surprisingly large amount of dead skin cells and fragments into the environment [9]. As analytical capabilities are improved and focused on the characterization of this fraction (and the living fraction), detailed biochemical information within the aerosol will be identified. This has the potential to significantly impact fields ranging from biochemical forensics and biodiversity studies to medical profiling and environmental studies.
Accurately characterizing bioaerosol to differentiate its source appears to be fighting against the basic concept that many biological structures and metabolic pathways are common to all humans (and many other species) resulting in apparently common biochemical profiles. However, and importantly, structures and pathways exhibit the extensive polymorphisms and divergent post-translational modifications (PTMs) that reflect individual genetics, familial and tribal background, personal history, living environment and health. This variability yields an array of biochemical fingerprints indicative of these polymorphisms and modifications. Analogous to facial-recognitions and more recent technologies such as “odorprint,” the idea is not to rely on a specific feature (or compound) for recognition, but to obtain multi-component signatures that reflect individuality [10]. Furthermore, it is possible that the concentration, degradation, and type of aerosolized material of interest is dependent on time from human presence, and potentially this information can be used to time-stamp human and individual occupancy. By exploiting the capabilities of current and emerging analytical technology it is likely that interpretable patterns can be generated.
The purpose of this review is to discuss the role of bioaerosols, with a focus on human-origin bioparticles, as carriers of biochemical information. Additionally, we discuss analysis needs and challenges based on the current state of knowledge for the study of biological aerosols. This work is not intended to provide an exhaustive review of previous studies on aerosol and bioaerosols, but instead, to provide examples of how analytical chemistry performed in the field and in the laboratory can shed new light in our understanding and analysis of unexploited biochemical fingerprints contained in aerosolized human debris.
2. Sources of Molecular “Signatures” in Bioaerosols
The concept of detecting unique molecular signatures from a biological source is predicated on the premise that a unique pattern exists—whether it can be detected or not. Several lines of reasoning and sources of information suggest that, indeed, this signature is generated and released to our surroundings. A variety of biochemical processes are distinctive to each individual and enough material is released into the environment to potentially be detectable. Skin cells are jettisoned in gram quantities daily and remain suspended as an aerosol for hours to days [9]. Presumably, each of these skin cells retains a biochemical fingerprint of the originator. Here, we review current knowledge about the sources of molecular signatures that can most likely be found in skin debris.
2.1 Skin Cells: Sloughing Processes and Biochemical Signature
Aerosolized skin cells, along with other forms of animal debris (e.g. dander), are a type of non-viable bioaerosol. They are generated from viable organisms then released into the air spontaneously, as a consequence of environmental conditions, or some other mechanical disturbance. The existence of aerosolized skin in airborne particulate matter has been recognized for more than three decades [11, 12], and it continues to be investigated and better understood in more recent years [8, 13].
Skin flakes comprise a substantial proportion of the recognizable particles of indoor air. This type of debris is a also major constituent of house dust which is constantly re-aerosolized allowing the skin flakes to re-circulate in the air mass [13]. Bahadori and coworkers reported mean concentrations of such dust in the breathing zone (44 ± 3μgm−3) is more than twice that in the ambient air [14]. Popular culture notes the large amount of sloughed cells with an urban myth that suggests bed mattresses double in mass over ten years from dead skin cells and dust mites, although more accurately the true attributed weight increase is approximately 20% [15]. The role these particles play has also been a major concern in the sick building syndrome [16].
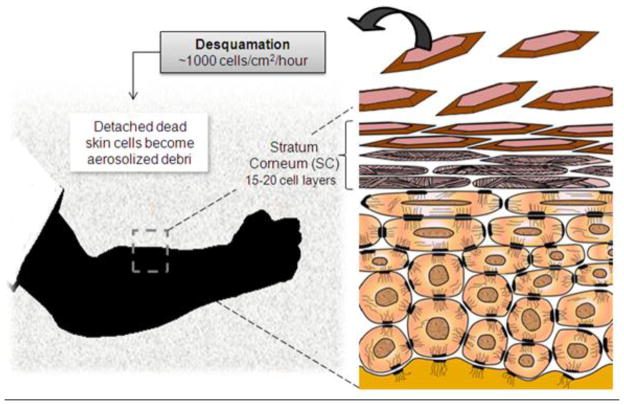
Aerosolized skin cells are the result of the continuous regeneration of the epidermis. This structure is the external, uppermost multilayer compartment of the skin where cornification (or keratinization, culminating in cell death) occurs resulting in spontaneous detachment (desquamation) of corneocytes [9,17–19]. The cornification process is the highly organized differentiation of keratinocytes going from a proliferating cell type in the basal layer of the epidermis to an association of flattened, corneocytes in the outermost layer (stratum corneum, SC) [17]. The SC consists of approximately fifteen layers from which cells are continuously discharged into the environment (Figure 2). The released corneocytes are dead cells, but form the physical layer that protects the skin.
Figure 2.
Diagram of the epidermal desquamation process [9]. Humans shed approximately billions of cells per day. Each skin flake is the product of a program of differentiation that ends on easily-detachable corneocytes. As desquamation occurs, dead skin cells settle slowly in the air providing an unexploited opportunity to obtain biochemical information unique to their source from aerosolized samples.
Through the desquamation process, a single human sheds approximately one gram of aerosolized skin flakes daily, releasing an estimated 107 particles per person per day [13, 20]. The average size of these particles is much smaller than the interweave pores of the majority of clothing fabrics allowing the skin flakes to move freely through clothing and be released into the airstreams [21, 22]. Earlier studies described the majority of particles circulating in the air as small (less than 1–2 μm in diameter), while the majority of skin cells freshly emitted by humans are larger with diameters of 5–15 μm [11, 23]. Each shed skin flake contains a complex mixture of proteins, lipids, small peptides and other biomolecules that is characteristic of its specific source [13].
The product of epidermal desquamation as a source of biochemical information has been largely ignored. Furthermore, aerosolized skin cells have not been considered signature carriers of the individual from which they originate. As a result, there has been little interest in the biomolecules associated with human skin (as well as animals)—how these molecules differ with age, conditions and identity of the source, and their evolution post-desquamation. Some studies have addressed ethnic differences in the desquamation process, although, results remain inconclusive and the majority of skin properties studied (e.g. water content, pH gradients) are not applicable to the analysis of aerosolized skin cells [24]. Nevertheless, examples of biomolecular profiling from shed material (e.g. bulk amino acid of detached feathers for bird speciation [25]) demonstrate the information that can be gleaned through detached bioparticles.
A trivial argument can be made that unique biomolecular systems result in unique biophysical structures and these structures can be used for fingerprinting purposes. This is the underlying assumption to many microscopic and histological strategies. In terms of an analytical approach to extract the necessary molecular information, the structure of corneocytes alone may initially be used to categorize the source. Skin cells obtained by stripping methods (removing layers of cells in adhesive coated tape), as well other traditional techniques (e.g. detergent scrubs), show that the geometry of corneocytes can be correlated with the type of skin epidermis [26, 27], its age [28, 29], and its health [30]. These correlations are for dermatological treatment [28, 29, 31] and their applicability to the proposed system of bioaerosol fingerprinting is not known. Other studies show that women shed larger skin cells than men, and that size increases with age [26]. Some studies have addressed environmental effects (e.g. solar exposure) [32] on corneocytes. However, many inconsistencies exist among the reports [9]. Further, there are no studies of corneocyte geometry changes from cells collected from air samples. The extent to which corneocyte geometry assessment will be valuable to bioaerosol fingerprinting is unknown, but it does provide a valuable line of inquiry.
2.2 DNA
Since the mid-1980s advanced technology has allowed the DNA-typing of biological material to become the most powerful tool for identification purposes. The DNA profile from an individual is largely unique and identification can be made from one profile only. For DNA typing, short tandem repeats (STRs) polymorphism provides the basis of personal identification [33–35].
Traditionally, DNA-typing has been a routinely-used tool in forensics for the analysis of biological fluids, tissues and uprooted hair. More recent studies have reported DNA typing from fingerprints and skin debris left by even a single skin contact on objects and clothes [35–37]. Van Oorschot and Jones reported that substantial transfer of material (approximately 1–75 ng of DNA) occurs during initial contact [37]. Kisilevly and Wickenheiser profiled DNA of skin cells transferred through handling [38]. They reported that the amount of DNA transferred to a substrate depends on the handler, with some individuals being “good” epithelial cell donors (sloughers), while others individuals are poor epithelial cell donors (non-sloughers). Obviously, the probabilities of obtaining a full DNA profile are maximized with the former kind [38, 39]. Schulz and Reichert reported preliminary tests showing successful DNA typing in archived fingerprints that have been manipulated using soot powder, magnetic powder, and scotch tape [36, 40]. Reports such as this demonstrate the ability for DNA profiling in samples that have stored and relatively contaminated. DNA traces, such as the ones left in fingerprints, can also be easily wiped or brushed off from surfaces [35], suggesting they can also be easily aerosolized.
Further, the DNA typing of a single human dandruff particle was demonstrated by Herber and Herold [41]. Dandruff can be a constituent of bioaerosols and is derived from the horny layer of the skin where the cells do not completely differentiate and its aggregates contain nuclei. This is in distinct contrast to fully cornified cells (emitted skin flakes) where the nuclei completely disappears and no nuclear fragments or remnants remain [42]. In their study, Herber and Herold reported an estimated range of 0.8–1.5 ng DNA per dandruff. Additionally, they were able to obtain successful STR analysis for 90% of their samples. In loosely related results, DNA profiling for the identification of viruses and bacteria from bioaerosol samples has been reported in the literature. These results support the general idea that emitted skin (and other human-related biota) particles can serve as a source of a biochemical signature utilizing DNA analysis.
Limiting this line of reasoning are fingerprint DNA-typing studies showing that more than 1 ng of DNA (equivalent to 200 cells) is required (DNA typing in single cells has been demonstrated, but only for the buccal cell type[43]). Picogram levels of DNA have also been reported to provide satisfactory DNA typing. However, the analysis of minute sample sizes is highly complicated by contamination issues [44], which is a significant concern for aerosolized samples. Degradation and environmental effects will further limit the usefulness of DNA as an information source. Currently, there are no reports of DNA typing being applied to aerosolized human skin cells for identification purposes. Further, considering that fully cornified cells do not contain any DNA [42]; the probability of finding useful amounts of DNA is not favorable.
2.3 Protein Variants and Polymorphisms
Cellular proteins (along with other biomolecules) contained within aerosolized skin debris can presumably be used for identification purposes, but unlike other identifiers, such as DNA and fingerprints, can also give information on the individual’s state of health, where she or he has been living, and in which environmental conditions. Since protein sequences are linked to gene sequences, proteins can be considered a more characteristic biomarker of an individual than other type of molecules (e.g. lipids). Polymorphisms in DNA coding regions are precisely reflected in the polymorphisms of proteins and their derivatives. The same information that allows for DNA analysis to reflect genealogy, family and individuality may thus be obtained from specific proteins.
In humans, the SC contains 75–80% proteins (dry weight) [45]. Protein analysis in the SC has been performed mainly in order to address desquamation abnormalities. Furthermore, variability in the expression of SC proteins may or may not exist among various individuals, but in either case, the extensive literature documenting protein and DNA polymorphisms [46, 47] suggest that differences in expression are likely and therefore proteins make good candidates for the obtaining of molecular signatures. Of specific interest are the reports regarding the inter-variation shown for epidermal keratin proteins [48] that exist in dead skin cells.
Protein profiling has already been demonstrated with automated systems capable of detecting aerosolized bacterial cells and spores [49, 50]. In a moderately warm and humid environment the larger human aerosol settle and are digested first by the fungus Aspergillus Repens and then by dust mites [51]. Thus, the concentration and type of aerosolized proteins are dependent on time from human presence, and potentially this information can be used to time-stamp human and individual occupancy.
Protein variants among populations are already an area of high interest to emerging fields such as personalized medicine [52]. Beyond protein quantification, the search for disease biomarkers has heavily involved the study of protein polymorphism as well as posttranslational modifications such as oxidation, glycosylation, and truncation within the products of a single gene between healthy and unhealthy individuals. Borges and coworkers state that these type of modifications “extend the diversity of human gene products dramatically beyond 20,000–25,000 genes in the human genome” [53]. Quantifying these variants by means of proteomics and mass spectroscopy (MS) methods has elucidated the immense diversity of proteins (and protein modifications) in samples such as human plasma. Along with other studies, these assessments have lead the field of population proteomics [53–56]. Pioneers in this field have already proposed the creation of protein-diversity databases in which protein variants are indexed relative to age, sex, race, geographical region, disease, as well as other useful metrics [57]. As the efforts towards expanding the knowledge of protein variability in humans (and other organisms) continue, it is expected that other fields such as fingerprinting technologies based on biomolecular profiles undergo a parallel progress. Unlike other areas, protein fingerprinting would not require the complete isolation and characterization of low abundance proteins or variants. Instead, the sole acquisition of protein profiles generated by these variabilities can become the basis of obtaining a biochemical pattern for database generation and identification purposes.
Keratin Polymorphisms
Skin cells consist of more than 80% keratins cross linked to other cornified proteins [42]. Aside from being abundant in skin debris, these are the most appropriate protein target when seeking individual variability. Keratins consist of more than 20 polypeptides (K1–K20) that are classified into relatively acidic Type I (K9– K20) and neutral- to-basic Type II (K1–K8) keratins [58]. All epithelial cells typically express at least one Type I and one Type II keratin. For example, K4 and K13 are characteristic of the buccal mucosa while K1 and K10 are found on the dry surface of the skin [59].
Keratin is the major non-aqueous component (wt/wt) of the SC. Dead skin cells mostly consist of keratin intermediate filaments (KIFs) which are keratin structures that form the cytoskeleton of all cells [17]. As skin flakes are spontaneously released into air stream as a result of the desquamation process, human keratins become part of the bioaerosol in areas where humans are (were) present. The epithelial human keratin K10, derived from shed human skin and its associated bacteria, has recently been determined by Fox and coworkers as the most abundant protein in airborne dust of both occupied and unoccupied school rooms [60]. In addition, keratins have also been identified as a common contaminant in protein analysis such as gel electrophoresis and MS. It is believed that the source of these keratins is the laboratory air in which skin particles can be pervasive [60].
Heterogeneity in keratin structures has been reported for both animal [61] and human [48, 62] subjects. Polymorphisms in keratin genes give rise to protein heterogeneity in various types of epithelium including epidermis. More than a decade ago, Mischke and Wild identified these polymorphisms as important factors that could impact forensics sciences [62]. Even though their study mainly emphasized keratin polymorphism from epidermis samples obtained by means of surgery, they also reported inter-individual variation in the processed keratins from the layers of the SC.
Mischke and Wild showed that polymorphic keratins were present in human epidermis and some were identified among the individuals for specific constituent protein subunits, but this pattern does not correlate with sex, age, or ethnic origin [62]. In their study, they concluded that keratin polymorphisms can generally be expresses in all human epithelial capable of expressing keratins 1, 4, 5, and 10 [62]. As previously mentioned, keratins 1 and 10 are expressed in the dry surface of the skin. Therefore, keratin polymorphisms are expected to exist in the dead skin cells that eventually detach from the skin surface to become aerosolized human particulates in the environment.
In 1992, Korge and coworkers reported that human K10 is more polymorphic than was previously thought. Their results confirmed that the human K10 intermediate filament protein is polymorphic in amino acid sequence and in size. This was determined by using PCR amplification followed by sequence analysis on DNA. They observed variations in the V2 subdomain near the C-terminus in glycine-rich sequences with variations of as much as 114 base pairs (38 amino acids), with all individuals having one or two variants. The K10 polymorphism is restricted to insertions and deletions of the glycine-rich quasipeptide repeats that form the glycine-loop motif in the terminal domain. They also reported that the polymorphisms can be described by simple allelic variations that segregate by normal Mendelian mechanisms [48].
To some extent keratin proteins have been neglected because cytosolic proteins present less difficulty for analysis [63]. However and clearly, there are potential benefits from understanding keratins and their polymorphisms (and variants), not only for diagnostic tools and other already recognized applications, but pattern generation. Some of the potential benefits of keratin analysis for the application proposed here include their abundance in aerosolized material, the evidence of polymorphism, as well as their structural robustness in comparison with other protein material. The robustness of keratins may in fact be the key factor to obtaining crucial information from aerosolized particles in environment where other type of biochemical “stamps” would be highly degraded. It is important to note that the studies mentioned above did not particularly use aerosolized skin cells as their samples, but most certainly traces of these signatures remain after the desquamation process. The development of a bioaerosol fingerprinting technology heavily relies on understanding how these keratin polymorphisms remain in skin cells post-desquamation and finding strategies for practical detection from aerosolized human debris.
2.4 Other Classes of Molecular Targets
Aside from DNA and proteins, individualized information from aerosolized human debris may also be obtained from SC lipids or more exotic sources such as skin bacteria.
Lipids components were first used for general identification purposes of skin flakes approximately three decades ago. Analysis of surface fat material in recovered airborne particulate matter allowed the identification of skin particles in material that was previously dismissed as “dust of unknown origin” [12]. Regional variations in SC lipids have been addressed by many studies [64]. However, the lipid makeup of dead skin cells post-desquamation is not well-known. Consequently, inter-individual variability in lipid content from skin flakes has not been addressed; where understanding how this composition varies may be used to generate additional molecular fingerprints for aerosolize material. Presumably, lipids in aerosolized skin cells may not be as readily available in comparison to other biomolecules, since the mixture of ceramides, cholesterol and fatty acids is primarily located in the intercellular spaces of the SC [65] rather than within the individual corneocytes.
The human skin also harbors complex microbial ecosystems that appear to be unique to each individual, making them a potential source for a biochemical fingerprint. However, little is known in detail about its species composition. Recently, Gao et al. reported the variability of bacteria species existing in the surface of the skin using PCR-based methods [66]. Biota in superficial human skin expresses great diversity among individuals, with a few conserved and well represented genera, but otherwise low level interpersonal consensus. The importance of this study in the context of this work is that once the individual variabilities in skin biota composition are better understood it can potentially be another source for pattern recognition. This idea of using bacteria as a source of information is also supported by the results obtained by Tham and Zuraimi, who concluded that the main contributors of viable bacteria in indoor environments are in fact humans [23].
It is important to keep in mind that environmental conditions such as temperature, humidity, and light exposure as well as host factors including genotype, health, gender, immune status, and cosmetic use may affect microbial composition, population size and community structure.[66] These factors may increase the complexity of the biota and its variability, but also suggests a better, more information-rich source of patterns—if interpretable.
3. Molecular Profile Success Stories
Specific cases involving the use of molecular profiles for identification or differentiation offer some insight into molecular fingerprinting strategies. For example, profiles of mammalian and reptilian keratins (horn, hoof, and tortoiseshell) have been used to differentiate a broad range of sea turtles and bovid species. These studies involved the use of Fourier-transform Raman spectroscopy, diffuse reflectance Fourier-transform spectroscopy (DRIFT), and discriminant analysis to distinguish and identify the species-specific keratins [67]. Spectral library searches allowed for a comparison of the unknown to a set of possible matches (species population) [68]. Furthermore, the level of statistical confidence for each specific assignment could be calculated.
DNA-typing has allowed the detection and identification of viruses and bacteria in bioaerosol samples. Peccia and Hernandez have reviewed the emerging technologies incorporating PCR-based approaches with aerosol science for the identification, characterization and quantification of microorganisms for both indoor and outdoor environments [69]. Aside from microorganism detection in large-scale aerosol collections [70, 71], the identification of virus and bacteria from aerosolized DNA has also been demonstrated in smaller sampling devices. Pyankov and coworkers reported a personal sampler that could rapidly detect viable airborne microorganisms [72]. The identification of the influenza and vaccinia viruses [73] as well as the mumps and measles viruses [74] from bioaerosol samples has also been reported. Targeted PCR analysis is not the most appropriate approach for obtaining molecular fingerprints from skin flakes emitted by humans. However, these analyses overcame substantial contamination in terms of a large number of microorganisms and other material commonly found in ambient air. This demonstrates that large and complex backgrounds such as these do not preclude molecular pattern recognition strategies from being successful [72–74].
Field fractionation, capillary electrophoresis as well as electrophoresis technologies on microfluidic formats have been used to resolve a variety of microorganisms, and smaller particles such as viruses and DNA fragments [75–78]. An autonomous microfluidic device has also been demonstrated for the detection of aerosolized bacterial and spores based on protein profiles [50]. Similar technology has been also reported by Fruetel and coworkers for the protein profiling of viruses, however, not in aerosol samples [49]. These technologies and other protocols could be applied to obtaining protein profiles of the aereosolized human debris of interest [79].
Another powerful example of molecular fingerprinting from complex samples is the detection of airborne signatures of humans based on volatile organic compounds (VOCs) of human scent. Through this technology the distinctive odor characteristics that allows canine scent discrimination are exploited to differentiate primary odors among human subjects. The “primary odor” contains the constituents that are persistent over time regardless of diet or environmental factors (including exogenous sources such as lotions and soaps) [80]. Using the volatile organics that makeup the human scent as means for medical diagnosis as well as markers of genetic individuality has been demonstrated within the past decade [80–82]. A variety of extraction techniques (e.g. solid-phase microextraction) have been combined with GC-MS technologies and pattern-recognition strategies in search for quantitative differences in qualitatively similar profiles. Extensive surveys of odor-producing VOCs including acids, alcohols, aldehydes, hydrocarbons, esters, ketones and nitrogen-containing compounds showed reproducible and individualized profile based on relative ratio of these components [82]. Similarly to other identification systems, they stated how these profiles could be stored in a searchable database for their use as a biometric measure. By performing comparisons via Spearman Rank Correlations and narrowing the compounds considered for their profiles they were able to obtain a greater degree of both individualization and discrimination [82]. Even though the existences of valid VOCs signatures have been demonstrated, this strategy has significant limitations. Airborne signatures rely upon volatile compounds which are relatively in small number, can be confused with background compounds, disperse rapidly and have poor detection limits.
4. Bioaerosols Analysis and Pattern Recognition Strategies: Needs and Challenges
When considering which of the various biomaterials described in previous sections for detection and identification, it is important to understand what information can be collected from each type (Table 1). For example, the detection of bio-terrorism threats focuses on the collection and analysis of bacterial and viral particles specifically those that maintain their pathogenicity. Targeted biomolecules should be chosen for ease of detection in order to maximize unique identification and detectability. With identification made either through detection of a single component or by comparing a collection of components.
Table 1.
Possible sources for biochemical signatures in bioaerosols from various sample types
| Sample Type | Skin Debris | Fur/Hair | Feathers | Scales | Insect Debris | Plant Debris |
|---|---|---|---|---|---|---|
| DNA | occasionally, truncated | only in root-bulb | yes | occasionally, truncated | yes | yes |
| RNA | yes | not beyond root-bulb | unlikely | unlikely | yes | yes |
| Proteins | yes, structural (e.g. keratin) | yes, structural (e.g. keratin) | yes, structural (e.g. keratin) | yes, structural (e.g. keratin) | structural (e.g. chitin) | yes, structural |
| Nucleic Acids | small amounts | not beyond root-bulb | small amounts | small amounts | yes | small amounts |
| Lipids | yes, large amounts | no | no | no | yes | yes |
| Glycosylated Species | yes | no | no | no | likely | likely |
| Amino Acids | yes | yes | yes | yes | yes | yes |
The first hurdle to utilizing the information borne in aerosolized biomolecules and cells is to capture sufficient amounts of material to generate statistically distinguishable and reproducible patterns for classification. Steps towards this goal have been taken by research described in previous sections [83–86].
However, a plethora of complicating organism-specific and environmental variables will require refinement of sampling and detection techniques. For example, the concentration of target proteins in the air range largely depending on the environment measured. Current measurements of total aerosolized protein range in concentration from 0.1 to 3 μgm−3 (up to 8 μgm−3 for physically induced samples) in the presence of humans, and can vary across in 0 to 2 μgm−3 for outdoor situations with limited human presence [8, 69, 87]. Within this amount of total protein are thousands of fractions of different individual species and mixtures with varying magnitudes of individual concentration. The detection limits for individual molecular species of proteins is conservatively in the 100 pg range depending on the protein purification and detection strategy used. Detection schemes focusing on the DNA in certain sample types (e.g. buccal) use as few as a single cell to functionally create a DNA profile [43]. These limits of detection illuminate some of the challenging aspects of accurately detecting biomolecular sub-fractions using current technology. Expanding and improving current methods will lead to a richer suite of probe biomolecules being detectable, while the necessary concentrations for detection are reduced.
Analyses strategies for bioaerosols have been extensively reviewed in the literature [1, 83, 88–92]. Traditionally, bioaerosols are analyzed though culture, microscopy, biochemistry, immunochemistry, and flow cytometry. Other technologies have also been used such as a fluorescent aerodynamic particle sizer (FLAPS), fluorescence in situ hybridization (FISH) [78], laser-induced fluorescence (LIF) [93] to a lesser extent. Technologies based on spectroscopic techniques have also been used for the characterization of bioaerosols such as Raman spectroscopy [94, 95] and Fourier transformer infrared spectroscopy [96]. Emerging capillary electrophoresis approaches are also promising avenues for generating biomolecular profiles in aerosolized human debris [97, 98].
The use of MS has already been reported for the analysis of aerosolized material of biological origin [99–103], including the Bioaerosol Mass Spectrometer (BMAS) developed for the detection of microorganisms [104, 105]. Ariya and coworkers recently reviewed the compounds used to identify the specific bioaerosol sources that were separated and detected by GC/MS strategies [1]. They report that detection limits for individual compounds are typically in the low pgm−3 for GC and LC-MS and that even overlapping compounds in very complex mixtures could be successfully interpreted [1]. The signatures used to detect microorganisms correspond to the mass spectrum -predominantly made of peptide and proteins- that is acquired for a particular organism under a variety of conditions, which can then be matched to the predicted masses of organisms with sequenced genomes [106, 107].
To generate a pattern related to the molecular constituents of bioaerosols, spectrometric, spectroscopic, and/or separation (including molecular recognition) strategies can be used. The underlying principle is to be very inclusive towards any technique or approach that can help differentiate one sample from another. This can take the form of separated chromatographic or electrophoretic peaks, molecular recognition strategies, differing absorption or emissions at varying wavelengths or mass spectrometric analysis. The undertaking of discerning complex patterns for identification is well developed within the data mining community. However, as with any pattern recognition, the accuracy of the analytical technique can impact interpretation—essentially defining the number of values that can be assigned. A rough estimate of the necessary protein concentrations can be generated using a standard assessment of signal to noise. Approximately setting three times the detection limit as the accuracy and the dynamic range at four orders of magnitude suggests there are possibly 107 unique signatures available from current strategies. This can also be described as having n number of vectors (fractions) having m discernable values giving n × m unique solutions for truly random projections. However, real world sample will not give this high a number of signatures, since it is not a random system—this is simply an estimate of the order magnitude that can be obtained. Pattern recognition strategies can be viewed as an integral means of combing environmentally and biologically complex samples and distilling them into statistically relevant signatures for the source of the material.
For example, chemometric techniques applied to bacterial taxonomy[108] and geographical sourcing of medicinal plants[109] was aided through the classification of data into groups via related strategies. Chemometric characterization of aerosol bacteria by LIF has also utilized various approaches including principal component analysis, linear discriminant analysis and hierarchical cluster analysis to classify the microorganisms according to family, morphology and gram-test [110]. These reports exemplify the power of coupling analytical techniques with effective mathematical post-processing to achieve molecular profiles differentiation among aerosolized samples.
The ability to combine sample collection, analytical treatment, and pattern recognition will define how much information can be garnered. While the specificity of the identification desired dictates the analytical refinement required. For instance, is it sufficient in certain applications to discern the simple presence of any person? Conversely, it may be desirable to find a specific signature in the presence of other family and clan members, among other complicating signatures.
A more in-depth understanding of bioaerosol material is still required and performance of specific analytics remains to be established to allow separation and detection of molecular patterns.
Focusing on human detection various samplers, personal and static, could find use in specific settings. For examples, for defense applications a static sampler could be used in remote areas to monitor the presence and movement of adversary troops. While personal samplers may be more appropriate if a new setting needs to be screened. Application could include rescue mission for trapped miner in which a a more portable device will be more beneficial to search for aerosolized human fingerprints in potential sites were individuals could be trapped. Both types of samplers will require the subsequent separation of the debris of interest from other background particulates. Potential methods include the application of dielectrophoretic separation to quickly asses the bioparticle portions of interest in a given sample prior to further fractionation [111–113].
5. Envisioned Application of Technology
The future capabilities of this technology to perform gross identification would build upon the current possibilities of determining the presence of humans by expanding to distinguishing different types of organisms. In addition to determining their presence, looking at the degree of material degradation might also be exploited to determine how much time has passed since the occupation.
We envision a few different types of devices utilizing varying levels of technology and supplying various capabilities—from portable, relatively information-poor devices, to fixed information-rich systems. Some exciting applications of this technology include active and passive TTL (tag, track and locate), check points (military) and airport security, forensics, healthcare, personnel identification, non-invasive biological and environmental monitoring, archeology and paleontology, anti-poaching, and rescue missions.
6. Summary and Outlook
It may be premature to conclude whether aerosolized human and animal debris along with the excess protein content in the air can be used to identify individual sources of emitted particles. However, the combination of the current findings reported here in concert with the unexploited information that exists in aerosolized human and animal cells are certainly encouraging with regards to their use as “calling cards” left by the biological sources (humans or animals present in the area of air sampling).
The concept described throughout this review provides new direction and opportunities for research. Research should be directed at the identification of the biomolecules within the aerosolized skin cells and other aerosolized debris that provide maximized differentiation among sources. Strategies to exploit the information from these molecules must be examined and improved in order to take full advantage of the biochemical signatures contained within the aerosolized flakes. Understanding transformations and aging processes of bioaerosols needs to be further improved in order to exploit the opportunities for the timestamp aspects of a bioaerosol fingerprinting technology.
There is a critical need for synergism between the various available techniques for sampling, separation and analysis to achieve the necessary depth in obtaining information from human-emitted bioaerosol particles. This combined effort will allow us to be in a much better position for the development of novel state of the art technologies for the complete analysis of aerosolized cells.
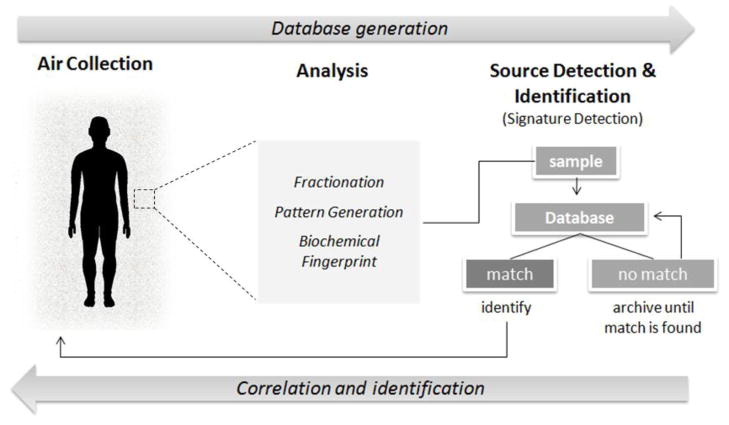
Figure 1.
Overall Schematic of Bioaerosol Fingerprinting Technology. The proposed technology consists of the development of devices for collection and analysis of bioaerosol signatures as a new “fingerprinting” technology. Target bioaerosol particles such as human-emitted dead skin cells, pervasively exist as suspended aerosolized particulates that contain biochemical information unique to their source. This information, most likely in the form of DNA and protein polymorphisms, could be exploited to obtain a biochemical signature from a location of interest. Signal analysis involving biometric comparison of signatures to a database can provide insights into potential and/or actual presence of individuals in a particular area. The development of such technology will require integration and refinement of existing but cutting-edge technologies for aerosol sample collection and automated biochemical analysis.
Acknowledgments
Partial support for this work was provided by NIH 1 R21 EB010191-01A1, Arizona Applied NanoTechnology Sensors (ASU Vice President for Research Office), and Bioaerosols for Pattern Recognition (ASU Vice President for Research Office)
References
- 1.Ariya PA, Sun J, Eltouny NA, Hudson ED, Hayes CT, Kos G. Physical and chemical characterization of bioaerosols-Implications for nucleation processes. Int Rev Phys Chem. 2009;28(1):1–32. [Google Scholar]
- 2.Tyndall J. Essays on the Floating-Matter of the Air in Relation to Putrefaction and Infection. New York: D. Appleton and Company; 1884. [Google Scholar]
- 3.Hoefs J. 2006 Stable Isotope Geochemistry. 5. Berlin, Germany: Springer; 2003. [Google Scholar]
- 4.Rogge WF, Medeiros PM, Simoneit BRT. Organic marker compounds for surface soil and fugitive dust from open lot dairies and cattle feedlots. Atmos Environ. 2006;40(1):27–49. [Google Scholar]
- 5.Rogge WF, Medeiros PM, Simoneit BRT. Organic marker compounds in surface soils of crop fields from the San Joaquin Valley fugitive dust characterization study. Atmos Environ. 2007;41(37):8183–8204. [Google Scholar]
- 6.Belan BD, Borodulin AI, Marchenko YV, Ol’kin SE, Panchenko MV, P’Yankov OV, Safatov AS, Buryak GA. Study of atmospheric aerosol protein components variability above forest areas in the south of western Siberia. Dokl Akad Nauk. 2000;374(6):827–829. [PubMed] [Google Scholar]
- 7.Hock N, Schneider J, Borrmann S, Rompp A, Moortgat G, Franze T, Schauer C, Poschl U, Plass-Dulmer C, Berresheim H. Rural continental aerosol properties and processes observed during the Hohenpeissenberg aerosol characterization experiment. Atmos Chem Phys. 2008;8:603–623. [Google Scholar]
- 8.Jaenicke R. Abundance of cellular material and proteins in the atmosphere. Science. 2005;308:73. doi: 10.1126/science.1106335. [DOI] [PubMed] [Google Scholar]
- 9.Milstone LM. Epidermal desquamation. J Dermatol Sci. 2004;36(3):131–140. doi: 10.1016/j.jdermsci.2004.05.004. [DOI] [PubMed] [Google Scholar]
- 10.Face Recognition Home Page. Available from: http://www.face-rec.org/
- 11.Clark RP. Skin scales among airborn particles. J Hyg. 1974;72(1):47. doi: 10.1017/s0022172400023196. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Clark RP, Shirley SG. Identification of skin in airborne particulate matter. Nature. 1973;246(5427):39–40. doi: 10.1038/246039a0. [DOI] [PubMed] [Google Scholar]
- 13.Tovey ER, Kemp AS, Almqvist C, Sharland A, Marks GB. Do immune responses to inhaled skin flakes modulate the expression of allergic disease? Clin Exp Allergy. 2007;37(8):1199–1203. doi: 10.1111/j.1365-2222.2007.02770.x. [DOI] [PubMed] [Google Scholar]
- 14.Bahadori TH, Suh H, Koutrakis P. Issues in human particulate exposure assessment: Relationship between outdoor, indoor, and personal exposures. Hum Ecol Risk Assess. 1999;5(3):459–470. [Google Scholar]
- 15.Discovery Channel Videos, DJDSW. [cited 2011 June 6th]; Available from: http://dsc.discovery.com/videos/dirty-jobs-dead-skin-weight.html.
- 16.Laumbach RJ, Kipen HM. Bioaerosols and sick building syndrome: particles, inflammation, and allergy. Curr Opin Allergy Clin Immunol. 2005;5(2):135–139. doi: 10.1097/01.all.0000162305.05105.d0. [DOI] [PubMed] [Google Scholar]
- 17.Candi E, Schmidt R, Melino G. The cornified envelope: a model of cell death in the skin. Nat Rev Mol Cell Bio. 2005;6(4):328–340. doi: 10.1038/nrm1619. [DOI] [PubMed] [Google Scholar]
- 18.Fuchs E. Keratins and the skin. Annu Rev Cell Dev Bio. 1995;11:123–153. doi: 10.1146/annurev.cb.11.110195.001011. [DOI] [PubMed] [Google Scholar]
- 19.Sun TT, Green H. Differentiation of epidermal keratinocyte in cell-culture - formation of cornified envelope. Cell. 1976;9(4):511–521. doi: 10.1016/0092-8674(76)90033-7. [DOI] [PubMed] [Google Scholar]
- 20.Rothman S. In Physiology and biochemistry of the skin. University of Chicago Press; 1954. [Google Scholar]
- 21.Lidwell OM, Mackintosh CA, Towers AG. Evaluation of fabrics in relation to their use as protective garments in nursing and surgery. 2. dispersal of skin organisms in a test chamber. J Hyg. 1978;81(3):453–469. doi: 10.1017/s002217240002533x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Mackintosh CA, Lidwell OM, Towers AG. Dimensions of skin fragments dispersed into air during activity. J Hyg. 1978;81(3):471. doi: 10.1017/s0022172400025341. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Tham KW, Zuraimi MS. Size relationship between airborne viable bacteria and particles in a controlled indoor environment study. Indoor Air. 2005;15:48–57. doi: 10.1111/j.1600-0668.2005.00303.x. [DOI] [PubMed] [Google Scholar]
- 24.Wesley NO, Maibach HI. Racial (ethnic) differences in skin properties - The objective data. Am J Clin Dermatol. 2003;4(12):843–860. doi: 10.2165/00128071-200304120-00004. [DOI] [PubMed] [Google Scholar]
- 25.Zhongwu S, Xueliang G, Hongfei Z, Qing W, Bing B. Analysis on keratin amino acids in the feather of cranes and storks and the application in species indentification. Scientia Silvae Sinicae. 2008;44(3):102–106. [Google Scholar]
- 26.Plewig G. Regional differences of cell sizes in human stratum corneum. 2. effects of sex and age. J Invest Dermatol. 1970;54(1):19. doi: 10.1111/1523-1747.ep12551488. [DOI] [PubMed] [Google Scholar]
- 27.Plewig G, Marples RR. Regional differences of cell sizes in human stratum corneum. J Invest Dermatol. 1970;54(1):13. doi: 10.1111/1523-1747.ep12551482. [DOI] [PubMed] [Google Scholar]
- 28.Grove GL, Kligman AM. Age-associated changes in human epidermal-cell renewal. J Gerontol. 1983;38(2):137–142. doi: 10.1093/geronj/38.2.137. [DOI] [PubMed] [Google Scholar]
- 29.Guz NV, Gaikwad RM, Dokukin ME, Sokolov I. A novel in vitro stripping method to study geometry of corneocytes with fluorescent microscopy: example of aging skin. Skin Res Technol. 2009;15(4):379–383. doi: 10.1111/j.1600-0846.2009.00369.x. [DOI] [PubMed] [Google Scholar]
- 30.Fluhr JW, Pelosi A, Lazzerini S, Dikstein S, Berardesca E. Differences in corneocyte surface area in pre- and post-menopausal women - Assessment with the noninvasive videomicroscopic imaging of corneocytes method (VIC) under basal conditions. Skin Pharmacol Appl. 2001;14:10–16. doi: 10.1159/000056384. [DOI] [PubMed] [Google Scholar]
- 31.Leveque JL, Corcuff P, Derigal J, Agache P. In vivo Studies Of The Evolution Of Physical-Properties Of The Human-Skin With Age. Int J Dermatol. 1984;23(5):322–329. doi: 10.1111/j.1365-4362.1984.tb04061.x. [DOI] [PubMed] [Google Scholar]
- 32.Corcuff P, Leveque JL. Corneocyte changes after acute UV irradiation and chronic solar exposure. Photodermatology. 1988;5(3):110–115. [PubMed] [Google Scholar]
- 33.Edwards A, Hammond HA, Jin L, Caskey CT, Chakraborty R. Genetic-variation at 5 trimeric and tetrameric tandem repeat loci In 4 human-population groups. Genomics. 1992;12(2):241–253. doi: 10.1016/0888-7543(92)90371-x. [DOI] [PubMed] [Google Scholar]
- 34.Kimpton CP, Gill P, Walton A, Urquhart A, Millican ES, Adams M. Automated DNA profiling employing multiplex amplification of short tandem repeat loci. PCR Methods Appl. 1993;3(1):13–22. doi: 10.1101/gr.3.1.13. [DOI] [PubMed] [Google Scholar]
- 35.Van Hoofstat DEO, Deforce DLD, De Pauw IPH, Van den Eeckhout EG. DNA typing of fingerprints using capillary electrophoresis: Effect of dactyloscopic powders. Electrophoresis. 1999;20(14):2870–2876. doi: 10.1002/(SICI)1522-2683(19991001)20:14<2870::AID-ELPS2870>3.0.CO;2-V. [DOI] [PubMed] [Google Scholar]
- 36.Schulz MM, Reichert W, Wehner HD, Mattern R. An already archived latent fingerprint as a DNA source for STR typing in a murder case. Arch Kriminol. 2004;213(5–6):165–170. [PubMed] [Google Scholar]
- 37.VanOorschot RAH, Jones MK. DNA fingerprints from fingerprints. Nature. 1997;387(6635):767–767. doi: 10.1038/42838. [DOI] [PubMed] [Google Scholar]
- 38.Kisilevsky AE, Wickenheiser RA. DNA PCR profiling of skin cells transferred through handling. Edmonton, Alberta: 1999. [Google Scholar]
- 39.Wickenheiser RA. Trace DNA: A review, discussion of theory, and application of the transfer of trace quantities of DNA through skin contact. J Forensic Sci. 2002;47(3):442–450. [PubMed] [Google Scholar]
- 40.Schulz MM, Reichert W. Archived or directly swabbed latent fingerprints as a DNA source for STR typing. Forensic Sci Int. 2002;127(1–2):128–130. doi: 10.1016/s0379-0738(02)00092-0. [DOI] [PubMed] [Google Scholar]
- 41.Herber B, Herold K. DNA typing of human dandruff. J Forensic Sci. 1998;43(3):648–656. [PubMed] [Google Scholar]
- 42.Lippens S, Denecker G, Ovaere P, Vandenabeele P, Declercq W. Death penalty for keratinocytes: apoptosis versus cornification. Cell Death Differ. 2005;12:1497–1508. doi: 10.1038/sj.cdd.4401722. [DOI] [PubMed] [Google Scholar]
- 43.Findlay I, Taylor A, Quirke P, Frazier R, Urquhart A. DNA fingerprinting from single cells. Nature. 1997;389(6651):555–556. doi: 10.1038/39225. [DOI] [PubMed] [Google Scholar]
- 44.Gill P, Whitaker J, Flaxman C, Brown N, Buckleton J. An investigation of the rigor of interpretation rules for STRs derived from less than 100 pg of DNA. Forensic Sci Int. 2000;112(1):17–40. doi: 10.1016/s0379-0738(00)00158-4. [DOI] [PubMed] [Google Scholar]
- 45.Williams AC, Barry BW. Skin absorption enhancers. Criti Rev Ther Drug. 1992;9(3–4):305–353. [PubMed] [Google Scholar]
- 46.Ayala FJ. Genetic variation in natural-populations- problem of electrophoretically cryptic alleles. Proc Natl Acad Sci USA-Biol. 1982;79(2):550–554. doi: 10.1073/pnas.79.2.550. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Ramshaw JAM, Coyne JA, Lewontin RC. The sensitivity of gel electrophoresis as a detector of genetic variation. Genetics. 1979;93(4):1019–1037. doi: 10.1093/genetics/93.4.1019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Korge BP, Gan SQ, McBride OW, Mischke D, Steinert PM. Extensive size polymorphism of the human keratin-10 chain resides in the C-terminal V2 subdomain due to variable numbers and sizes of glycine loops. Proc Natl Acad Sci USA. 1992;89(3):910–914. doi: 10.1073/pnas.89.3.910. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Fruetel JA, West JAA, Debusschere BJ, Hukari K, Lane TW, Najm HN, Ortega J, Renzi RF, Shokair I, VanderNoot VA. Identification of viruses using microfluidic protein profiling and bayesian classification. Anal Chem. 2008;80(23):9005–9012. doi: 10.1021/ac801342m. [DOI] [PubMed] [Google Scholar]
- 50.Stachowiak JC, Shugard EE, Mosier BP, Renzi RF, Caton PF, Ferko SM, de Vreugde JLV, Yee DD, Haroldsen BL, VanderNoot VA. Autonomous microfluidic sample preparation system for protein profile-based detection of aerosolized bacterial cells and spores. Anal Chem. 2007;79(15):5763–5770. doi: 10.1021/ac070567z. [DOI] [PubMed] [Google Scholar]
- 51.Collof MJ. Dust mites. CSIRO Publishing & Springer; Dordrecht, The Netherlands: 2009. [Google Scholar]
- 52.Anderson NL, Anderson NG. The human plasma proteome - history, character, and diagnostic prospects. Mol Cell Proteomics. 2002;1(11):845–867. doi: 10.1074/mcp.r200007-mcp200. [DOI] [PubMed] [Google Scholar]
- 53.Borges CR, Rehder DS, Jarvis JW, Schaab MR, Oran PE, Nelson R. Full-length characterization of proteins in human populations. Clin Chem. 2010;56(2):202–211. doi: 10.1373/clinchem.2009.134858. [DOI] [PubMed] [Google Scholar]
- 54.Anderson NL, Polanski M, Pieper R, Gatlin T, Tirumalai RS, Conrads TP, Veenstra TD, Adkins JN, Pounds JG, Fagan R, Lobley A. The human plasma proteome - A nonredundant list developed by combination of four separate sources. Mol Cell Proteomics. 2004;3(4):311–326. doi: 10.1074/mcp.M300127-MCP200. [DOI] [PubMed] [Google Scholar]
- 55.Nedelkov D, Kiernan UA, Niederkofler EE, Tubbs KA, Nelson RW. Investigating diversity in human plasma proteins. Proc Natl Acad Sci USA. 2005;102(31):10852–10857. doi: 10.1073/pnas.0500426102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Nedelkov D, Phillips DA, Tubbs KA, Nelson RW. Investigation of human protein variants and their frequency in the general population. Mol Cell Proteomics. 2007;6(7):1183–1187. doi: 10.1074/mcp.M700023-MCP200. [DOI] [PubMed] [Google Scholar]
- 57.Nedelkov D, Tubbs KA, Niederkofler EE, Kiernan UA, Nelson RW. High-throughput comprehensive analysis of human plasma proteins: A step toward population proteomics. Anal Chem. 2004;76(6):1733–1737. doi: 10.1021/ac035105+. [DOI] [PubMed] [Google Scholar]
- 58.Moll R, Franke WW, Schiller DL, Geiger B, Krepler R. The catalog of human cytokeratins - patterns of expression in normal epithelia, tumors and cultured-cells. Cell. 1982;31(1):11–24. doi: 10.1016/0092-8674(82)90400-7. [DOI] [PubMed] [Google Scholar]
- 59.Morley SM. Keratin and the skin: past, present and future. QJ Med. 1997;90:433–435. doi: 10.1093/qjmed/90.7.433. [DOI] [PubMed] [Google Scholar]
- 60.Fox K, Castanha E, Fow A, Feigley C, Salzberg D. Human K10 epithelial keratin is the most abundant protein in airborne dust of both occupied and unoccupied school rooms. J Environ Monitor. 2008;10:55–59. doi: 10.1039/b714802j. [DOI] [PubMed] [Google Scholar]
- 61.Brush AH. Tissue specific protein heterogeneity in keratin structures. Biochem Syst Ecol. 1986;14(5):547–551. [Google Scholar]
- 62.Mischke D, Wild G. Polymorphic keratins in human epidermis. J Invest Dermatol. 1987;88(3):191–197. doi: 10.1111/1523-1747.ep12525329. [DOI] [PubMed] [Google Scholar]
- 63.Plowman JE. The proteomics of keratin proteins. J Chromatogr B. 2007;849:181–189. doi: 10.1016/j.jchromb.2006.07.055. [DOI] [PubMed] [Google Scholar]
- 64.Lampe MA, Burlingame AL, Whitney J, Williams ML, Brown BE, Roitman E, Elias PM. Human stratum-corneum lipids - characterization and regional variations. J Lipid Res. 1983;24(2):120–130. [PubMed] [Google Scholar]
- 65.Dayan N. In: Skin aging handbook- an integrated approach to biochemistry and product development. Dayan N, editor. William Andrew Publishing; 2008. [Google Scholar]
- 66.Gao Z, Tseng CH, Pei ZH, Blaser MJ. Molecular analysis of human forearm superficial skin bacterial biota. Proc Natl Acad Sci USA. 2007;104(8):2927–2932. doi: 10.1073/pnas.0607077104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Edwards HGM, Hunt DE, Sibley MG. FT-Raman spectroscopic study of keratotic materials: horn, hoof and tortoiseshell. Spectrochim Acta A. 1998;54(5):745–757. doi: 10.1016/s1386-1425(98)00013-4. [DOI] [PubMed] [Google Scholar]
- 68.Espinoza EO, Baker BW. The analysis of sea turtle and bovid keratin artefacts using drift spectroscopy and discriminant analysis. Archaeometry. 2007;49(4):685–698. [Google Scholar]
- 69.Peccia J, Hernandez M. Incorporating polymerase chain reaction-based identification, population characterization, and quantification of microorganisms into aerosol science: A review. Atmos Environ. 2006;40(21):3941–3961. doi: 10.1016/j.atmosenv.2006.02.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Negrin MM, Del Panno MT, Ronco AE. Study of bioaerosols and site influence in, the La Plata area (Argentina) using conventional and DNA (fingerprint) based methods. Aerobiologia. 2007;23(4):249–258. [Google Scholar]
- 71.Maron PA, Lejon DPH, Carvalho E, Bizet K, Lemanceau P, Ranjard L, Mougel C. Assessing genetic structure and diversity of airborne bacterial communities by DNA fingerprinting and 16S rDNA clone library. Atmos Environ. 2005;39(20):3687–3695. [Google Scholar]
- 72.Pyankov OV, Agranovshi IE, Pyankov O, Mokhonova E, Mokhonova V, Safatov AS, Khromykh AA. Using a bioaerosol personal sampler in combination with real-time PCR analysis for rapid detection of airborne viruses. Environ Microbiol. 2007;9(4):992–1000. doi: 10.1111/j.1462-2920.2006.01226.x. [DOI] [PubMed] [Google Scholar]
- 73.Agranovski IE, Safatov AS, Sergeev AA, Pyankov OV, Petrishchenko VA, Mikheev MV, Sergeev AN. Rapid detection of airborne viruses by personal bioaerosol sampler combined with the PCR device. Atmos Environ. 2006;40(21):3924–3929. [Google Scholar]
- 74.Agranovski IE, Safatov AS, Agafonov AP, Pyankov OV, Sergeev AN. Monitoring of airborne mumps and measles viruses in a hospital. Clean. 2008;36(10–11):845–849. [Google Scholar]
- 75.Lee H, Williams SKR, Wahl KL. Analysis of whole bacterial cells by flow field-flow fractionation and matrix-assisted laser desorption/ionization time-of-flight mass spectrometry. Anal Chem. 2003;75(11):2746–2752. doi: 10.1021/ac020698u. [DOI] [PubMed] [Google Scholar]
- 76.Desai MJ, Armstrong DW. Separation, identification, and characterization of microorganisms by capillary electrophoresis. Microbiol Mol Bio Rev. 2003;67(1):38. doi: 10.1128/MMBR.67.1.38-51.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Ebersole RC, McCormick RM. Separation and isolation of viable bacteria by capillary zone electrophoresis. Nat BioTechnol. 1993;11(11):1278–1282. doi: 10.1038/nbt1193-1278. [DOI] [PubMed] [Google Scholar]
- 78.Ho J. Future of biological aerosol detection. Anal Chimi Acta. 2002;457(1):125–148. [Google Scholar]
- 79.Renzi RF, Stamps J, Horn BA, Ferko S, VanderNoot VA, West JAA, Crocker R, Wiedenman B, Yee D, Fruetel JA. Hand-held microanalytical instrument for chip-based electrophoretic separations of proteins. Anal Chem. 2005;77(2):435–441. doi: 10.1021/ac049214f. [DOI] [PubMed] [Google Scholar]
- 80.Curran AM, Rabin SI, Furton KG. Analysis of the uniqueness and persistence of human scent. Forensic Sci Comm [online] 2005;7(2) [Google Scholar]
- 81.Curran AM, Prada PA, Furton KG. The differentiation of the voltile organic signatures of individuals through SPME-GC/MS of characteristic human scent compounds. J Forensic Sci. 2010;55(1):50–57. doi: 10.1111/j.1556-4029.2009.01236.x. [DOI] [PubMed] [Google Scholar]
- 82.Curran AM. The frequency of occurrence and discriminatory power of compounds found in human scent across a population determined by SPME-GEMS. J Chromatogr B. 2007;846(1–2):86–97. doi: 10.1016/j.jchromb.2006.08.039. [DOI] [PubMed] [Google Scholar]
- 83.Griffiths WD, Decosemo GAL. The assessment of bioaerosols - a critical review. J Aerosol Science. 1994;25(8):1425–1458. [Google Scholar]
- 84.Lacey J, Venette J. Bioaerosols handbook. CRC Press/Lewis Publishers Inc.; CRC Press/Lewis Publishers; 1995. pp. 407–471. [Google Scholar]
- 85.Levetin E. Methods for aeroallergen sampling. Curr Allergy Asthma R. 2004;4(5):376–383. doi: 10.1007/s11882-004-0088-z. [DOI] [PubMed] [Google Scholar]
- 86.Muilenberg ML. Sampling devices. Immunol Allergy Clin. 2003;23(3):337. doi: 10.1016/s0889-8561(03)00030-4. [DOI] [PubMed] [Google Scholar]
- 87.Staton SJR, Castillo JA, Taylor TJ, Herckes P, Hayes MA. Detecting a “Naked” Person in the Desert: Indentifying Environmental from Bioaerosol Material using HPLC. unpublished. [Google Scholar]
- 88.Burge HA. Monitoring for airborn allergens. Ann Allergy. 1992;69(1):9–18. [PubMed] [Google Scholar]
- 89.Buttner MP, Willeke K, Grinsphun SA. In: Sampling and analysis of airborn microorganisms. Hurst CJ, editor. ASM Press; Washington, DC: 2002. pp. 814–826. [Google Scholar]
- 90.Eduard W, Heederik D. Methods for quantitative assessment of airborne levels of noninfectious microorganisms in highly contaminated work environments. Am Ind Hyg Assoc J. 1998;59(2):113–127. doi: 10.1080/15428119891010370. [DOI] [PubMed] [Google Scholar]
- 91.Menetrez MY, Foarde KK, Esch RK, Schwartz TD, Dean TR, Hays MD, Cho SH, Betancourt DA, Moore SA. An evaluation of indoor and outdoor biological particulate matter. Atmos Environ. 2009;43(34):5476–5483. [Google Scholar]
- 92.Pohlker C, Huffman JA, Poschl U. Autofluorescence of atmospheric bioaerosols- fluorescent biomolecules ans potential interferences. Atmos Meas Tech Discuss. 2011;4:5857–5933. [Google Scholar]
- 93.Pan YL, Pinnick RG, Hill SC, Rosen JM, Chang RK. Single-particle laser-induced-fluorescence spectra of biological and other organic-carbon aerosols in the atmosphere: Measurements at New Haven, Connecticut, and Las Cruces, New Mexico. J Geophys Res. 2007;112(D24) [Google Scholar]
- 94.Rosch P, Harz M, Peschke KD, Ronneberger O, Birkhardt H, Schule A, Schmauz G, Lankers M, Hofer S, Thiele H, Motzkus HW, Popp J. On-line monitoring and identification of bioaerosols. Anal Chem. 2006;78(7):2163–2170. doi: 10.1021/ac0514974. [DOI] [PubMed] [Google Scholar]
- 95.Sengupta A, Brar N, Davis EJ. Bioaerosol detection and characterization by surface-enhanced Raman spectroscopy. J Colloid Interface Sci. 2007;309(1):36–43. doi: 10.1016/j.jcis.2007.02.015. [DOI] [PubMed] [Google Scholar]
- 96.Ben-David A, Ren H. Detection, identification, and estimation of biological aerosols and vapors with a Fourier-transform infrared spectrometer. Appl Optics. 2003;42(24):4887–4900. doi: 10.1364/ao.42.004887. [DOI] [PubMed] [Google Scholar]
- 97.Chen KP, Pacheco JR, Staton SRJ, Hayes MA. Insulator-based dielectrophoretic separation of small particles in a sawtooth channel. Electrophoresis. 2009;30(9):1441–1448. doi: 10.1002/elps.200800833. [DOI] [PubMed] [Google Scholar]
- 98.Meighan MM, Keebaugh MW, Quihuis AM, Kenyon SM, Hayes MA. Electrophoretic exclusion for the selective transport of small molecules. Electrophoresis. 2009;30(21):3786–3792. doi: 10.1002/elps.200900340. [DOI] [PubMed] [Google Scholar]
- 99.Kleefsman I, Stowers MA, Verheijen PJT, Van Wuijckhuijse AL, Kientz CE, Marijnissen JCM. Bioaerosol analysis by single particle mass spectrometry. KONA. 2007;24:85–90. [Google Scholar]
- 100.Canagaratna MR, Jayne JT, Jimenez JL, Allan JD, Alfarra MR, Zhang Q, Onasch TB, Drewnick F, Coe H, Middlebrook A, Delia A, Williams LR, Trimborn AM, Northway MJ, DeCarlo PF, Kilb CE, Davidovits P, Worsnop DR. Chemical and microphysical characterization of ambient aerosols with the aerodyne aerosol mass spectrometer. Mass Spectrom Rev. 2007;26(2):185–222. doi: 10.1002/mas.20115. [DOI] [PubMed] [Google Scholar]
- 101.Kim JK, Jackson SN, Murray KK. Matrix-assisted laser desorption/ionization mass spectrometry of collected bioaerosol particles. Rapid Commun Mass Spectrom. 2005;19(12):1725–1729. doi: 10.1002/rcm.1982. [DOI] [PubMed] [Google Scholar]
- 102.Stowers MA, van Wuijckhuijse AL, Marijnissen JCM, Kientz CE, Ciach T. Fluorescence preselection of bioaerosol for single-particle mass spectrometry. Appl Optics. 2006;45(33):8531–8536. doi: 10.1364/ao.45.008531. [DOI] [PubMed] [Google Scholar]
- 103.van Wuijckhuijse AL, Stowers MA, Kleefsman WA, van Baar BLM, Kientz CE, Marijnissen JCM. Matrix-assisted laser desorption/ionisation aerosol time-of-flight mass spectrometry for the analysis of bioaerosols: development of a fast detector for airborne biological pathogens. J Aerosol Sci. 2005;36(5–6):677–687. [Google Scholar]
- 104.Srivastava A, Pitesky ME, Steele PT, Tobias HJ, Fergenson DP, Horn JM, Russell SC, Czerwieniec GA, Lebrilla CS, Gard EE, Frank M. Comprehensive assignment of mass spectral signatures from individual Bacillus atrophaeus spores in matrix-free laser desorption/ionization bioaerosol mass spectrometry. Anal Chem. 2005;77(10):3315–3323. doi: 10.1021/ac048298p. [DOI] [PubMed] [Google Scholar]
- 105.Steele PT, Tobias HJ, Fergenson DP, Pitesky ME, Horn JM, Czerwieniec GA, Russell SC, Lebrilla CB, Gard EE, Frank M. Laser power dependence of mass spectral signatures from individual bacterial spores in bioaerosol mass spectrometry. Anal Chem. 2003;75(20):5480–5487. doi: 10.1021/ac034419u. [DOI] [PubMed] [Google Scholar]
- 106.Anhalt JP, Fenselau C. Identification of bacteria using mass-spectrometry. Anal Chem. 1975;47(2):219–225. [Google Scholar]
- 107.Demirev PA, Fenselau C. Mass spectrometry for rapid characterization of microorganisms. Annu Rev Anal Chem. 2008;1:71–93. doi: 10.1146/annurev.anchem.1.031207.112838. [DOI] [PubMed] [Google Scholar]
- 108.Amiel C, Mariey L, Denis C, Pichon P, Travert J. FTIR spectroscopy and taxonomic purpose: Contribution to the classification of lactic acid bacteria. Lait. 2001;81(1–2):249–255. [Google Scholar]
- 109.Dharmaraj S, Jamaludin AS, Razak HM, Valliappan R, Ahman NA, Harn GL, Ismail Z. The classification of Phyllanthus niruri Linn. according to location by infrared spectroscopy. Vib Spectrosc. 2006;41(1):68–72. [Google Scholar]
- 110.Cabredo S, Parra A, Saenz C, Anzano J. Bioaerosols chemometric characterization by laser-induced fluorescence: Air sample analysis. Talanta. 2009;77(5):1837–1842. doi: 10.1016/j.talanta.2008.10.030. [DOI] [PubMed] [Google Scholar]
- 111.Pysher MD, Hayes MA. Electrophoretic and dielectrophoretic field gradient technique for separating bioparticles. Anal Chem. 2007;79(12):4552–4557. doi: 10.1021/ac070534j. [DOI] [PubMed] [Google Scholar]
- 112.Staton SJR, Chen KP, Taylor TJ, Pacheco JR, Hayes MA. Characterization of particle capture in a sawtooth patterned insulating electrokinetic microfluidic device. Electrophoresis. 2010;31(22):3634–3641. doi: 10.1002/elps.201000438. [DOI] [PubMed] [Google Scholar]
- 113.Meighan MM, Keebaugh MW, Quihuis AM, Kenyon SM, Hayes MA. Electrophoretic exclusion for the selective transport of small molecules. Electrophoresis. 2009;30(21):3786–3792. doi: 10.1002/elps.200900340. [DOI] [PubMed] [Google Scholar]