Abstract
Several MADS box gene lineages involved in flower development have undergone duplications that correlate with the diversification of large groups of flowering plants. In the APETALA1 gene lineage, a major duplication coincides with the origin of the core eudicots, resulting in the euFUL and the euAP1 clades. Arabidopsis FRUITFULL (FUL) and APETALA1 (AP1) function redundantly in specifying floral meristem identity but function independently in sepal and petal identity (AP1) and in proper fruit development and determinacy (FUL). Many of these functions are largely conserved in other core eudicot euAP1 and euFUL genes, but notably, the role of APETALA1 as an “A-function” (sepal and petal identity) gene is thought to be Brassicaceae specific. Understanding how functional divergence of the core eudicot duplicates occurred requires a careful examination of the function of preduplication (FUL-like) genes. Using virus-induced gene silencing, we show that FUL-like genes in opium poppy (Papaver somniferum) and California poppy (Eschscholzia californica) function in axillary meristem growth and in floral meristem and sepal identity and that they also play a key role in fruit development. Interestingly, in opium poppy, these genes also control flowering time and petal identity, suggesting that AP1/FUL homologs might have been independently recruited in petal identity. Because the FUL-like gene functional repertoire encompasses all roles previously described for the core eudicot euAP1 and euFUL genes, we postulate subfunctionalization as the functional outcome after the major AP1/FUL gene lineage duplication event.
The evolution of MADS box genes has featured numerous duplications and losses, affecting a single species or entire clades of land plants (Purugganan, 1997; Alvarez-Buylla et al., 2000; Becker and Theissen, 2003; Hileman et al., 2006). These transcription factors play key roles in plant development, most notably floral organ identity (Bowman et al., 1991, 1993; Coen and Meyerowitz, 1991). It has been speculated that duplications and subsequent diversification of MADS box genes may have been a factor in the evolution of morphological diversity in land plants and of angiosperms in particular (Alvarez-Buylla et al., 2000; Irish and Litt, 2005; Kaufmann et al., 2005; Hands et al., 2011). For example, within angiosperms, a cluster of duplications is correlated with the diversification of the core eudicots, a clade that encompasses the vast majority (roughly 75%) of extant flowering plants and that includes major model systems such as Arabidopsis (Arabidopsis thaliana), snapdragon (Antirrhinum majus), and tomato (Solanum lycopersicum; Fig. 1). Among the gene lineages affected by this event are key regulators of floral organ identity: APETALA1 (AP1; A function), APETALA3 (AP3; B function), and AGAMOUS (AG; C function; Kramer et al., 1998, 2004; Litt and Irish, 2003; Stellari et al., 2004; Zahn et al., 2005, 2006). Comparative functional studies of AP3 and AG orthologs in monocots, basal eudicots, and core eudicots suggest that, fundamentally, the functions of these genes in petal, stamen, and carpel identity (B and C function) were conserved before and after the core eudicot duplication (Ambrose et al., 2000; Lamb and Irish, 2003; de Martino et al., 2006; Drea et al., 2007; Kramer et al., 2007; Dreni et al., 2011; Hands et al., 2011; Sharma et al., 2011). However, we lack data with which to evaluate the effect of the core eudicot duplication on the function and evolution of the AP1 lineage; furthermore, the data that do exist have led to doubts about the conservation of the A function. In particular, we lack data from basal eudicot species that would allow us to evaluate whether the core eudicot duplication resulted in changes in the function of core eudicot AP1 lineage genes.
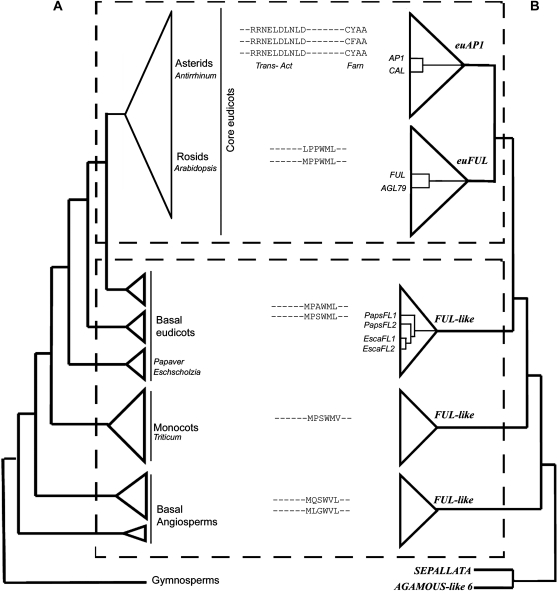
Figure 1.
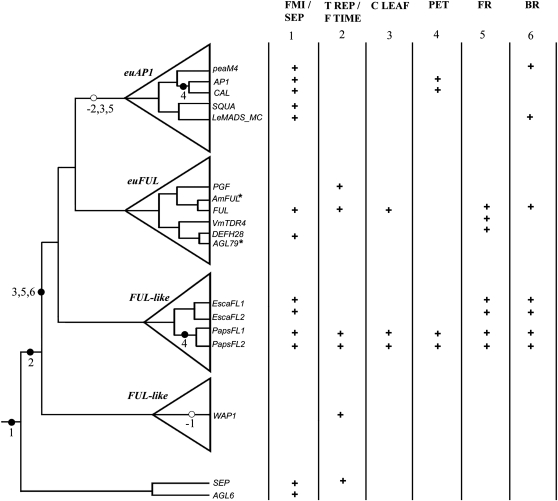
Simplified angiosperm phylogeny paired with the gene phylogeny of AP1/FUL homologs and their C-terminal motifs. This figure is based on Litt and Irish (2003) and N. Pabón-Mora and A. Litt (unpublished data). A, Simplified angiosperm phylogeny showing the five major groups of flowering plants (basal angiosperms, monocots, basal eudicots, rosids, and asterids) and indicating the phylogenetic positions of Papaver and Eschscholzia. The phylogenetic positions of Arabidopsis, Antirrhinum (snapdragon), and Triticum (wheat) are also shown. B, Simplified AP1/FUL gene lineage tree, showing the core eudicot gene duplication. The bottom dotted box indicates the groups in which FUL-like genes are found, which includes all taxa outside of the core eudicots. The top dotted box shows the euAP1 and euFUL genes in core eudicots. Arabidopsis AP1/FUL homologs (AP1, CAL, FUL, AGL79) are shown in the gene tree, as are PapsFL1, PapsFL2, EscaFL1, and EscaFL2. In the center are the C-terminal protein motifs typical of the gene groups depicted in B: the euAP1 transcriptional activation (Trans-Act) and farnesylation (Farn) motifs and the FUL-like motif characteristic of euFUL and FUL-like proteins.
As a result of the duplication, core eudicot species possess two types of AP1 lineage genes, euAP1 and euFUL (for FRUITFULL), and species outside of the core eudicots have only one type of gene (FUL-like genes; Litt and Irish, 2003; Preston and Kellogg, 2006; Shan et al., 2007; Litt and Kramer, 2010; Fig. 1). FUL-like and euFUL proteins possess a six-hydrophobic-amino-acid motif (FUL-like motif) of unknown function near the C terminus (Litt and Irish, 2003; Vandenbussche et al., 2003; Shan et al., 2007; Liu et al., 2010; Fig. 1). In contrast, the C terminus of euAP1 proteins has an acidic transcription activation motif that has been shown to activate transcription in a yeast system (Riechmann et al., 1996; Cho et al., 1999) and a farnesylation motif (CaaX) hypothesized to be important in mediating multiprotein complex formation (Yalovsky et al., 2000). The fact that new functional domains are found only in euAP1 proteins suggests that the functional capabilities they encode might be absent from FUL-like proteins.
Within the Brassicaceae, an additional duplication occurred in the euAP1 gene clade, producing the Arabidopsis paralogs CAULIFLOWER (CAL) and AP1 (Lowman and Purugganan, 1999; Alvarez-Buylla et al., 2006). AP1 and CAL are expressed in floral meristems and in developing sepal and petal primordia (Mandel et al., 1992; Kempin et al., 1995; Ferrándiz et al., 2000; Blázquez et al., 2006); expression patterns of orthologs in other core eudicots are for the most part similar, although they may also include bracts and reproductive organs (Huijser et al., 1992; Hardenack et al., 1994; Berbel et al., 2001; Shchennikova et al., 2004; Sather and Golenberg, 2009). In strong Arabidopsis ap1 mutants, sepals are converted to bract-like structures, petals are absent, and the bract-like organs of the first whorl subtend secondary flowers in the second whorl (tertiary flowers can also form; Irish and Sussex, 1990; Mandel et al., 1992; Bowman et al., 1993). AP1 also forms a protein complex with SEUSS and LEUNIG, which negatively regulates the C-function gene AGAMOUS, restricting it to the inner two whorls (Sridhar et al., 2006). CAL is redundant with AP1 for the specification of floral meristem identity (Kempin et al., 1995; Ferrándiz et al., 2000). In other core eudicots, euap1 mutants such as squamosa (squa) in snapdragon (Huijser et al., 1992), Medicago truncatula proliferating inflorescence meristem (mtpim) (Benlloch et al., 2006), and macrocalyx (mc) in tomato (Vrebalov et al., 2002) lack proper floral meristem identity and produce a ramified inflorescence with fewer flowers. The flowers show leaf-like sepals; nevertheless, petal identity is unaffected. This suggests that other euAP1 orthologs function in proper floral meristem and sepal identity, features that are structurally and developmentally linked, but not in petal identity (Huijser et al., 1992; Theissen et al., 2000; Litt, 2007; Causier et al., 2010).
FUL is expressed in cauline leaves and inflorescence meristems and later in the carpel primordia and fruits (Gu et al., 1998). Arabidopsis has a second euFUL paralog, AGAMOUS-Like79 (AGL79), although its sequence is highly divergent and it appears to be expressed in roots (Parenicová et al., 2003). In other core eudicots, the expression of euFUL genes is broader, including leaves and cauline leaves (or bracts), inflorescence and floral meristems, all floral organs, fruits, and ovules (Hardenack et al., 1994; Immink et al., 1999; Wu et al., 2000; Müller et al., 2001; Calonje et al., 2004; Shchennikova et al., 2004; Sather and Golenberg, 2009). In Arabidopsis ful mutants, floral organ identity is not affected; however, FUL is redundant with AP1 and CAL in regulating floral meristem identity (Ferrándiz et al., 2000) and has a unique function in regulating cell differentiation during fruit development (Gu et al., 1998; Liljegren et al., 2000; Ferrándiz, 2002). ful mutants also have defects in cauline leaf development (Gu et al., 1998), and FUL has been shown to participate in regulating flowering time, axillary meristem activation, meristem determinacy, and plant longevity (Melzer et al., 2008). Functional studies of other euFUL genes are scarce. In petunia (Petunia hybrida), silencing of PETUNIA FLOWERING GENE (PFG) resulted in plants that remained vegetative (Immink et al., 1999).Overexpression of an Antirrhinum euFUL paralog, DEFICIENS-homolog28 (DEFH28), in Arabidopsis (Müller et al., 2001) and of the Nicotiana tabacum FUL (NtFUL) in tobacco (Nicotiana tabacum; Smykal et al., 2007) resulted in fruits with defective lignification that failed to dehisce. These data suggest that other euFUL genes may have the same dual roles as FUL: an early role in promoting the transition to reproductive meristems and a late role in proper fruit development.
Noncore eudicot FUL-like genes, like euFUL genes, are broadly expressed in vegetative and reproductive tissues (Yu and Goh, 2000; Gocal et al., 2001; Chen et al., 2007; Preston and Kellogg, 2007, 2008; Wu et al., 2007; Danilevskaya et al., 2008); however, less is known about their functions. Studies in cereals have shown that FUL-like genes are up-regulated in leaves and meristems in response to vernalization and may promote inflorescence initiation (Murai et al., 2003; Trevaskis et al., 2003, 2007; Preston and Kellogg, 2007, 2008). Function is only known for WHEAT APETALA1-Like (WAP1), one of three FUL-like paralogs of wheat (Triticum aestivum), which is required for proper phase transition after vernalization (Murai et al., 2003). The data available suggest that FUL-like genes may be important in the transition from vegetative to reproductive meristems, but because of the lack of functional studies of FUL-like genes from species outside of the core eudicots, this remains an untested hypothesis.
Here, we present data regarding the expression and function of FUL-like genes from California poppy (Eschscholzia californica) and opium poppy (Papaver somniferum ‘Persian White’). Both taxa belong to the Papaveraceae, an early-diverging family of basal eudicots; therefore, they are evolutionary intermediates between the distantly related monocots and core eudicots. Importantly, both taxa are amenable to functional analysis using virus-induced gene silencing (VIGS; Hileman et al., 2005; Drea et al., 2007; Wege et al., 2007; Orashakova et al., 2009). Differences in floral morphology, in addition to the close relationship of these two species, make a comparison between them a robust platform from which to assess the role of FUL-like genes in basal eudicots. We test the following two alternative hypotheses regarding the functional evolution of this gene lineage. (1) Postduplication euAP1 and euFUL genes retained an ancestral role in mediating the floral transition and in floral meristem identity but acquired new functions in perianth identity and fruit development, respectively. These new functions might be associated with changes in protein interactions and, in the euAP1 clade, with new sequence motifs. (2) Postduplication euFUL and euAP1 genes diverged functionally after the core eudicot duplication event without acquiring new functions. According to this hypothesis, the “new” motifs of the euAP1 proteins do not confer novel functions. Rather, noncore eudicot FUL-like proteins encode the same functions as euAP1 and euFUL proteins combined.
RESULTS
California poppy and opium poppy are annual herbs that grow vegetatively as rosettes. Because both species have terminal flowers, the reproductive meristem initially has an inflorescence character, forming cauline leaves subtending axillary buds. However, after producing four to six cauline leaves, the apical meristem becomes a terminal floral meristem. Whereas in opium poppy there is a single terminal flower and dormant axillary buds (only in the lowermost cauline leaves), in California poppy the reproductive axis develops into a multiple-flowered cymose inflorescence. Each cyme has three floral buds, one large terminal flower, and two smaller lateral flowers. There are three orders of branching in this species. Branches or flowers may develop from buds on the main axis (first order), from buds on the first order branches (second order), and from buds on the second order branches (third order; Supplemental Fig. S1A). Flowers are very similar in the two species, having two sepals, two alternate whorls of two petals, a large number of stamens, and two (in California poppy) to eight (in opium poppy) carpels. The two species differ in the morphology of the sepals and fruit. Opium poppy has free sepals and a superior ovary that develops into a capsule that releases the seeds though apical pores. In contrast, California poppy has fused sepals and a floral cup that surrounds the semi-inferior ovary, which becomes a longitudinally dehiscent capsule. In addition, the floral cup delimits the lower persistent versus the upper deciduous region of the sepals during anthesis. Developmental landmarks in inflorescence and flower morphology have been described for these species (Ronse De Craene and Smets, 1990; Becker et al., 2005; Drea et al., 2007).
Identification of Opium Poppy and California Poppy FUL-like Genes
Two FUL-like paralogs in opium poppy, PapsFL1 and PapsFL2 (for Papaver somniferum), had been identified previously (Litt and Irish, 2003). The full-length coding sequences share 78% nucleotide identity and 60% amino acid identity. Using degenerate primers designed to amplify AP1/FUL genes (following Litt and Irish [2003]), we also identified two FUL-like genes in California poppy, EscaFL1 and EscaFL2; these share 82% nucleotide identity and 80% amino acid identity. Opium poppy and California poppy FUL-like genes fall within the basal eudicot FUL-like gene clade, consistent with the phylogenetic position of these species, although the duplications appear to be independent (Fig. 1B; N. Pabón-Mora and A. Litt, unpublished data). Sequence analysis predicts that all the proteins from opium poppy and California poppy possess the conserved FUL-like C-terminal motif (Supplemental Fig. S2).
Expression of FUL-like Genes in Opium Poppy
Following the floral development stages defined by Drea et al. (2007), we evaluated FUL-like gene expression in floral organs at late preanthesis stages of development (stages P7, petal expansion initiated, and P8, petals fully expanded inside the sepals) and anthesis, as well as in fruits and leaves, using reverse transcription (RT)-PCR. The results (Fig. 2A) show that both PapsFL1 and PapsFL2 are broadly expressed before and at anthesis in all floral parts as well as in leaves and the fruit.
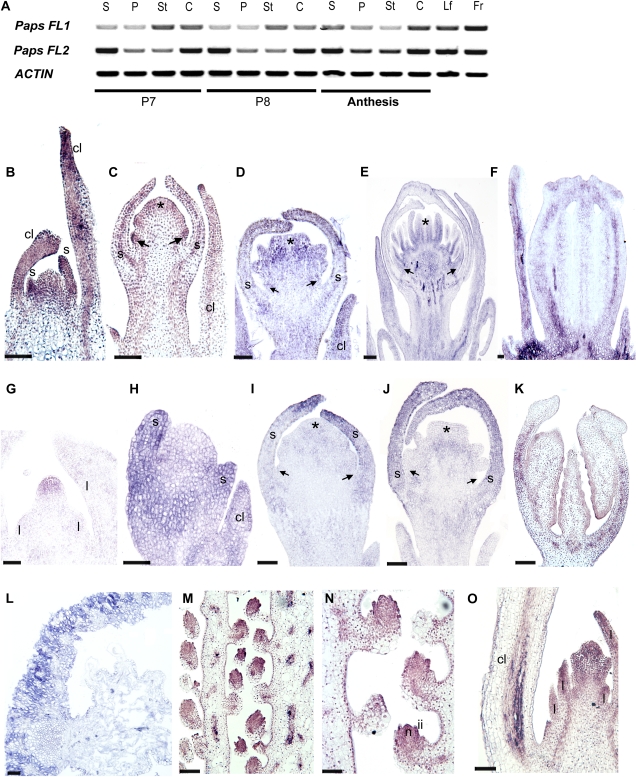
Figure 2.
Expression of PapsFUL-like genes at different developmental stages. A, RT-PCR results showing the expression of PapsFL1 and PapsFL2 in dissected floral organs at different floral bud stages and in flowers at anthesis as well as in fruits and leaves. Stages are based on Drea et al. (2007). At bud stage P7, petal primordia have not elongated; at bud stage P8, petals are fully expanded inside the floral bud. B to O, In situ mRNA hybridization. B to F, Expression of PapsFL1. G to K, Expression of PapsFL2. L to O, Expression common to PapsFL1 and PapsFL2 (L, PapsFL1 section; M–O, PapsFL2 sections). B and H, Early floral meristem; sepal primordia are starting to differentiate. C and I, Floral bud with large sepals protecting the incipient petal, stamen, and carpel primordia. D and J, Floral bud with overlapping sepals and fully differentiated petal, stamen, and carpel primordia. E, Floral bud with clearly defined anther and filament. F and K, Longitudinal section of the carpel in a preanthesis floral bud. G, Shoot apical meristem before the transition to flowering. L, Cross-section of the young fruit showing the fruit wall. M and N, Longitudinal section of the ovary showing placenta and ovules. O, Dormant axillary bud subtended by the lowermost cauline leaves. c, Carpel; cl, cauline leaf; fr, fruit; ii, inner integument; l, leaf primordia; lf, leaf; n, nucellus; p, petal; s, sepal; st, stamen. Arrows indicate petal primordia, and asterisks indicate carpel primordia. Bars = 50 μm (B–E, H–J, and O), 70 μm (G), 150 μm (F and K ), 100 μm (L), and 120 μm (M and N).
A detailed examination of expression during early floral development (stages P0–P6) was performed using in situ mRNA hybridization. The results show that PapsFL1 is expressed in developing cauline leaves throughout development (Fig. 2, B–D) but is absent from the vegetative meristem (data not shown). Expression is seen in the young floral meristem, especially in sepal primordia (Fig. 2B). During early development at stage P3 (petal, stamen, and carpel primordia are visible), PapsFL1 is expressed in all floral organ primordia (Fig. 2C) and in the floral pedicel, and this expression continues through stage P6 (Fig. 2, D and E). During stage P6, PapsFL1 expression is evident in the carpel wall (Fig. 2F) and the septa toward the tip of the carpel (data not shown). PapsFL2 is detected in the vegetative meristem (Fig. 2G), and it is also expressed in the developing cauline leaves during flower development, similar to PapsFL1 (Fig. 2H). Starting in the stage P3 young floral meristem, PapsFL2 expression becomes localized to the sepals (Fig. 2, H–J). It appears to be absent from petal, stamen, and carpel primordia during stages P4 (Fig. 2J) and P5 (data not shown), but it is detected again at P6 in the carpel wall (Fig. 2K). PapsFL1 and PapsFL2 show a common expression pattern in the fruit wall (Fig. 2L) and the ovules (Fig. 2, M and N), particularly in the inner integument and the nucellus at anthesis. In addition, both copies are also expressed in the dormant axillary meristems subtended by the lowermost cauline leaves (Fig. 2O). Hybridization with sense PapsFL1 and PapsFL2 probes showed no signal (Supplemental Fig. S3). An opium poppy PISTILLATA (PI) probe (Drea et al., 2007) was used as a positive control, and we detected its expression as described previously (Supplemental Fig. S3).
Expression of FUL-like Genes in California Poppy
Using RT-PCR, we evaluated FUL-like gene expression in California poppy floral organs at the same late preanthesis developmental stages as in opium poppy (stages P7 and P8) and at anthesis as well as in fruits and leaves. The results show that EscaFL1 and EscaFL2 are broadly expressed in all floral tissues at P7, P8, and anthesis, and both genes are expressed in fruits and leaves (Fig. 3A).
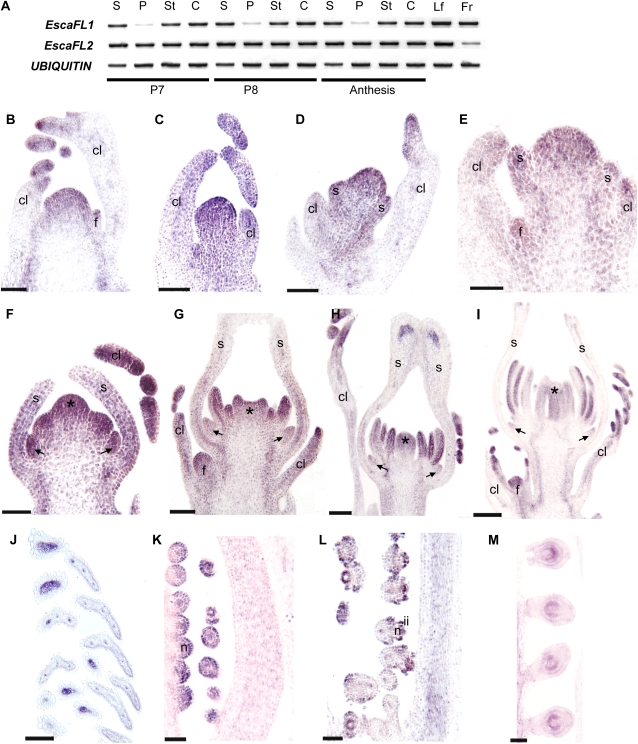
Figure 3.
Expression of EscaFUL-like genes at different developmental stages. A, RT-PCR results showing the expression of EscaFL1 and EscaFL2 in floral buds (P7 and P8) and different parts of the flower at anthesis as well as in leaves and fruits. B to M, In situ mRNA hybridization of EscaFL1 and EscaFL2. Expression of EscaFL1 and EscaFL2 is identical (B, E–H, and J–M, EcFL1 sections; C, D, G, I, and N, EscaFL2 sections). B, Inflorescence meristem protected by two highly dissected cauline leaves. C and D, Axillary inflorescence showing the terminal flower before sepal initiation (C) and at the beginning of sepal initiation (D). E, Axillary inflorescence showing a terminal flower with sepal primordia and a floral meristem axillary to the cauline leaf on the left. F, Floral bud with elongating sepals enclosing petal, stamen, and carpel primordia. G, Floral bud with fused sepals around petal, stamen, and carpel primordia. H, Floral bud with fully closed carpel wall. I, Floral bud with sporogenous tissue in the stamens. Sepals were forced open artificially in this sample. J, Tips of the highly dissected mature cauline leaves. K to M, Early stages of ovule development showing the nucellus (K), the initiation of the two integuments (L), and the fully formed bitegmic ovules (M). c, Carpel; cl, cauline leaf; f, axillary floral meristem; fr, fruit; ii, inner integument; im, inflorescence meristem; lf, leaf; n, nucellus; p, petal; s, sepal; st, stamen. Arrows indicate petal primordia, and asterisks indicate carpel primordia. Bars = 60 μm (B–F), 100 μm (G–J), and 50 μm (K–M).
In situ mRNA hybridization during early floral development (stages P0–P6) indicates that at the tissue level, expression patterns of EscaFL1 and EscaFL2 are identical. EscaFUL-like genes (both copies) are strongly expressed throughout the inflorescence meristem (Fig. 3B), in the provascular strands of the main reproductive axis (Fig. 3B), and in the primordia and growing tips of the cauline leaves (Fig. 3, B–J). EscaFUL-like genes are also expressed throughout the young floral meristem before (Fig. 3C) and during (Fig. 3, D and E) early sepal differentiation. During stage P3, California poppy FUL-like genes continue to be expressed in the sepal primordia, and expression begins in petal, stamen, and carpel primordia (Fig. 3F). Expression in the sepals decreases basipetally during stage P4, particularly after the apical fusion of the sepals (Fig. 3, G and H), and it remains only associated with the vasculature at the apex of the sepals (Fig. 3H). During stage P5, expression decreases in petals (Fig. 3, G–I) but remains strong in stamens and carpels (Fig. 3, H and I). During stage P6, the two genes are expressed in the carpel wall (Fig. 3L) and the developing ovule, particularly in the nucellus (Fig. 3, K–M) and during the initiation of the two integuments (Fig. 3L). Hybridization with the sense EscaFUL-like probes showed no signal, and EscaAG (the California poppy AG ortholog) gene expression was used as a positive control (Supplemental Fig. S3).
Silencing of PapsFUL-like Genes Using TRV-VIGS
To investigate the function of FUL-like genes in basal eudicots, we used VIGS, a transient posttranscriptional gene-silencing mechanism that promotes the degradation of target endogenous mRNAs in the plant (Dinesh-Kumar et al., 2003; Burch-Smith et al., 2004; Liu and Page, 2008). We used the bipartite Tobacco rattle virus, in which RNA1 (or TRV1) encodes the viral replicase and the RNA-dependent RNA polymerase and RNA2 (or TRV2) encodes the coat protein and has a multiple cloning site for insertion of the endogenous target sequence. This approach has been successfully implemented in poppies (Hileman et al., 2005; Drea et al., 2007; Wege et al., 2007; Yellina et al., 2010) and other Ranunculales species (Gould and Kramer, 2007; Kramer et al., 2007; Di Stilio et al., 2010). In order to specifically silence each FUL-like paralog in opium poppy, we generated TRV2 constructs carrying a short fragment of PapsFL1 or PapsFL2 (Supplemental Fig. S2). In addition, the two constructs were mixed together to silence both copies simultaneously.
PapsFL1
A total of 100 seedlings with zero to two true leaves were infiltrated with TRV1 and TRV2-PapsFL1 (Supplemental Fig. S2; Supplemental Table S1). Seedlings were grown until flowering, and cauline leaves, sepals, and fruits were screened using RT-PCR for the presence of TRV1 and TRV2 and for the levels of PapsFL1 transcript (Supplemental Fig. S4A). Initially, based on RT-PCR results, plants were classified as unsilenced (those treated with TRV2-PapsFL1 but showing no noticeable reduction in transcript) or showing mild, moderate, or strong down-regulation. However, similar mutant phenotypes were observed at all levels of down-regulation. Only the severity of the phenotypes and the number of flowers per plant displaying a specific phenotype increased as transcript levels were reduced. In 32% (n = 27) of the treated plants, there was down-regulation of PapsFL1, with no down-regulation of PapsFL2, demonstrating that the TRV2:PapsFL1 construct was gene specific (Supplemental Fig. S4A). The reduction of transcript abundance was confirmed using quantitative (q)RT-PCR (Supplemental Fig. S4B), and these experiments also suggest that there may be an increase of PapsFL2 transcript levels upon the down-regulation of PapsFL1. Plants were examined for novel phenotypes throughout development. Whereas wild-type opium poppy plants possess a single terminal flower (Fig. 4A), 78% of the plants with reduced levels of PapsFL1 transcript (n = 21) showed outgrowth of axillary branches after the terminal flower senesced and fruit development was initiated (Fig. 4B). The number of branches ranged from two to six (Fig. 4B). In 38% (n = 8) of the branching papsfl1 plants, the vegetative and cauline leaves showed apparent abnormal meristematic activity, resulting in broader, deeply lobed laminas accompanied by multiple midveins (Fig. 4, D and E). In addition, in contrast to wild-type cauline leaves, which decrease in size from the base to the apex of the inflorescence stem (Fig. 4A), the papsfl1 cauline leaves were more uniform in size and larger than the wild type (Fig. 4B). Analysis of epidermal cells showed no abnormalities compared with the wild type (data not shown).
Figure 4.
Inflorescence and cauline leaf phenotypes in opium poppy plants treated with TRV2-PapsFL1 and TRV2-PapsFL2 separately. A, Wild-type opium poppy. B, Plants down-regulated for papsfl1. C, Plants down-regulated for papsfl2. D to F, Leaf clearings of cauline leaves in wild-type opium poppy (D), papsfl1 plants (E), and papsfl2 plants (F). Two cauline leaves are presented in each panel to illustrate the variation of size and shape from larger, more basal cauline leaves (left) to smaller, more apical cauline leaves (right). White arrows indicate axillary branches. Bars = 5 cm (A–C) and 1 cm (D–F).
In 47% (n = 10) of the branching papsfl1 plants, the sepals of one to three flowers per plant exhibited apparent homeotic transformation to leaf-like organs. They were characterized by lobed margins and a waxy cuticle and remained attached to the base of the flower after anthesis and during fruit development (Fig. 5, B and G). This is in contrast to the early deciduous sepals of wild-type flowers (Fig. 5A), which lack a waxy cuticle and have entire margins (Fig. 5D). These leaf-like organs, furthermore, exhibited typical leaf cell types (Fig. 5, H and I), in contrast to the cell types characteristic of sepals (Fig. 5, E and F).
Figure 5.
Floral phenotypes in opium poppy plants treated with TRV2-PapsFL1 and TRV2-PapsFL2 separately. A, Wild-type opium poppy flower after deciduous sepals have fallen. B and C, Flowers showing persistent leaf-like sepals that remain attached to the base of the flower in papsfl1 plants (B) and papsfl2 plants (C). D, Wild-type sepals. E and F, SEM of the abaxial (E) and adaxial (F) wild-type sepal surfaces. G, Persistent papsfl1 leaf-like sepals during fruit development. H and I, SEM of the abaxial (H) and adaxial (I) leaf-like papsfl1 sepal surface. J, Persistent papsfl2 leaf-like sepals during fruit development. K and L, SEM of the abaxial (K) and adaxial (L) leaf-like papsfl2 sepal surface. M, Wild-type opium poppy petals. N and O, SEM of the abaxial (N) and adaxial (O) wild-type petal surfaces. P, papsfl1 flower showing small green patches on the abaxial surface of the outer petals. Q and R, SEM of the petaloid abaxial (Q) and adaxial (R) papsfl1 petal surfaces. S, papsfl2 flowers showing small green areas in the distal portion of the petal. T and U, SEM of the petaloid abaxial (T) and adaxial (U) papsfl2 petal surfaces. V, Wild-type immature opium poppy fruit (poricidal capsule formed by eight fused carpels). W, Cross-section of the wild-type fruit showing main vascular bundles of each placenta. X, papsfl1 fruit. Y, Cross-section of papsfl1 fruit. Z, papsfl2 fruit. AA, Cross-section of papsfl2 fruit. p, Placenta. Black arrows point to vascular traces in the pericarp, and white arrows point to green patches on the petals. Bars = 1.5 cm (A–C), 0.5 cm (D, G, J, P, V, X, and Z), 40 μm (E, F, H, I, K, L, N, O, Q, R, T, and U), 0.7 cm (M and S), and 500 μm (W, Y, and AA ).
In general, papsfl1 plants did not show defects in petal identity (Fig. 5, B, Q, and R) with the exception of occasional small green patches (Fig. 5P), the epidermis of which exhibited cells that appeared to be leaf or carpel like (data not shown). For the most part, the petal laminas exhibited the elongated rectangular cells typical of wild-type petal epidermis (Fig. 5, N and O). The identity of stamens and carpels was not affected in VIGS-treated plants (Supplemental Table S1). However, 23% (n = 5) of the papsfl1 plants exhibited carpel defects, such as asymmetrical elongation of the carpel wall resulting in bending of the carpel and the fruit, and premature rupture of the fruits before full seed maturation (Fig. 5, V and X). These fruits had lignification and placentation defects and irregularly thickened pericarps (Fig. 5, W and Y). Nevertheless, epidermal features of the fruit are unchanged (data not shown). None of these abnormal phenotypes were observed in plants treated identically but transformed with an empty TRV2 vector lacking the target sequence (Supplemental Table S1; data not shown).
PapsFL2
We infiltrated 80 seedlings of opium poppy with TRV1 and TRV2-PapsFL2. Seedlings were grown and screened as for PapsFL1. In 19% (n = 13) of the plants, there was down-regulation of PapsFL2 with no change or even an increase of PapsFL1 transcript abundance (Supplemental Fig. S4, C and D). As in papsfl1 plants, the extent of down-regulation was somewhat correlated with the severity of the mutant phenotype, but the types of defects were the same regardless of the degree of down-regulation. Phenotypes were similar to those observed for PapsFL1, including branched inflorescences (n = 10; Fig. 4C; Supplemental Table S1), abnormally broad, lobed cauline leaves (n = 5; Fig. 4, C and F), partial or total homeotic conversion of deciduous sepals to persistent leaf-like organs with leaf-like epidermal cells (n = 7; Fig. 5, C and J), and defects in carpel and fruit development (n = 4; Fig. 5Z), including anatomical defects and premature fruit rupture similar to what was observed in papsfl1 plants (Fig. 5AA). Petals were mostly unaffected (Fig. 5, S–U); however, they occasionally developed distal abaxial green areas (Fig. 5, C and S) with leaf- or carpel-like epidermal cells similar to what was seen in papsfl1 plants (data not shown).
PapsFL1-FL2
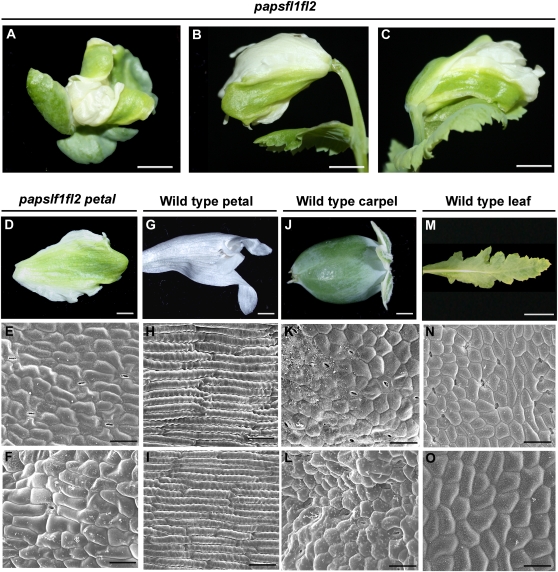
A total of 108 seedlings of opium poppy were infiltrated with TRV1, TRV2-PapsFL1, and TRV2-PapsFL2 in a 1:1:1 ratio. Seedlings were grown and screened as before. Some plants showed down-regulation of only one copy, as expected due to the heterogeneous systemic spread of the two different vectors. Plants showing down-regulation of only one gene were not studied further. A total of 13.8% (n = 15) of the plants showed down-regulation of both PapsFL1 and PapsFL2 (Supplemental Fig. S4, E and F; henceforth referred to as papsfl1-fl2). Of the double down-regulated plants, 11% (n = 12) showed a delay in reproductive transition, as evidenced by bolting that was 1 to 2 weeks later than in wild-type plants. To confirm this observation, the number of total leaves was counted. Untreated wild-type plants (n = 12) produced an average of 11 ± 0.20 leaves, and treated plants that showed no down-regulation produced an average of 11 ± 0.60 leaves (n = 8) before producing an inflorescence axis; in contrast, papsfl1-fl2 plants (n = 12) produced 18 ± 0.56 leaves (P < 0.001). Morphological phenotypes observed for papsfl1-fl2 plants were similar to those documented for the down-regulation of each gene independently and included branching in the inflorescence (n = 7), overgrowth and shape defects in cauline leaves (n = 5), leaf-like sepals (n = 5), and defects in carpel and fruit development (n = 5; data not shown). In addition, a novel morphological phenotype emerged when both copies were simultaneously down-regulated. Double papsfl1-fl2 plants showed large patches of green tissue on the abaxial and adaxial surfaces of the two outer petals, in some cases occupying up to 70% of the petal area (Fig. 6, A–D). Scanning electron microscopy (SEM) examination showed a loss of wild-type petal cell identity on both the abaxial (Fig. 6, E and H) and adaxial (Fig. 6, F and I) surfaces, replaced by apparent leaf- or carpel-like cell identity (Fig. 6, J–O). To test whether this phenotype was associated with the ectopic expression of C-function genes, we evaluated transcript levels of PapsAG1 and PapsAG2 in the green petals and carpels of papsfl1-fl2. PapsAG1 and -2 have been shown to specify stamen and carpel identity in opium poppy; down-regulation of both PapsAG homologs concurrently resulted in a homeotic transformation of the third and fourth whorl androecium and gynoecium into petaloid and sepaloid and organs, respectively (Hands et al., 2011). However, the leaf- or carpel-like tissue in the papsfl1-fl2 petals was not associated with the overexpression of PapsAG1 or PapsAG2 when compared with the wild-type petal (data not shown). Interestingly, the inner two petals in down-regulated flowers always showed wild-type morphology and epidermis.
Figure 6.
Floral phenotypes in opium poppy plants treated with TRV2-PapsFL1 and TRV2-PapsFL2 simultaneously. A to C, Double down-regulated plants showing large green areas in the two outer petals in opium poppy. A, Front view. B and C, Side views. D, Dissected papsfl1-papsfl2 green petal. E and F, SEM of the abaxial (E) and adaxial (F) papsfl1-papsfl2 petal surface. G, Dissected wild-type petal. H and I, SEM of the abaxial (H) and adaxial (I) wild-type petal surface. J, Dissected wild-type carpel. K and L, SEM of the abaxial (K) and adaxial (L) surface of young wild-type carpel. M, Dissected wild-type leaf. N and O, SEM of the abaxial (N) and adaxial (O) surface of wild-type leaf. Bars = 0.75 cm (A–C), 0.5 cm (D, G, J, and M), 50 μm (E, F, H, I, N, and O), and 30 μm (K and L).
Interactions among Opium Poppy FUL-like Proteins
Since the single papsfl1 and papsfl2 down-regulated plants showed identical phenotypes, we wanted to test whether PapsFL1 and PapsFL2 could potentially interact to function as a dimer. We used a yeast two-hybrid system to test for potential homodimerization and heterodimerization of PapsFL1 and PapsFL2 (Supplemental Fig. S5). Interaction between PapsFL1 and PapsFL2 is strong in both directions. PapsFL1 proteins form weak homodimers in the yeast system, but PapsFL2 proteins do not (Supplemental Fig. S5).
Silencing of EscaFUL-like Genes Using TRV-VIGS
To silence California poppy (Esca) FUL-like genes, we designed two different vectors, TRV2:EscaFL1 and TRV2:EscaFL2, carrying gene-specific inserts (Supplemental Fig. S2). Sixty seedlings with zero to two true leaves were infiltrated with TRV1 and TRV2:EscaFL1 and 60 with TRV1 and TRV2:EscaFL2. Seedlings were grown and screened as for the opium poppy experiments. A total of 40 plants showing abnormal phenotypes and 17 with wild-type appearance were screened. All 40 plants with abnormalities showed reduced transcript levels. However, independent of the TRV2 vector used, both EscaFL1 and EscaFL2 showed some degree of down-regulation (Supplemental Fig. S4, G and H; henceforth referred to as escafl1-fl2).
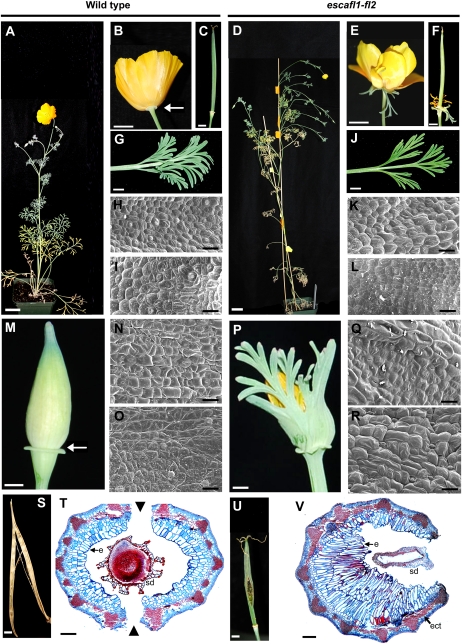
The phenotypes of escafl1-fl2 plants were similar to those observed in papsfl1 and papsfl2 plants. As with poppy, specific phenotypes were not correlated with the degree of down-regulation of FUL-like transcripts, but the severity of the phenotypes was somewhat correlated. escafl1-fl2 plants showed a significant increase in the number of first- and second order branches (P < 0.001 for both) compared with the wild type (Fig. 7, A and D; Table I; Supplemental Fig. S1). First order branches also grew longer, developing more leaves and associated axillary floral meristems. In contrast to opium poppy, cauline leaves in the down-regulated California poppy plants were identical in size, shape, and epidermal features to the wild type (Fig. 7, G–L). In addition, although escafl1-fl2 plants grew taller and had more branches, there was no observed difference in flowering time when compared with the wild-type plants growing side by side. In 100% (n = 40) of the escafl1-fl2 branched plants, one or both sepals of one to four flowers per inflorescence were partially or completely transformed into persistent leafy organs that remained attached to the base of the flower (Fig. 7, E and P) and fruit (Fig. 7F), unlike the wild-type deciduous sepals (Fig. 7, B, C, and M). Complete transformation was associated with the loss of the floral cup (Supplemental Fig. S6, J–L). These unfused organs possessed highly dissected margins characteristic of leaves, instead of the normal smooth-margined, fused calyx found in wild-type plants (Fig. 7, E, M, and 7P; Supplemental Fig. S6). They also have epidermal features similar to those of leaves (Fig. 7, Q, and R), in contrast to those of wild-type sepals (Fig. 7, N and O). escafl1-fl2 plants did not show defects in petal, stamen, or carpel identity (Fig. 7E; Supplemental Table S1). In addition, because leaf-like sepals were persistent at the base of the flower, in some escafl1-fl2 plants the other floral organs did not expand properly and remained trapped inside the leafy sepals (Supplemental Fig. S6).
Figure 7.
Floral phenotypes in California poppy plants treated with TRV2-EscaFL1 and TRV2- EscaFL2. A to C, Wild-type California poppy plant (A) and flower showing the persistent floral cup after sepal abscission (B) and fruit (C). D to F, Down-regulated escafl1-fl2 plant showing increased branching compared with the wild type (D) and leaf-like organs replacing the sepals and persisting after anthesis (E) and during fruit development (F). G, Wild-type cauline leaf. H and I, SEM of the abaxial (H) and adaxial (I) wild-type cauline leaf surface. J, Cauline leaves in escafl1-fl2 plants. K and L, SEM of the abaxial (K) and adaxial (L) escafl1-fl2 cauline leaf surface. M, Wild-type fused sepals before anthesis. N and O, SEM of the abaxial (N) and adaxial (O) wild-type sepal surface. P, Homeotic transformation of sepals into leaf-like organs. Q and R, SEM of the abaxial (Q) and adaxial (R) leaf-like escafl1-fl2 sepal surface. S, Dehisced wild-type fruit. T, Cross-section of the wild-type fruit before dehiscence showing the ring of lignified tissue and the two dehiscence zones. U, escafl1-fl2 fruit showing premature rupture of the fruit wall and exposure of the immature seeds. V, Cross-section of the escafl1-fl2 fruit showing the lignified ring interrupted by a thinner, weaker section of the pericarp through which the fruit ruptures. White arrows point to the abscission zone between the floral cup and the deciduous portion of the sepals; arrowheads point to the dehiscence zones in the fruit. e, Endocarp; ect, ectopic lignification; sd, seed. Bars = 3 cm (A and C), 0.5 cm (B, D–F, I, R, and T), 0.3 cm (L and O), 20 μm (G, H, J, K, M, N, P, and Q), and 0.1 mm (S and U).
Table I. Comparisons between number of first and second order branches in California poppy wild-type plants and escafl1-fl2 plants.
Values are means ± se. n = 18 for both groups. ANOVA (α = 0.005) first order branching P < 0.001; ANOVA (α = 0.005) second order branching P < 0.001.
| Branches | Wild Type | escafl1-fl2 |
| No. of first order branches | 5.4 ± 0.62 | 9.16 ± 0.5 |
| No. of second order branches | 3.16 ± 0.66 | 9.11 ± 1.04 |
Down-regulation of the two EscaFL paralogs in California poppy produced fruit defects similar to those seen in opium poppy. In 10% (n = 4) of the escafl1-fl2 plants, we observed shorter fruits and premature rupture of the carpel wall (Fig. 7, S–V). This may be the result of ectopic lignification of the pericarp and irregular growth of the innermost layer of the fruit (i.e. the inner endocarp; Fig. 7V). In the wild-type fruit, lignification is only associated with vascular traces, and the cells of the innermost layer of the fruit are slightly larger than cells in adjacent layers (Fig. 7T). In the down-regulated plants, lignification is also seen between vascular bundles, and the cells of the inner layers of the pericarp are dramatically elongated (Fig. 7V).
DISCUSSION
The Expression Domain of Basal Eudicot FUL-like Genes Includes the Domains of Both euFUL and euAP1 Homologs
In general, opium poppy and California poppy FUL-like genes have broad expression patterns throughout plant development (Figs. 2 and 3). After germination, FUL-like genes in both species are expressed in leaves. Once the plant has transitioned to reproduction, transcripts are found throughout the inflorescence and floral meristems, in the growing tips of cauline leaves, and in the axillary meristems. Expression of FUL-like genes is maintained during the early differentiation of all floral organs (stages P3 and P4), with the exception of PapsFL2 expression, which is restricted to sepals. At later stages (stages P5 and P6), EscaFL1 and EscaFL2 expression becomes localized to stamens and carpels. Both opium poppy and California poppy FUL-like genes are expressed in the carpel wall and ovules and during fruit development.
These expression patterns are consistent with those reported for FUL-like genes from basal angiosperms, monocots, and basal eudicots as well as those for core eudicot euFUL genes. Expression of these genes tends to be broad, reported often in leaves, bracts, or cauline leaves, inflorescence and floral meristems, most or all floral organs, fruits, and ovules (Fornara et al., 2004; Litt, 2007; Chen et al., 2008; Danilevskaya et al., 2008; Preston and Kellogg, 2008; Sather and Golenberg, 2009). In contrast, the expression patterns of euAP1 genes are very different; euAP1 expression is mainly restricted to floral meristems and floral organs, particularly sepals and petals, with expression also reported in some species in bracts, carpels, and ovules (Bowman et al., 1993; Hardenack et al., 1994; Ferrándiz et al., 2000; Berbel et al., 2001; Shchennikova et al., 2004; Sather and Golenberg, 2009). Our data and previously published data indicate that expression of FUL-like genes is present at all the developmental stages and in all the spatial domains that have been reported for the euFUL and euAP1 genes. Thus, the preduplication expression domain of AP1/FUL genes includes a broad range of stages and organs; after the core eudicot duplication, the euFUL genes maintained this broad expression, whereas euAP1 expression became restricted to a narrower domain consisting mainly of the floral meristem, sepals, and petals. This differential repression (or the lack of up-regulation) in vegetative organs and reproductive floral organs may account for the functional differences of euAP1 genes when compared with the euFUL and FUL-like genes.
FUL-like Genes Function Pleiotropically during Plant Development
We have shown that down-regulation of FUL-like genes in the two Papaveraceae species results in changes in inflorescence architecture and defects in floral meristem identity, as shown by the homeotic transformation of sepals into leaf-like organs (Figs. 5, D, G, and J, and 7, M and P). In addition, fruit development is abnormal and fruits rupture prematurely (Figs. 5, V–AA, and 7, S–V). These phenotypes are consistent with functions commonly described for other AP1/FUL homologs. Arabidopsis AP1, CAL, and FUL as well as euAP1 orthologs in other species such as snapdragon, tomato, pea (Pisum sativum), and M. truncatula have been shown to be involved redundantly in the determination of proper floral meristem identity (Huijser et al., 1992; Berbel et al., 2001; Taylor et al., 2002; Vrebalov et al., 2002; Benlloch et al., 2006). However, because the redundancy of euAP1 genes with euFUL genes has not been investigated except in Arabidopsis, it is unclear whether proper floral meristem identity is a common role for other euFUL genes. In addition, the fact that homeotic conversion of sepals into leaf-like organs in Eschscholzia results in the loss of the floral cup suggests that the differentiation of this structure is dependent on correct floral meristem and sepal identity. FUL plays a role in maintaining proper carpel wall growth during fruit development in Arabidopsis, and experiments overexpressing the euFUL gene DEFH28 from snapdragon in Arabidopsis (Müller et al., 2001) or silencing euFUL copies such as VmTR4 from bilberry (Vaccinium myrtillus; Jaakola et al., 2010) and Solanum lycopersicum MADS-box protein7 (SlMBP7) from tomato (R. Meyer, N. Pabón-Mora, and A. Litt, unpublished data) suggest that other euFUL genes also play a role in fruit development. We have shown that FUL-like genes in Papaveraceae also play a role in proper fruit development similar to that of FUL and other euFUL genes, including ectopic lignification of the mesocarp (Gu et al., 1998; Müller et al., 2001; Smykal et al., 2007; Jaakola et al., 2010); this and other abnormalities could be responsible for premature rupture of the fruit wall. Thus, our data from Papaveraceae show that their FUL-like genes (1) function in proper floral meristem and sepal identity, similar to AP1 and other euAP1 genes, and (2) are required for proper fruit wall growth and cell differentiation, similar to euFUL genes.
We have further shown that down-regulation of both FUL-like copies in opium poppy results in additional phenotypes that were not observed in doubly down-regulated California poppy plants (Fig. 6). This may be a consequence of the occurrence of different gene duplication events in Papaver and Eschscholzia (N. Pabón-Mora and A. Litt, unpublished data). Double papsfl1-fl2 mutant plants exhibit delayed flowering, and a similar role in the reproductive transition has been reported for FUL-like and euFUL genes (Immink et al., 1999; Murai et al., 2003). However, down-regulation of California poppy FUL-like genes does not appear to affect flowering time. Because this phenotype is not universal for down-regulated Papaveraceae FUL-like genes, and because all down-regulated opium poppy plants eventually did flower, it is likely that FUL-like genes are redundant with other key genes that control the transition to flowering, as has been suggested for FUL in Arabidopsis (Ferrándiz et al., 2000).
In addition, papsfl1-fl2 double mutant plants exhibit mosaic outer petals with large green patches and abnormal epidermal cell identity (Fig. 6, A–I). This suggests that in opium poppy, FUL-like genes, similar to Arabidopsis AP1, are required for the proper specification of floral meristem and perianth identity. The loss of sepal identity is linked to the loss of floral identity; likewise, the loss of petal identity may be linked to the loss of floral identity or may be an independent function of FUL-like genes. No other AP1/FUL homolog besides AP1 has been shown to be an A-function gene as defined in the ABC model of floral organ identity (Coen and Meyerowitz, 1991), specifying both sepal and petal identity (Berbel et al., 2001; Taylor et al., 2002; Murai et al., 2003; Benlloch et al., 2006; Melzer et al., 2008). Nonetheless, two major differences can be noted in the functions of opium poppy FUL-like genes and Arabidopsis AP1 in the second whorl: (1) in opium poppy, only the outermost whorl of petals is affected, whereas in Arabidopsis, all petals, which are in a single whorl, are affected; and (2) the second whorl organs of opium poppy consist of mosaic organs with both petal- and leaf-like or carpel-like epidermal cells, suggesting a homeotic transformation, whereas in Arabidopsis, the second whorl petals are absent and instead the second whorl consists of ectopic meristems in the axils of the leaf-like first whorl organs (Irish and Sussex, 1990; Bowman et al., 1993). Therefore, although in both cases the down-regulation of AP1/FUL lineage genes results in the loss of sepal and petal identity, the phenotype in the second whorl is different enough to suggest differences in the developmental pathways leading to the loss of petal identity.
In addition to specifying floral organ identity, A-function and C-function genes are also expected to repress each other, thereby defining the boundary between the outer sterile and inner reproductive organs of the flower (Coen and Meyerowitz, 1991; Bowman et al., 1993; Gustafson-Brown et al., 1994). In Arabidopsis, AP1 plays a role in regulating AG expression through the formation of a complex with SEUSS and LEUNIG (Sridhar et al., 2004, 2006); however, loss of AP1 function does not result in ectopic expression of AG (Bowman et al., 1991; Gustafson-Brown et al., 1994; Yanofsky, 1995) or carpeloid characteristics in the first whorl (Irish and Sussex, 1990; Coen and Meyerowitz, 1991; Ferrándiz et al., 2000). In wild-type opium poppy flowers, AG homologs are expressed at low levels in sepals and petals (Hands et al., 2011); thus, C-class gene expression is not restricted to the inner two whorls as in Arabidopsis. We showed that in opium poppy plants in which both FUL-like paralogs were down-regulated, potential carpel-like epidermal cells are observed in the transformed sectors of second whorl organs (Fig. 6, D–L). In addition, we also showed that in these green petal sectors, PapsAG1 and PapsAG2 expression does not increase in comparison with wild-type petals, suggesting that misregulation of PapsAG is not responsible for the change in epidermal identity. These data suggest that although the opium poppy FUL-like genes specify sepal and petal identity, they do not appear to regulate C-function gene expression, one of the components of the A function specified by the ABC model (Coen and Meyerowitz, 1991). Carpel-like cell identity may be the result of persistent wild-type expression of C-class genes in the second whorl after the loss of FUL-like gene function.
Opium Poppy FUL-like Proteins Form Heterodimers in a Yeast System
Identical phenotypes were observed when either of the two PapsFUL-like genes was down-regulated; this is in agreement with the fact that PapsFL1 and PapsFL2 interact strongly in a yeast system and suggests that they might act in planta as a heterodimer that is critical for proper floral meristem identity, sepal identity, the repression of axillary meristem growth, and fruit development (Figs. 4 and 5). In addition, new phenotypes emerged when both PapsFL1 and PapsFL2 were simultaneously down-regulated, suggesting that they redundantly regulate flowering time and petal identity (Fig. 6). The fact that the flowering-time phenotype is only manifest when both copies are down-regulated suggests that the two paralogs may function interchangeably in forming complexes with other proteins important in this process, such as SHORT VEGETATIVE PHASE (SVP)/AGAMOUS-Like24 (AGL24) and SUPPRESSOR OF OVEREXPRESSION OF CONSTANS1 (SOC1) orthologs, as has been suggested for FUL and AP1 (de Folter et al., 2005; Gregis et al., 2006). Likewise, in petal development, the opium poppy FUL-like proteins could be interacting with PapsAP3 and PapsPI in a fashion similar to the higher order protein complexes that AP1 (and other euAP1 genes) forms and that are thought to control floral organ identity in Arabidopsis and snapdragon (Davies et al., 1996; Egea-Cortines et al., 1999; Honma and Goto, 2001; Theissen, 2001).
The Core Eudicot Duplication in the AP1/FUL Gene Lineage Resulted in Subfunctionalization in euFUL and euAP1 Genes
The available information on the evolution of the AP1/FUL gene lineage suggests a scenario in which a major duplication event, accompanied by protein sequence divergence of one resulting paralagous clade, coincides with the diversification of a large group of flowering plants, the core eudicots. Because basal eudicot FUL-like genes represent the AP1/FUL gene type prior to the core eudicot gene duplication, the data presented here allow us to gain an understanding of functional evolution in the core eudicot euFUL and euAP1 genes. Our data show that FUL-like genes in both species in the Papaveraceae are important in axillary meristem dormancy and in proper floral meristem and sepal identity; in addition, FUL-like genes exert a role in fruit development by promoting normal development of the fruit wall during fruit maturation (Figs. 5, V–AA, and 7, S–V). Finally, our data from opium poppy suggest that FUL-like genes may also regulate cauline leaf development and flowering time and may be important for proper petal identity (Figs. 4, D–F, and 6, A–I). These functions have been differentially retained in euAP1 and euFUL genes, with the former functioning in floral meristem and sepal identity and the latter in the reproductive transition, floral meristem identity, fruit development, and axillary meristem activation (Irish and Sussex, 1990; Bowman et al., 1993; Gu et al., 1998; Ferrándiz et al., 2000; Berbel et al., 2001; Blázquez et al., 2006; Melzer et al., 2008; Jaakola et al., 2010). Therefore, our data suggest that the functions reported for euFUL and euAP1 genes in core eudicots are each part of the original functional repertoire of preduplication FUL-like genes and that, following the core eudicot duplication, the AP1/FUL gene lineage underwent subfunctionalization (Fig. 8). Some functions were retained redundantly in both euAP1 and euFUL genes, such as specification of floral meristem identity; however, most functions appear to have been parceled out to one or the other paralog.
Figure 8.
Optimization and mapping of functions recorded for AP1/FUL homologs. Based on the available data, we hypothesize that the ancestral functions in the AP1/FUL gene lineage include floral meristem and sepal identity (1), because these are functions that are shared with the sister SEPALLATA and AGL6 gene lineages. Transition to the reproductive meristem (2) appears to be ancestral just to the AP1/FUL lineage. Before the diversification of the Papaveraceae, the genes acquired functions in cauline leaf development (3), branching (6), and fruit development (5), although the acquisition of these functions could have happened earlier than is shown here. After the diversification of the core eudicots, some of these functions (1 and 6) were retained by both the euFUL and the euAP1 clades, whereas others (2, 3, and 5) were exclusively retained by members of the euFUL clade. A role in petal identity (4) appears to have been independently acquired in opium poppy and Arabidopsis. Asterisks indicate that no functional data are available. FMI/SEP, Floral meristem identity and sepal identity; T REP/F TIME, transition to reproductive meristems/flowering time; C LEAF, cauline leaf development; PET, petal identity; FR, fruit development; BR, branching. Black circles indicate gain of function (also numbered on the right), and white circles indicate loss of function. + symbolizes that the function has been recorded for that gene.
Our data also suggest that that the acquisition of different and characteristic motifs at the C terminus of euAP1 proteins is not correlated with the acquisition of novel core eudicot-specific functions. Evidence regarding the importance of the euAP1 motifs for proper protein function is inconsistent. Yalovsky et al. (2000) showed that an AP1 protein with a mutated farnesylation motif was unable to completely recapitulate the normal overexpression phenotype for AP1; this suggests that farnesylation is required for proper protein function. However, chimeric AP1 proteins carrying the C-terminal domain of AGAMOUS, which lacks a farnesylation or a transcription activation domain, were able to produce a typical overexpression AP1 phenotype in Arabidopsis (Krizek and Meyerowitz, 1996). In addition, PEAM4 (the pea euAP1 protein), which lacks a farnesylation motif (Berbel et al., 2001), and monocot FUL-like genes can complement the Arabidopsis ap1 mutant (Chen et al., 2008), suggesting that these proteins with different C-terminal motifs can provide the same functions as AP1 in an Arabidopsis background. Our data also suggest that the novel C-terminal motifs of euAP1 proteins, even though they have been shown to be functional (farnesylation and transcription activation), do not in fact result in novel roles for these proteins. Nonetheless, we know that AP1 and FUL each has unique functional roles in flower development; these data raise the possibility that these roles may be determined by differences in regulation rather than sequence. Further studies that explore the regulation of AP1 and FUL in Arabidopsis and the complementation of ap1 and ful mutants with FUL-like genes could help address some of these remaining questions.
In addition, a novel function, specification of petal identity, appears to have been acquired in parallel in different angiosperm lineages. To date, this has only been observed in Arabidopsis (Irish and Sussex, 1990; Coen and Meyerowitz, 1991; Castillejo et al., 2005) and now in Papaver. The absence of a canonical A-function gene, controlling the identity of the two outer floral whorls, from any other core eudicot has raised questions regarding the universality of this element of the ABC model (Gutierrez-Cortines and Davies, 2000; Theissen et al., 2000; Shepard and Purugganan, 2002; Smyth, 2005; Litt, 2007; Causier et al., 2010; Litt and Kramer, 2010; Wollmann et al., 2010). It has been suggested that this role is unique to Brassicaceae, possibly due to Brassicaceae-specific duplications and functional diversification (Lowman and Purugganan, 1999). We demonstrate that FUL-like genes in opium poppy behave to a large degree as A-function genes, suggesting that AP1/FUL homologs have been independently coopted in the determination of proper petal identity across different groups of flowering plants.
MATERIALS AND METHODS
Plant Material and Growth Conditions
Opium poppy (Papaver somniferum ‘Persian White’) seeds were obtained from J.L. Hudson. Seeds were germinated at 14 h of light/10 h of dark at 22°C. Seedlings were grown in the same conditions. California poppy (Eschscholzia californica) seeds were obtained from Seed Empire. Seeds were germinated at constant light at 25°C. Seedlings were grown in the same conditions.
Cloning of FUL-like Genes
Partial sequences of opium poppy FUL-like genes were obtained from GenBank (accession nos. AY306177 and AY306178). These sequences were used to design primers for 3′ RACE. Total RNA was prepared from dissected organs using TRIZOL reagent (Invitrogen) and was DNaseI (Roche) treated to remove residual genomic DNA. Five micrograms was used as a template for cDNA synthesis with SuperScript III reverse transcriptase (Invitrogen). The resulting cDNA was diluted 1:10. 3′ RACE was performed using the FirstChoice RLM-RACE Kit (Applied Biosystems) according to the manufacturer’s instructions to get the complete nucleotide sequence at the end of the C-terminal domain and the 3′ untranslated region (UTR) of PapsFL1 and PapsFL2. Degenerate primers (Litt and Irish, 2003) were used to amplify California poppy FUL-like genes. PCR products were cloned into pCR2.1-TOPO (Invitrogen) and sequenced.
RT-PCR
Expression of FUL-like genes was assayed in all floral organs at stages P7, P8, and anthesis and in leaves and fruits. Total RNA was prepared from dissected organs as described above from three different individuals. Three micrograms of RNA was used as a template for cDNA synthesis (as described above). The resulting cDNA was diluted 1:10 and amplified using locus-specific primers (all primers are listed in Supplemental Table S2). Reactions for FUL-like genes were run for 29 cycles at an annealing temperature of 55°C. Reactions for ACTIN or UBIQUITIN (used as a loading control) were run for 31 cycles at an annealing temperature of 56°C or 47°C, respectively. PCR products were run on a 1.2% agarose gel stained with ethidium bromide and digitally photographed using an Ultraviolet Products DigiDoc-it Darkroom equipped with a Cannon PC1089 digital camera.
In Situ Hybridization and Anatomy
Developing shoot apical meristems in the vegetative and reproductive stages were collected from wild-type plants of opium poppy and California poppy and fixed under vacuum in freshly prepared FAA (50% ethanol, 3.7% formaldehyde, and 5% glacial acetic acid). Samples were prepared and sectioned at 10 μm according to standard methods (Langdale, 1993) on a Microm HM3555 rotary microtome. DNA templates for RNA probe synthesis were obtained by PCR amplification of 400- to 550-bp fragments. To ensure specificity, the probe templates included 200 to 210 bp of the 3′ UTR and approximately 300 bp of the coding region. Because EscaFL1 and EscaFL2 were more similar to each other than PapsFL1 and PapsFL2, the DNA templates for California poppy included 500 bp of the variable K and C domains of EscaFL1 and EscaFL2 (Supplemental Fig. S2; Supplemental Table S2); nonetheless, we cannot rule out the possibility that the probes could cross-hybridize. Digoxigenin-labeled RNA probes were prepared using T7 polymerase (Roche), murine RNase inhibitor (New England Biolabs), and RNA labeling mix (Roche) according to each manufacturer’s protocol. RNA in situ hybridization was performed according to Ferrándiz et al. (2000), optimized for each species: slides with opium poppy sections were hybridized overnight at 55°C, whereas California poppy slides were hybridized overnight at 47°C. Probe concentration was identical for all the experiments, including the antisense control hybridizations. In situ hybridized sections were subsequently dehydrated and permanently mounted in Permount (Fisher).
For fruit anatomy, material was fixed, embedded, and sectioned as above, but slides were directly stained with Johansen’s safranin and 0.5% Astra Blue in 2% tartaric acid. All sections were digitally photographed using a Zeiss Axioplan microscope equipped with a Nikon DXM1200C digital camera.
Leaf clearings were made following Ellis et al. (2009). Macroscopic photographs of leaves and flowers were taken using an EOS Canon Rebel XS digital camera or a Nikon DXM1200F digital camera adapted to a Nikon SMZ1500 stereoscope.
TRV-VIGS
TRV1 and TRV2 vectors were provided by V. Irish (Yale University) and E. Kramer (Harvard University). A 630-bp fragment of PapsFL1 (Supplemental Fig. S2) was amplified including a portion of the K domain, the C domain, and a portion of the 3′ UTR from floral bud cDNA using primers that added EcoRI and XbaI restriction sites to the respective 5′ and 3′ ends of the PCR product (Supplemental Table S2). A 590-bp fragment of PapsFL2 (Supplemental Fig. S2) was amplified including a portion of the K domain, the C domain, and a portion of the 3′ UTR adding BamHI to the 3′ end and using a naturally occurring XbaI site in the 5′ end of the PCR fragment. The PCR products were cloned into the pCR2.1-TOPO plasmid vector (Invitrogen), then digested with EcoRI (Roche) and XbaI (Roche) for PapsFL1 and with XbaI and BamHI (Roche) for PapsFL2. Fragments were ligated into the similarly digested TRV2 vector using T4 DNA ligase (New England Biolabs) according to the manufacturer’s protocol. This created two TRV2 constructs: TRV2-PapsFL1 and TRV2-PapsFL2. A construct carrying fragments of both PapsFL1 and PapsFL2 was also designed but proved to be ineffective; therefore, a mixture of both TRV2 vectors was used to silence both copies simultaneously (see below).
A similar strategy was used to create TRV2-EscaFL1 and TRV2-EscaFL2. A 508-bp fragment of EscaFL1 and a 500-bp fragment of EscaFL2 (Supplemental Figure S2) excluding the MADS domain and the first 15 amino acids of the K domain were amplified from floral bud cDNA using primers that added KpnI and SacI restriction sites to the 5′ and 3′ ends, respectively, of the PCR products. Cloning, digestion, and ligation were as for TRV2-PapsFL1 and TRV-PapsFL2. All vectors (TRV1, TRV2-PapsFL1, TRV2-PapsFL2, TRV2-EscaFL1, TRV2-EscaFL2, and TRV2-empty) were sequenced and separately transformed into Agrobacterium tumefaciens strain EHA105.
Agrobacterium growth and plant infiltration methods followed Drea et al. (2007), Wege et al. (2007), and Orashakova et al. (2009) and were modified as follows. Agrobacterium was resuspended to an optical density at 600 nm of 2.0 in 5% Suc. For infiltration, TRV2-target gene was mixed independently with TRV1 in equal volumes. For opium poppy, 100 seedlings were transformed with TRV1 and TRV2-PapsFL1, 80 seedlings were transformed with TRV1 and TRV2-PapsFL2, and 108 were transformed with TRV1 and TRV2-PapsFL1 and TRV2-PapsFL2. For California poppy, 60 seedlings were transformed with TRV1 and TRV2-EscaFL1 and 60 seedlings were transformed with TRV1 and TRV2-EscaFL2. Forty seedlings per species were transformed with TRV1 and TRV2-empty. Twenty wild-type seedlings per species were grown side by side with the transformed plants as a control. To inoculate, incisions were made in the hypocotyls of each seedling with a needle and a drop of the Agrobacterium suspension was placed on the wound.
Screening in Opium Poppy
The treatment resulted in 5% to 20% mortality of opium poppy plants in each group of transformants. Four to 5 weeks after transformations, tissue from the youngest leaves, cauline leaves, sepals, and fruits was collected from all opium poppy plants, including those treated with the experimental construct or the empty TRV2 vector and the wild type. Eighty-five poppy plants treated with TRV1 + TRV2-PapsFL1, 69 poppy plants treated with TRV1 + TRV2-PapsFL2, 20 plants treated with TRV1 + TRV2-empty, and three wild-type plants were evaluated for target gene expression. RNA extraction, cDNA synthesis, and RT-PCR were performed as described above for RT-PCR.
The levels of PapsFL1 and PapsFL2 mRNA relative to ACTIN were determined in wild-type plants and in VIGS-treated plants using semiquantitative RT-PCR. PapsFL1 and PapsFL2 primers amplify a region outside the fragments used in the TRV constructs (Supplemental Table S2). Aliquots of PCR products were removed every two cycles from cycle 25 to 35 and were run on a gel to identify the linear range of amplification. These were determined to be 28 to 32 cycles for ACTIN, 28 to 30 cycles for PapsFL1, and 30 to 32 cycles for PapsFL2. PCR reactions were conducted using 1:20 dilutions of template cDNA with 31 cycles of amplification for ACTIN and 29 cycles of amplification for PapsFL1 and PapsFL2. The products were separated by electrophoresis on a 1% agarose gel containing 0.1 mg L−1 ethidium bromide. Relative amounts of PapsFL1 and PapsFL2 were compared in the treated plants that showed silencing phenotypes versus the TRV2-empty vector-treated plants and the wild-type plants. Gels were photographed as described above. Reactions were repeated two additional times.
To screen for the presence of the vector, cDNA was synthesized from each RNA sample using a reverse primer in the vector sequence. The presence of both TRV1 and TRV2 in cDNA samples from VIGS-treated plants and control untreated plants was assessed by RT-PCR using vector-specific primers (Hileman et al., 2005).
Screening in California Poppy
The treatment resulted in 3% to 11% mortality of California poppy plants in each group of transformants. To test for down-regulation and the presence of the vector, tissue was collected as for opium poppy from three wild-type plants and transformed plants that showed phenotypic changes: 30 plants treated with TRV2-EscaFL1 and 17 plants treated with TRV2-EscaFL2 (total n = 47). RNA was prepared as described above for all the samples. The relative mRNA levels of EscaFL1 and EscaFL2 were determined in wild-type and VIGS-treated plants using semiquantitative RT-PCR as described above for opium poppy (Supplemental Table S2). ACTIN levels were variable between different sample tissues collected for California poppy, so UBIQUITIN was used instead as a control. The linear range of amplification from these loci was 30 to 32 cycles for UBIQUITIN and 28 to 30 cycles for EscaFL1 and EscaFL2. PCR products were prepared using 1:20 dilutions of template cDNA with 31 cycles of amplification for UBIQUITIN and 29 cycles of amplification for EscaFL1 and EscaFL2. Gels were photographed as described above. Reactions were repeated two additional times.
qRT-PCR
To confirm down-regulation and to better evaluate the reduction of transcript in the down-regulated plants, a subset of samples that by semiquantitative RT-PCR showed mild, moderate, and strong down-regulation was subjected to qRT-PCR. qRT-PCR was carried out using the 7300 qPCR system and SDS software (Applied Biosystems). Primers used for the quantification of transcripts were designed using Primer Express version 3.0 (Applied Biosystems; Supplemental Table S2). qRT-PCR was performed in total volumes of 25 μL containing 12.5 μL of FastStart Universal SYBR Green (Roche), 4 μL of diluted cDNA, 2.5 μL of each PCR primer (1 μm), and 3.5 μL of deionized water. PCR conditions were 2 min at 50°C, 10 min at 95°C, followed by 40 cycles of denaturation at 95°C for 15 s and annealing-extension at 60°C for 2 min. Three genes, Elf1a, ACTIN, and GADPH, were tested as endogenous controls; GAPDH showed consistency and expression in the appropriate range across different tissues and therefore was used as the constitutive reference transcript (Yellina et al., 2010). The level of FUL-like mRNA in down-regulated leaf tissue was analyzed relative to the wild type using the 2−ΔΔCt method (Livak and Schmittgen, 2001). se is reported for three technical replicates of each sample.
Yeast Two-Hybrid Analysis
Full-length PapsFL1 and PapsFL2 were amplified from floral bud cDNA using primers that added NdeI and EcoRI restriction sites to the respective 5′ and 3′ ends of the PCR product (Supplemental Table S2). Each full coding sequence was fused with the GAL4 binding domain in the pGBKT7 vector (Clontech) and the GAL4 activation domain in the pGADT7 vector (Clontech). Single constructs were transformed into yeast strain AH109. All constructs show minimal autoactivation upon the addition of 2.5 mm 3-Amino-1,2,4-triazole. To test interactions, pairwise combinations of constructs were transformed into AH109. Homodimerization and heterodimerization were tested by growing colonies on selective medium (synthetic drop-out). Growth was measured after 3 d and after 6 d. Interactions between each vector and an empty vector were used as negative controls.
SEM
For SEM studies, leaves, cauline leaves, floral buds, dissected floral organs, and fruits from wild-type and down-regulated plants of opium poppy and California poppy were fixed under vacuum in FAA and stored in 70% ethanol. For analysis, material was dehydrated through an ethanol series and critical point dried using a Samdri 790 CPD. Material was mounted on aluminum stubs with adhesive tabs (Electron Microscopy Sciences), sputter coated with gold palladium in a Hummer 6.2 sputter coater, and examined and photographed at 10 kV in a JEOL JSM-5410 LV scanning electron microscope.
Sequence data from this article can be found in the GenBank/EMBL data libraries under accession numbers AY306177, AY306178, HM592297, and HM592298.
Supplemental Data
The following materials are available in the online version of this article.
Supplemental Figure S1. Diagram showing differences in first, second, and third order branching in California poppy wild-type and escafl1fl2 plants.
Supplemental Figure S2. Amino acid alignment of the opium poppy and California poppy FUL-like proteins and primer locations.
Supplemental Figure S3. In situ hybridization controls.
Supplemental Figure S4. Locus-specific RT-PCR and qRT-PCR on cDNA prepared from organs of VIGS-treated plants.
Supplemental Figure S5. Protein interactions between PapsFL1 and PapsFL2 as determined by growth on selective synthetic drop-out medium.
Supplemental Figure S6. Range of variation of the leaf-like sepal phenotype in escafl1-fl2 California poppy plants.
Supplemental Table S1. Summary of phenotypes identified using VIGS to silence poppy FUL-like genes individually and simultaneously.
Supplemental Table S2. List of primers.
Supplementary Material
Acknowledgments
We thank Vivian Irish and Elena Kramer for sharing vectors and Elena Kramer for comments on the manuscript and discussion of the data. We thank Conny Bartholmes, Mike Baxter, Amelia Clements, Lynn Holappa, Rachel Meyer, Chiara Mizzoti, Sara Simonini, and Emilio Tobón for technical assistance and Sinead Drea for primer sequences used in generating the in situ probe for PapsPI. We thank two anonymous reviewers for their helpful comments and Andrew and Judith Economos for support.
References
- Alvarez-Buylla ER, García-Ponce B, Garay-Arroyo A. (2006) Unique and redundant functional domains of APETALA1 and CAULIFLOWER, two recently duplicated Arabidopsis thaliana floral MADS-box genes. J Exp Bot 57: 3099–3107 [DOI] [PubMed] [Google Scholar]
- Alvarez-Buylla ER, Pelaz S, Liljegren SJ, Gold SE, Burgeff C, Ditta GS, Ribas de Pouplana L, Martínez-Castilla L, Yanofsky MF. (2000) An ancestral MADS-box gene duplication occurred before the divergence of plants and animals. Proc Natl Acad Sci USA 97: 5328–5333 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ambrose BA, Lerner DR, Ciceri P, Padilla CM, Yanofsky MF, Schmidt RJ. (2000) Molecular and genetic analyses of the silky1 gene reveal conservation in floral organ specification between eudicots and monocots. Mol Cell 5: 569–579 [DOI] [PubMed] [Google Scholar]
- Becker A, Gleissberg S, Smyth DR. (2005) Floral and vegetative morphogenesis in California poppy (Eschscholzia californica Cham.). Int J Plant Sci 166: 537–555 [Google Scholar]
- Becker A, Theissen G. (2003) The major clades of MADS-box genes and their role in the development and evolution of flowering plants. Mol Phylogenet Evol 29: 464–489 [DOI] [PubMed] [Google Scholar]
- Benlloch R, d’Erfurth I, Ferrandiz C, Cosson V, Beltrán JP, Cañas LA, Kondorosi A, Madueño F, Ratet P. (2006) Isolation of mtpim proves Tnt1 a useful reverse genetics tool in Medicago truncatula and uncovers new aspects of AP1-like functions in legumes. Plant Physiol 142: 972–983 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Berbel A, Navarro C, Ferrándiz C, Cañas LA, Madueño F, Beltrán JP. (2001) Analysis of PEAM4, the pea AP1 functional homologue, supports a model for AP1-like genes controlling both floral meristem and floral organ identity in different plant species. Plant J 25: 441–451 [DOI] [PubMed] [Google Scholar]
- Blázquez MA, Ferrándiz C, Madueño F, Parcy F. (2006) How floral meristems are built. Plant Mol Biol 60: 855–870 [DOI] [PubMed] [Google Scholar]
- Bowman JL, Alvarez J, Weigel D, Meyerowitz EM, Smyth DR. (1993) Control of flower development in Arabidopsis thaliana by APETALA1 and interacting genes. Development 119: 721–743 [Google Scholar]
- Bowman JL, Smyth DR, Meyerowitz EM. (1991) Genetic interactions among floral homeotic genes of Arabidopsis. Development 112: 1–20 [DOI] [PubMed] [Google Scholar]
- Burch-Smith TM, Anderson JC, Martin GB, Dinesh-Kumar SP. (2004) Applications and advantages of virus-induced gene silencing for gene function studies in plants. Plant J 39: 734–746 [DOI] [PubMed] [Google Scholar]
- Calonje M, Cubas P, Martínez-Zapater JM, Carmona MJ. (2004) Floral meristem identity genes are expressed during tendril development in grapevine. Plant Physiol 135: 1491–1501 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Castillejo C, Romera-Branchat M, Pelaz S. (2005) A new role of the Arabidopsis SEPALLATA3 gene revealed by its constitutive expression. Plant J 43: 586–596 [DOI] [PubMed] [Google Scholar]
- Causier B, Schwarz-Sommer Z, Davies B. (2010) Floral organ identity: 20 years of ABCs. Semin Cell Dev Biol 21: 73–79 [DOI] [PubMed] [Google Scholar]
- Chen D, Guo B, Hexige S, Zhang T, Shen D, Ming F. (2007) SQUA-like genes in the orchid Phalaenopsis are expressed in both vegetative and reproductive tissues. Planta 226: 369–380 [DOI] [PubMed] [Google Scholar]
- Chen MK, Lin IC, Yang CH. (2008) Functional analysis of three lily (Lilium longiflorum) APETALA1-like MADS box genes in regulating floral transition and formation. Plant Cell Physiol 49: 704–717 [DOI] [PubMed] [Google Scholar]
- Cho S, Jang S, Chae S, Chung KM, Moon YH, An G, Jang SK. (1999) Analysis of the C-terminal region of Arabidopsis thaliana APETALA1 as a transcription activation domain. Plant Mol Biol 40: 419–429 [DOI] [PubMed] [Google Scholar]
- Coen ES, Meyerowitz EM. (1991) The war of the whorls: genetic interactions controlling flower development. Nature 353: 31–37 [DOI] [PubMed] [Google Scholar]
- Danilevskaya ON, Meng X, Selinger DA, Deschamps S, Hermon P, Vansant G, Gupta R, Ananiev EV, Muszynski MG. (2008) Involvement of the MADS-box gene ZMM4 in floral induction and inflorescence development in maize. Plant Physiol 147: 2054–2069 [DOI] [PMC free article] [PubMed]
- Davies B, Egea-Cortines M, de Andrade Silva E, Saedler H, Sommer H. (1996) Multiple interactions amongst floral homeotic MADS box proteins. EMBO J 15: 4330–4343 [PMC free article] [PubMed] [Google Scholar]
- de Folter S, Immink RGH, Kieffer M, Parenicová L, Henz SR, Weigel D, Busscher M, Kooiker M, Colombo L, Kater MM, et al. (2005) Comprehensive interaction map of the Arabidopsis MADS box transcription factors. Plant Cell 17: 1424–1433 [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Martino G, Pan I, Emmanuel E, Levy A, Irish VF. (2006) Functional analyses of two tomato APETALA3 genes demonstrate diversification in their roles in regulating floral development. Plant Cell 18: 1833–1845 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dinesh-Kumar SP, Anandalakshmi R, Marathe R, Schiff M, Liu Y. (2003) Virus induced gene silencing. In E Grotewold, ed, Methods in Molecular Biology. Humana Press, Totowa, NJ, pp 287–294 [DOI] [PubMed]
- Di Stilio VS, Kumar RA, Oddone AM, Tolkin TR, Salles P, McCarty K. (2010) Virus-induced gene silencing as a tool for comparative functional studies in Thalictrum. PLoS ONE 5: e12064. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Drea S, Hileman LC, de Martino G, Irish VF. (2007) Functional analyses of genetic pathways controlling petal specification in poppy. Development 134: 4157–4166 [DOI] [PubMed] [Google Scholar]
- Dreni L, Pilatone A, Yun D, Erreni S, Pajoro A, Caporali E, Zhang D, Kater MM. (2011) Functional analysis of all AGAMOUS subfamily members in rice reveals their roles in reproductive organ identity determination and meristem determinacy. Plant Cell 23: 2850–2863 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Egea-Cortines M, Saedler H, Sommer H. (1999) Ternary complex formation between the MADS-box proteins SQUAMOSA, DEFICIENS and GLOBOSA is involved in the control of floral architecture in Antirrhinum majus. EMBO J 18: 5370–5379 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ellis B, Daly DC, Hickey LJ, Johnson KR, Mitchell JD, Wilf P, Wing SL. (2009) Manual of Leaf Architecture. Cornell University Press, Ithaca, NY
- Ferrándiz C. (2002) Regulation of fruit dehiscence in Arabidopsis. J Exp Bot 53: 2031–2038 [DOI] [PubMed] [Google Scholar]
- Ferrándiz C, Gu Q, Martienssen R, Yanofsky MF. (2000) Redundant regulation of meristem identity and plant architecture by FRUITFULL, APETALA1 and CAULIFLOWER. Development 127: 725–734 [DOI] [PubMed] [Google Scholar]
- Fornara F, Parenicová L, Falasca G, Pelucchi N, Masiero S, Ciannamea S, Lopez-Dee Z, Altamura MM, Colombo L, Kater MM. (2004) Functional characterization of OsMADS18, a member of the AP1/SQUA subfamily of MADS box genes. Plant Physiol 135: 2207–2219 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gocal GF, King RW, Blundell CA, Schwartz OM, Andersen CH, Weigel D. (2001) Evolution of floral meristem identity genes: analysis of Lolium temulentum genes related to APETALA1 and LEAFY of Arabidopsis. Plant Physiol 125: 1788–1801 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gould B, Kramer EM. (April 12, 2007) Virus-induced gene silencing as a tool for functional analyses in the emerging model plant Aquilegia (columbine, Ranunculaceae). Plant Methods 3: http://dx.doi.org/10.1186/1746-4811-3-6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gregis V, Sessa A, Colombo L, Kater MM. (2006) AGL24, SHORT VEGETATIVE PHASE, and APETALA1 redundantly control AGAMOUS during early stages of flower development in Arabidopsis. Plant Cell 18: 1373–1382 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gu Q, Ferrándiz C, Yanofsky MF, Martienssen R. (1998) The FRUITFULL MADS-box gene mediates cell differentiation during Arabidopsis fruit development. Development 125: 1509–1517 [DOI] [PubMed] [Google Scholar]
- Gustafson-Brown C, Savidge B, Yanofsky MF. (1994) Regulation of the Arabidopsis floral homeotic gene APETALA1. Cell 76: 131–143 [DOI] [PubMed] [Google Scholar]
- Gutierrez-Cortines ME, Davies B. (2000) Beyond the ABCs: ternary complex formation in the control of floral organ identity. Trends Plant Sci 5: 471–476 [DOI] [PubMed] [Google Scholar]
- Hands P, Vosnakis N, Betts D, Irish VF, Drea S. (2011) Alternate transcripts of a floral developmental regulator have both distinct and redundant functions in opium poppy. Ann Bot (Lond) 107: 1557–1566 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hardenack S, Ye D, Saedler H, Grant S. (1994) Comparison of MADS box gene expression in developing male and female flowers of the dioecious plant white campion. Plant Cell 6: 1775–1787 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hileman LC, Drea S, Martino G, Litt A, Irish VF. (2005) Virus-induced gene silencing is an effective tool for assaying gene function in the basal eudicot species Papaver somniferum (opium poppy). Plant J 44: 334–341 [DOI] [PubMed] [Google Scholar]
- Hileman LC, Sundstrom JF, Litt A, Chen M, Shumba T, Irish VF. (2006) Molecular and phylogenetic analyses of the MADS-box gene family in tomato. Mol Biol Evol 23: 2245–2258 [DOI] [PubMed] [Google Scholar]
- Honma T, Goto K. (2001) Complexes of MADS-box proteins are sufficient to convert leaves into floral organs. Nature 409: 525–529 [DOI] [PubMed] [Google Scholar]
- Huijser P, Klein J, Lönnig WE, Meijer H, Saedler H, Sommer H. (1992) Bracteomania, an inflorescence anomaly, is caused by the loss of function of the MADS-box gene squamosa in Antirrhinum majus. EMBO J 11: 1239–1249 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Immink RGH, Hannapel DJ, Ferrario S, Busscher M, Franken J, Lookeren Campagne MM, Angenent GC. (1999) A petunia MADS box gene involved in the transition from vegetative to reproductive development. Development 126: 5117–5126 [DOI] [PubMed] [Google Scholar]
- Irish VF, Litt A. (2005) Flower development and evolution: gene duplication, diversification and redeployment. Curr Opin Genet Dev 15: 454–460 [DOI] [PubMed] [Google Scholar]
- Irish VF, Sussex IM. (1990) Function of the apetala-1 gene during Arabidopsis floral development. Plant Cell 2: 741–753 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jaakola L, Poole M, Jones MO, Kämäräinen-Karppinen T, Koskimäki JJ, Hohtola A, Häggman H, Fraser PD, Manning K, King GJ, et al. (2010) A SQUAMOSA MADS box gene involved in the regulation of anthocyanin accumulation in bilberry fruits. Plant Physiol 153: 1619–1629 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaufmann K, Melzer R, Theissen G. (2005) MIKC-type MADS domain proteins: structural modularity, protein interactions and network evolution in land plants. Gene 347: 183–198 [DOI] [PubMed]
- Kempin SA, Savidge B, Yanofsky MF. (1995) Molecular basis of the cauliflower phenotype in Arabidopsis. Science 267: 522–525 [DOI] [PubMed] [Google Scholar]
- Kramer EM, Dorit RL, Irish VF. (1998) Molecular evolution of genes controlling petal and stamen development: duplication and divergence within the APETALA3 and PISTILLATA MADS-box gene lineages. Genetics 149: 765–783 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kramer EM, Holappa L, Gould B, Jaramillo MA, Setnikov D, Santiago PM. (2007) Elaboration of B gene function to include the identity of novel floral organs in the lower eudicot Aquilegia. Plant Cell 19: 750–766 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kramer EM, Jaramillo MA, Di Stilio VS. (2004) Patterns of gene duplication and functional evolution during the diversification of the AGAMOUS subfamily of MADS box genes in angiosperms. Genetics 166: 1011–1023 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krizek BA, Meyerowitz EM. (1996) Mapping the protein regions responsible for the functional specificities of the Arabidopsis MADS domain organ-identity proteins. Proc Natl Acad Sci USA 93: 4063–4070 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lamb RS, Irish VF. (2003) Functional divergence within the APETALA3/PISTILLATA floral homeotic gene lineages. Proc Natl Acad Sci USA 100: 6558–6563 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Langdale JA. (1993). In situ hybridization. In M Freeling, V Walbot, eds, The Maize Handbook. Springer-Verlag, New York, pp 165–180
- Liljegren SJ, Ditta G, Eshed Y, Savidge B, Bowman J, Yanofsky MF. (2000) SHATTERPROOF MADS-box genes control seed dispersal in Arabidopsis. Nature 404: 766–770 [DOI] [PubMed] [Google Scholar]
- Litt A. (2007) An evaluation of A-function: evidence from the APETALA 1 and APETALA 2 gene lineages. Int J Plant Sci 168: 73–91 [Google Scholar]
- Litt A, Irish VF. (2003) Duplication and diversification in the APETALA1/FRUITFULL floral homeotic gene lineage: implications for the evolution of floral development. Genetics 165: 821–833 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Litt A, Kramer EM. (2010) The ABC model and the diversification of floral organ identity. Semin Cell Dev Biol 21: 129–137 [DOI] [PubMed] [Google Scholar]
- Liu C, Zhang J, Zhang N, Shan H, Su K, Zhang J, Meng Z, Kong H, Chen Z. (2010) Interactions among proteins of floral MADS-box genes in basal eudicots: implications for evolution of the regulatory network for flower development. Mol Biol Evol 27: 1598–1611 [DOI] [PubMed] [Google Scholar]
- Liu E, Page JE. (January 22, 2008) Optimized cDNA libraries for virus-induced gene silencing (VIGS) using tobacco rattle virus. Plant Methods 4: http://dx.doi.org/10.1186/1746-4811-4-5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Livak KJ, Schmittgen TD. (2001) Analysis of relative gene expression data using real-time quantitative PCR and the 2−ΔΔCT method. Methods 25: 402–408 [DOI] [PubMed] [Google Scholar]
- Lowman AC, Purugganan MD. (1999) Duplication of the Brassica oleracea APETALA1 floral homeotic gene and the evolution of domesticated cauliflower. J Hered 90: 514–520 [DOI] [PubMed] [Google Scholar]
- Mandel MA, Gustafson-Brown C, Savidge B, Yanofsky MF. (1992) Molecular characterization of the Arabidopsis floral homeotic gene APETALA1. Nature 360: 273–277 [DOI] [PubMed] [Google Scholar]
- Melzer S, Lens F, Gennen J, Vanneste S, Rohde A, Beeckman T. (2008) Flowering-time genes modulate meristem determinacy and growth form in Arabidopsis thaliana. Nat Genet 40: 1489–1492 [DOI] [PubMed] [Google Scholar]
- Müller BM, Saedler H, Zachgo S. (2001) The MADS-box gene DEFH28 from Antirrhinum is involved in the regulation of floral meristem identity and fruit development. Plant J 28: 169–179 [DOI] [PubMed] [Google Scholar]
- Murai K, Miyamae M, Kato H, Takumi S, Ogihara Y. (2003) WAP1, a wheat APETALA1 homolog, plays a central role in the phase transition from vegetative to reproductive growth. Plant Cell Physiol 44: 1255–1265 [DOI] [PubMed] [Google Scholar]
- Orashakova S, Lange M, Lange S, Wege S, Becker A. (2009) The CRABS CLAW ortholog from California poppy (Eschscholzia californica, Papaveraceae), EcCRC, is involved in floral meristem termination, gynoecium differentiation and ovule initiation. Plant J 58: 682–693 [DOI] [PubMed] [Google Scholar]
- Parenicová L, de Folter S, Kieffer M, Horner DS, Favalli C, Busscher J, Cook HE, Ingram RM, Kater MM, Davies B, et al. (2003) Molecular and phylogenetic analyses of the complete MADS-box transcription factor family in Arabidopsis: new openings to the MADS world. Plant Cell 15: 1538–1551 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Preston JC, Kellogg EA. (2006) Reconstructing the evolutionary history of paralogous APETALA1/FRUITFULL-like genes in grasses (Poaceae). Genetics 174: 421–437 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Preston JC, Kellogg EA. (2007) Conservation and divergence of APETALA1/FRUITFULL-like gene function in grasses: evidence from gene expression analyses. Plant J 52: 69–81 [DOI] [PubMed] [Google Scholar]
- Preston JC, Kellogg EA. (2008) Discrete developmental roles for temperate cereal grass VERNALIZATION1/FRUITFULL-like genes in flowering competency and the transition to flowering. Plant Physiol 146: 265–276 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Purugganan MD. (1997) The MADS-box floral homeotic gene lineages predate the origin of seed plants: phylogenetic and molecular clock estimates. J Mol Evol 45: 392–396 [DOI] [PubMed] [Google Scholar]
- Riechmann JL, Krizek BA, Meyerowitz EM. (1996) Dimerization specificity of Arabidopsis MADS domain homeotic proteins APETALA1, APETALA3, PISTILLATA, and AGAMOUS. Proc Natl Acad Sci USA 93: 4793–4798 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ronse De Craene L, Smets E. (1990) The systematic relationship between Begoniaceae and Papaveraceae: a comparative study of their floral development. Bull Jard Bot Natl Belg 60: 229–273
- Sather DN, Golenberg EM. (2009) Duplication of AP1 within the Spinacia oleracea L. AP1/FUL clade is followed by rapid amino acid and regulatory evolution. Planta 229: 507–521 [DOI] [PubMed] [Google Scholar]
- Shan H, Zhang N, Liu C, Xu G, Zhang J, Chen Z, Kong H. (2007) Patterns of gene duplication and functional diversification during the evolution of the AP1/SQUA subfamily of plant MADS-box genes. Mol Phylogenet Evol 44: 26–41 [DOI] [PubMed] [Google Scholar]
- Sharma B, Guo C, Kong H, Kramer EM. (2011) Petal-specific subfunctionalization of an APETALA3 paralog in the Ranunculales and its implications for petal evolution. New Phytol 191: 870–883 [DOI] [PubMed] [Google Scholar]
- Shchennikova AV, Shulga OA, Immink R, Skryabin KG, Angenent GC. (2004) Identification and characterization of four Chrysanthemum MADS-box genes, belonging to the APETALA1/FRUITFULL and SEPALLATA3 subfamilies. Plant Physiol 134: 1632–1641 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shepard KA, Purugganan MD. (2002) The genetics of plant morphological evolution. Curr Opin Plant Biol 5: 49–55 [DOI] [PubMed] [Google Scholar]
- Smykal P, Gennen J, De Bodt S, Ranganath V, Melzer S. (2007) Flowering of strict photoperiodic Nicotiana varieties in non-inductive conditions by transgenic approaches. Plant Mol Biol 65: 233–242 [DOI] [PubMed] [Google Scholar]
- Smyth DR. (2005) Morphogenesis of flowers: our evolving view. Plant Cell 17: 330–341 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sridhar VV, Surendrarao A, Gonzalez D, Conlan RS, Liu Z. (2004) Transcriptional repression of target genes by LEUNIG and SEUSS, two interacting regulatory proteins for Arabidopsis flower development. Proc Natl Acad Sci USA 101: 11494–11499 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sridhar VV, Surendrarao A, Liu Z. (2006) APETALA1 and SEPALLATA3 interact with SEUSS to mediate transcription repression during flower development. Development 133: 3159–3166 [DOI] [PubMed] [Google Scholar]
- Stellari GM, Jaramillo MA, Kramer EM. (2004) Evolution of the APETALA3 and PISTILLATA lineages of MADS-box-containing genes in the basal angiosperms. Mol Biol Evol 21: 506–519 [DOI] [PubMed] [Google Scholar]
- Taylor SA, Hofer JMI, Murfet IC, Sollinger JD, Singer SR, Knox MR, Ellis TH. (2002) PROLIFERATING INFLORESCENCE MERISTEM, a MADS-box gene that regulates floral meristem identity in pea. Plant Physiol 129: 1150–1159 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Theissen G. (2001) Development of floral organ identity: stories from the MADS house. Curr Opin Plant Biol 4: 75–85 [DOI] [PubMed] [Google Scholar]
- Theissen G, Becker A, Di Rosa A, Kanno A, Kim JT, Münster T, Winter KU, Saedler H. (2000) A short history of MADS-box genes in plants. Plant Mol Biol 42: 115–149 [PubMed] [Google Scholar]
- Trevaskis B, Bagnall DJ, Ellis MH, Peacock WJ, Dennis ES. (2003) MADS box genes control vernalization-induced flowering in cereals. Proc Natl Acad Sci USA 100: 13099–13104 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Trevaskis B, Hemming MN, Dennis ES, Peacock WJ. (2007) The molecular basis of vernalization-induced flowering in cereals. Trends Plant Sci 12: 352–357 [DOI] [PubMed] [Google Scholar]
- Vandenbussche M, Theissen G, Van de Peer Y, Gerats T. (2003) Structural diversification and neo-functionalization during floral MADS-box gene evolution by C-terminal frameshift mutations. Nucleic Acids Res 31: 4401–4409 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vrebalov J, Ruezinsky D, Padmanabhan V, White R, Medrano D, Drake R, Schuch W, Giovannoni J. (2002) A MADS-box gene necessary for fruit ripening at the tomato ripening-inhibitor (rin) locus. Science 296: 343–346 [DOI] [PubMed] [Google Scholar]
- Wege S, Scholz A, Gleissberg S, Becker A. (2007) Highly efficient virus-induced gene silencing (VIGS) in California poppy (Escholzia californica): an evaluation of VIGS as a strategy to obtain functional data from non-model plants. Ann Bot (Lond) 100: 641–649 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wollmann H, Mica E, Todesco M, Long JA, Weigel D. (2010) On reconciling the interactions between APETALA2, miR172 and AGAMOUS with the ABC model of flower development. Development 137: 3633–3642 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu H, Su H, Hu J. (2007) The identification of A-, B-, C-, and E-class MADS-box genes and implications for perianth evolution in the basal eudicot Trochodendron aralioides (Trochodendraceae). Int J Plant Sci 168: 775–799 [Google Scholar]
- Wu Y, Li HQ, Zhang JS, Zheng Z, Xue S, Li Y. (2000) Molecular cloning and characterization of two tobacco MADS-box genes. Sex Plant Reprod 13: 163–169 [Google Scholar]
- Yalovsky S, Rodríguez-Concepción M, Bracha K, Toledo-Ortiz G, Gruissem W. (2000) Prenylation of the floral transcription factor APETALA1 modulates its function. Plant Cell 12: 1257–1266 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yanofsky MF. (1995) Floral meristems to floral organs: genes controlling early events in Arabidopsis flower development. Annu Rev Plant Physiol Plant Mol Biol 46: 167–188 [Google Scholar]
- Yellina AL, Orashakova S, Lange S, Erdmann R, Leebens-Mack J, Becker A. (December 1, 2010) Floral homeotic C function genes repress specific B function genes in the carpel whorl of the basal eudicot California poppy (Eschscholzia californica). Evodevo 1: http://dx.doi.org/10.1186/2041-9139-1-13 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu H, Goh CJ. (2000) Identification and characterization of three orchid MADS-box genes of the AP1/AGL9 subfamily during floral transition. Plant Physiol 123: 1325–1336 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zahn LM, Kong H, Leebens-Mack JH, Kim S, Soltis PS, Landherr LL, Soltis DE, Depamphilis CW, Ma H. (2005) The evolution of the SEPALLATA subfamily of MADS-box genes: a preangiosperm origin with multiple duplications throughout angiosperm history. Genetics 169: 2209–2223 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zahn LM, Leebens-Mack JH, Arrington JM, Hu Y, Landherr LL, dePamphilis CW, Becker A, Theissen G, Ma H. (2006) Conservation and divergence in the AGAMOUS subfamily of MADS-box genes: evidence of independent sub- and neofunctionalization events. Evol Dev 8: 30–45 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.