Abstract
Brassinosteroids (BRs) are a unique class of plant steroid hormones that orchestrate myriad growth and developmental processes. Although BRs have long been known to protect plants from a suite of biotic and abiotic stresses, our understanding of the underlying molecular mechanisms is still rudimentary. Aiming to further decipher the molecular logic of BR-modulated immunity, we have examined the dynamics and impact of BRs during infection of rice (Oryza sativa) with the root oomycete Pythium graminicola. Challenging the prevailing view that BRs positively regulate plant innate immunity, we show that P. graminicola exploits BRs as virulence factors and hijacks the rice BR machinery to inflict disease. Moreover, we demonstrate that this immune-suppressive effect of BRs is due, at least in part, to negative cross talk with salicylic acid (SA) and gibberellic acid (GA) pathways. BR-mediated suppression of SA defenses occurred downstream of SA biosynthesis, but upstream of the master defense regulators NONEXPRESSOR OF PATHOGENESIS-RELATED GENES1 and OsWRKY45. In contrast, BR alleviated GA-directed immune responses by interfering at multiple levels with GA metabolism, resulting in indirect stabilization of the DELLA protein and central GA repressor SLENDER RICE1 (SLR1). Collectively, these data favor a model whereby P. graminicola coopts the plant BR pathway as a decoy to antagonize effectual SA- and GA-mediated defenses. Our results highlight the importance of BRs in modulating plant immunity and uncover pathogen-mediated manipulation of plant steroid homeostasis as a core virulence strategy.
To effectively combat invasion by microbial pathogens, plants have evolved a plethora of sophisticated mechanisms providing several strategic layers of constitutive and inducible defense responses. Many of these responses are regulated by an array of cross-communicating signal transduction pathways within which plant hormones fulfill central roles. Salicylic acid (SA), jasmonic acid (JA), and ethylene (ET) are the archetypal defense hormones and their importance in the hard wiring of the plant innate immune system is well established (Grant and Jones, 2009; Robert-Seilaniantz et al., 2011). Although there are exceptions, SA is predominantly associated with resistance to biotrophic pathogens, whereas necrotrophic pathogens are usually deterred by JA/ET-driven defenses. Moreover, interaction between these two types of defenses is mostly antagonistic, giving precedence to the notion that plant innate immunity follows in essence a binary model with SA and JA/ET having opposing influences (Pieterse et al., 2009).
Recently, abscisic acid (ABA), gibberellic acid (GA), cytokinins, and auxins emerged as critical regulators of plant-microbe interactions as well. Although their significance is less well understood, mounting evidence suggests these hormones influence disease outcomes by positively or negatively interfering with the SA-JA-ET backbone of the immune signaling circuitry (Pieterse et al., 2009). Such interplay or cross talk between individual hormone conduits is thought to enable plants to flexibly tailor their inducible defense arsenal to the type of invader encountered and to utilize their resources in a cost-efficient manner (Verhage et al., 2010). However, exciting new developments suggest that hormone cross talk may also be exploited by pathogens to shut down effective defenses through negative network connections (Robert-Seilaniantz et al., 2011). A classic example reflecting this situation is the production by some Pseudomonas syringae strains of a phytotoxin called coronatine that structurally resembles JA derivatives. Actively secreted in the host, coronatine is assumed to hyperactivate JA signaling, thereby counteracting SA-dependent defenses and facilitating bacterial invasion (Brooks et al., 2005; Cui et al., 2005; Melotto et al., 2006).
Brassinosteroids (BRs) are one of the latest growth regulators to be implicated in plant immunity. Discovered nearly 40 years ago, BRs are polyhydroxylated steroid hormones with important roles in regulating myriad physiological and developmental processes, including seed germination, skotomorphogenesis, flowering, and senescence (Clouse and Sasse, 1998). Over the past decade, molecular genetic studies using Arabidopsis (Arabidopsis thaliana) and rice (Oryza sativa) as model plants have identified numerous genes involved in BR biosynthesis and gene regulation. Coupled with more recent biochemical approaches, these studies have provided fascinating insights into the various aspects of plant steroid signaling, ranging from BR perception at the cell surface to activation of transcription factors in the nucleus (Kim and Wang, 2010). According to current concepts, BRs directly bind to the extracellular domain of the receptor-like kinase BRASSINOSTEROID INSENSITIVE1 (BRI1; She et al., 2011), thereby inducing a series of biochemical responses, including heterodimerization of BRI1 with, and activation of, another receptor kinase, BRI1-ASSOCIATED KINASE1 (BAK1; Li et al., 2002; Yun et al., 2009), phosphorylation of BRI1-interacting signaling kinases (Tang et al., 2008), and activation of the protein phosphatase BRI1 SUPPRESSOR PROTEIN1 (Kim et al., 2009). These events eventually culminate in inhibition of the shaggy-like kinase BRI1-INSENSITIVE2 (Vert and Chory, 2006) and resultant activation of the transcription factors BRASSINAZOLE-RESISTANT1 (BZR1) and BRI1-EMS-SUPPRESSOR1 (BES1)/BZR2 that orchestrate downstream gene expression (Sun et al., 2010; Yu et al., 2011).
Besides their critical role in growth regulation, BRs are well known to protect plants from a broad palette of environmental stresses, including low and high temperatures, drought, salinity, and insect herbivory (Bajguz and Hayat, 2009). Throughout the past decade, various BR-induced molecular changes that are related to stress tolerance have been identified. These include enhanced expression of stress-responsive genes (Kagale et al., 2007), protection of the translational machinery (Dhaubhadel et al., 2002), potentiated accumulation of osmoprotectants (Divi and Krishna, 2009), NADPH-oxidase-mediated accumulation of hydrogen peroxide (Xia et al., 2009b), and enhanced photosynthesis efficiency (Xia et al., 2009a). Recently, however, Divi et al. (2010) uncovered yet another mode of BR action during abiotic stress. Using mutant and transgenic Arabidopsis, they demonstrated that 24-epibrassinolide (BL)-induced tolerance to salt and temperature stress is reliant on the SA master regulatory protein NONEXPRESSOR OF PATHOGENESIS-RELATED GENES1 (NPR1), implicating a crucial role of the SA signaling pathway in BR-mediated stress responses.
Interestingly, BRs may also regulate plant responses to pathogen attack. For instance, exogenous application of BR lowers susceptibility of rice to fungal blast and bacterial blight diseases, and activates resistance of tobacco (Nicotiana tabacum) to Tobacco mosaic virus (TMV), P. syringae, and the powdery mildew fungus Oidium sp. (Nakashita et al., 2003). In keeping with these data, there is ample evidence from both field experiments and greenhouse trials demonstrating the protective effects of exogenous BRs against a fairly broad range of fungal, viral, and bacterial pathogens exhibiting diverse parasitic habits (Bajguz and Hayat, 2009). Together with the significantly increased BR levels in TMV-infected tobacco and the immune-suppressive effect of the BR biosynthesis inhibitor brassinazole (BRZ; Nakashita et al., 2003), these findings draw important inferences tagging BRs as powerful activators of broad-spectrum disease resistance in plants. However, in contrast to the relative wealth of information on BR’s function in the plant’s developmental program, relatively little is known about the molecular mechanisms underpinning BR-modulated plant immunity. Also, it remains to be resolved if, and how, BRs adjust and coordinate immune responses to soilborne pathogens.
Aiming to further decipher the molecular logic of BR-modulated immunity, we have analyzed the role of BRs during progression of rice infection by the root oomycete Pythium graminicola. Unlike most other experimentally tractable model plants, rice is a staple food for more than half the world and a model for monocots, which include cereal crops and biofuel grasses (Jung et al., 2008). P. graminicola, on the other hand, has recently been earmarked as one of the driving factors behind the progressive yield decline frequently observed in aerobic rice fields (Van Buyten et al., 2012). In contrast to the prevailing view that BRs boost plant innate immunity, our results provide compelling evidence that P. graminicola exploits endogenous BRs as virulence factors and hijacks the host BR machinery to inflict disease. Through genetic, physiological, and pathological analyses, we further show that BRs steer their detrimental effects on Pythium resistance, at least in part, through antagonistic cross talk with SA and GA. While challenging the common assumption that BRs positively influence plant defense responses, these data add substantial breadth to our understanding of the immune-regulatory role of BRs and reveal several heretofore-unknown aspects of BR pathway cross talk and signal integration.
RESULTS
BRs Suppress Basal Immunity of Rice to the Root Pathogen P. graminicola
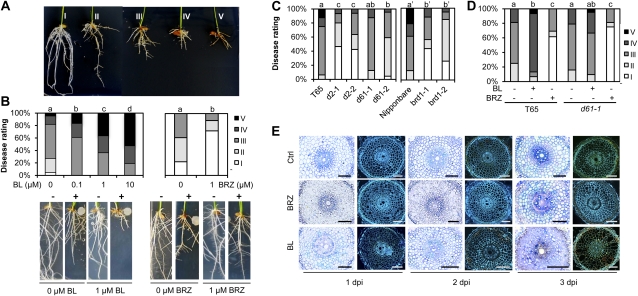
In a first attempt to elucidate the role of BRs in governing root immunity to P. graminicola, we tested the effect of exogenously administered BL, a biologically active BR (Nakashita et al., 2003). To this end, rice seeds (cv Nipponbare) were germinated and grown on agar plates containing various concentrations of BL. Because rice plants are susceptible to Pythium infection for only a few days after planting (Chun and Schneider, 1998), 3-d-old seedlings were tested for their susceptibility to the virulent strain PB912 132. Within 2 to 3 d post inoculation (dpi), roots of solvent-treated control plants developed typical brown necrotic patches, a symptom accompanied by strong reduction of seminal root length compared to noninoculated controls (Fig. 1A). Surprisingly, feeding plants with increasing concentrations of BL favored subsequent infection with P. graminicola, resulting in more extensive necrosis and stunting of inoculated roots as compared to the respective mock-inoculated controls (Fig. 1B). In contrast, treatment with 1 μm BRZ, a triazole that reversibly and specifically inhibits BR biosynthesis (Asami et al., 2000), led to a substantial reduction in disease severity (Fig. 1B). Importantly, neither BL nor BRZ had a significant impact on the in vitro growth of P. graminicola, implicating the involvement of plant-mediated responses (data not shown).
Figure 1.
BRs suppress root immunity in rice against the ooymycete P. graminicola. A, Illustration of the I-to-V disease severity scale used for disease evaluation. For more details on the different disease classes, see “Materials and Methods.” B, Disease promoting and reducing effect of exogenously administered BL and BRZ, respectively. Seeds (cv Nipponbare) were germinated on Gamborg B5 medium containing different concentrations of BL or BRZ, and, 3 d post imbibition, inoculated with 0.6-cm mycelial plugs of virulent P. graminicola PB912 132. Representative pictures were taken 7 dpi. Minus sign (−) = mock control, plus sign (+) = infected. Data represent one of three experiments with very similar results. Different letters indicate statistically significant differences (Mann Whitney; n ≥ 15; α = 0.05). C, Resistance of BR-deficient d2-1, d2-2, brd1-1, and brd1-2 and BR-insensitive d61-1 and d61-2. D, Effectiveness of exogenously administered BL and BRZ in wild-type T65 and BR-insensitive d61-1. For graphs C through D, statistical analysis was performed on pooled data from three independent experiments (Mann Whitney; n ≥ 32; α = 0.05). E, Microscopic analysis of early infection events in control, BL (1 μm), and BRZ (1 μm) pretreated Nipponbare roots inoculated with P. graminicola PB912 132. Left section: colonization of 5 μm root sections as visualized by trypan blue staining and bright-field microscopy. Right section: autofluorescence of representative root sections stained with Calcofluor white M2R (UV light excitation). Bars = 100 μm.
To gauge the physiological significance of these findings, we also tested several mutants that are either deficient in or insensitive to BR. As shown in Figure 1C, disease tests with the BR-deficient mutants brd1-1, brd1-2, d2-1, and d2-2 (Mori et al., 2002; Hong et al., 2003) revealed enhanced resistance as compared to the respective wild types, while complementing d2-2 with exogenous BL restored susceptibility to wild-type levels (Supplemental Fig. S1). In contrast, no strong differences in overall disease severity could be observed between wild-type T65 plants and the BR-insensitive mutants d61-1 and d61-2 (Fig. 1C), both of which carry a loss-of-function mutation in the rice BR receptor gene OsBRI1 (Yamamuro et al., 2000). One possible interpretation of this finding is that BR signaling plays a subordinate role in the P. graminicola resistance machinery. However, given the strong negative feedback regulation of BR biosynthesis, it is equally possible that an increase in resistance resulting from BR insensitivity is masked by the elevated levels of endogenous BRs in d61 (Yamamuro et al., 2000). To discriminate between these possibilities, we sought to supply wild-type and signal-defective d61-1 seedlings with 1 μm BRZ to lower the endogenous BR content and observe any effect on pathogen resistance. Interestingly, despite the higher basal levels of BR in the mutant line, BRZ treatment was equally effective in d61-1 and the wild type, suggesting that both BR biosynthesis and BR signaling serve higher susceptibility to Pythium attack (Fig. 1D).
To further discriminate the immune-regulatory role of BRs, samples of BL- and BRZ-treated Nipponbare roots were collected at various times after inoculation and analyzed for pathogen colonization and cellular reactions using a combination of bright-field and epifluorescence microscopy (Fig. 1E). Regardless of BL or BRZ treatment, numerous hyphae were found to be present along the longitudinal axis of the root within 12 h post inoculation. Following penetration (Supplemental Fig. S2A), primary hyphae differentiated rapidly into bulbous invading hyphae, giving rise to a dense network that, by 2 dpi, penetrated all inner cell layers of the root, including the cortex, the endodermis, and the vascular tissue (Fig. 1E; Supplemental Fig. S2B). Similar to what has been reported for the rice blast pathogen Magnaporthe oryzae (Kankanala et al., 2007), invading hyphae became more bulbous prior to crossing the cell wall and constricted dramatically, resulting in a thin invasive peg at the point of passage (Supplemental Fig. S2C). In control plants, early fungal progression, i.e. prior to 2 dpi, was characterized by successive invasions of root cells with no apparent loss of cell viability. In BL-supplemented plants, however, this apparent biotrophic phase was considerably shortened, with necrosis-related autofluorescence being detectable from as early as 24 h post inoculation. BL-treated seedlings were further characterized by dramatically enhanced pathogen growth relative to that seen in nontreated controls (Fig. 1E). Comparing control and BRZ-treated seedlings, no marked differences could be observed in pathogen proliferation, despite there being a substantial difference in overall disease severity between these treatments. However, BRZ treatment caused a significant delay in the onset of tissue necrotization, with the large majority of colonized cells remaining void of autofluorescence until as late as 3 dpi.
Temporal Dynamics of BR Biosynthesis and Signaling in Response to P. graminicola Attack
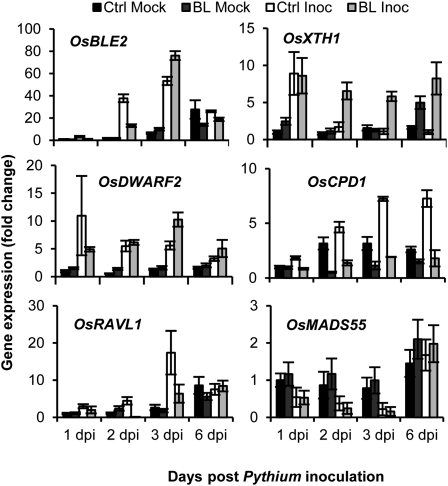
Recent advances in plant immunity research have provided fascinating insights into the ingenious ways by which microbial pathogens modify plant hormone homeostasis to subdue host immune responses and enforce a successful infection (Robert-Seilaniantz et al., 2011). To assess whether P. graminicola similarly coopts the BR pathway to tap into the rice signaling infrastructure, we used quantitative reverse transcription (qRT)-PCR to monitor the steady-state mRNA levels of various BR-responsive, biosynthetic, and regulatory genes in roots of inoculated Nipponbare plants grown in the presence or absence of BL. As shown in Figure 2, expression of the BL-inducible gene BRASSINOLIDE ENHANCED2 (OsBLE2; Yang et al., 2003) responded strongly to Pythium infection, and peaked at 3 dpi at approximately 55 times the levels found in noninoculated controls. At this time point, pathogen-induced transcription of OsBLE2 was even more pronounced in BL-pretreated plants, as was the expression of the BR-responsive endoglucan transglucosylase OsXTH1(Duan et al., 2006). Reaching a maximum at 1 dpi in control-inoculated roots, BL pretreatment caused OsXTH1 transcript levels to remain high throughout the course of the infection, suggesting a positive correlation between BR-inducible gene expression and overall susceptibility to P. graminicola. In a similar vein, Pythium inoculation entailed a strong up-regulation of the BR biosynthesis genes OsDWARF2 and CONSTITUTIVE PHOTOMORPHOGENESIS AND DWARFISM1 (OsCPD1), with mRNA levels peaking at 7 to 10 times the levels found in noninoculated controls. These results were rather unexpected given the apparent activation of BR signaling in Pythium-infected roots and the well-established negative feedback regulation of BR biosynthesis (Wang et al., 2002; He et al., 2005). Although not evident in the case of OsDWARF2, feedback inhibition was clearly seen for OsCPD1, with BL treatment strongly down-regulating expression of the gene relative to that seen in the respective mock controls. These outcomes suggest that P. graminicola coordinately affects the expression of genes involved in both BR signaling and biosynthesis, thereby bypassing feedback regulatory mechanisms. In line with this notion, pathogen-induced expression of OsBLE2 and OsXTH1 was greatly attenuated in signal-defective d61-1, whereas expression of the BR biosynthesis genes OsCPD1 and OsDWARF2 was comparable to or even higher than in the corresponding wild-type T65 (Supplemental Fig. S3).
Figure 2.
Effect of BL pretreatment on BR-response, -biosynthesis, and -regulatory genes in P. graminicola-inoculated rice roots. For details on BL treatments (1 μm) and Pythium bioassays see legend to Figure 1. Transcript levels were normalized using actin as an internal reference and expressed relative to the normalized expression levels in mock-inoculated control plants at 1 dpi. Data are means ± sd of two technical and two biological replicates from a representative experiment, each biological replicate representing a pooled sample from at least six individual plants.
In addition to the increased transcription of BR-biosynthetic and -response genes in P. graminicola-infected roots, we also found transcriptional alterations of key regulatory genes, including RAV-LIKE1 (OsRAVL1). Consistent with its pivotal role in maintaining basal activity of both BR signaling and biosynthesis (Il Je et al., 2010), OsRAVL1 mRNA levels responded strongly to pathogen inoculation, showing an approximately 15-fold induction over noninoculated controls. In the inoculated BL treatment, however, OsRAVL1 expression was less pronounced, presumably due to the already strong activation of BR signaling in this system. In contrast, both control and BL-pretreated roots exhibited a strong pathogen-specific down-regulation of the BR-negative regulator MADS-box PROTEIN55 (OsMADS55; Lee et al., 2008), suggesting a putative mechanism by which P. graminicola elevates BR signal processing. While demonstrating that successful infection of rice by P. graminicola is associated with major transcriptional reprogramming of various BR-associated genes, these data raise the possibility that P. graminicola usurps the host BR machinery to induce a state of susceptibility. In compliance with this concept, genome-wide transcriptome analysis of PB912 132 inoculated Nipponbare roots revealed more than one-third of all BR-associated genes (68 out of 192) represented on the array to be significantly altered after Pythium attack (Supplemental Table S1), with examples including the major BR biosynthetic genes OsDWARF (Mori et al., 2002) and OsDWARF2, the putative BR receptor gene BRI1-LIKE 3 (Nakamura et al., 2006), and the negative signaling regulator MADS-box PROTEIN1 (Duan et al., 2006). Moreover, analysis of the proximal 1-kb promoter region of all 4,381 genes significantly up-regulated by P. graminicola identified a significant overrepresentation of various cis-elements known to be responsive to BRs (Table I). These included two E-box elements (CANNTG), various motifs containing the BR-response element (BRRE), and the G-box CACGTG, which contains two inverted repeats of the core BRRE sequence CGTG.
Table I. BR-responsive cis-elements significantly enriched in P. graminicola-up-regulated rice genes.
| Cis-Element | Sequence | P Value | Reference |
| BRREa | CGTGCA | 2.40E-03 | He et al. (2005) |
| CGTGGC | 2.29E-02 | He et al. (2005) | |
| ACGTGG | 1.19E-02 | He et al. (2005) | |
| CCGTGC | 8.65E-03 | He et al. (2005) | |
| G-box | CACGTG | 5.56E-03 | Sun et al. (2010) |
| E-boxb | CAGCTG | 2.25E-02 | Yin et al. (2005); Il Je et al. (2010) |
| CACATG | 1.42E-02 | Yin et al. (2005); Il Je et al. (2010) |
CGTG = BRRE core sequence.
E-box consensus sequence = CANNTG.
BRs Antagonize Salicylate-Mediated Immunity against P. graminicola
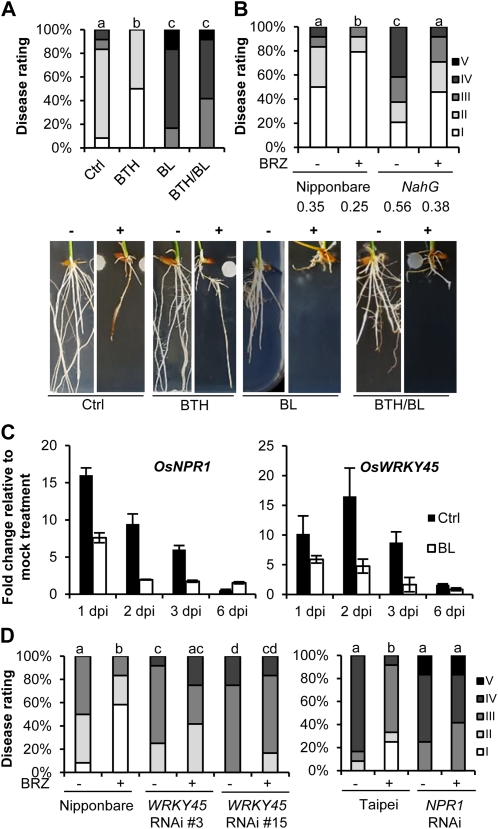
Mounting evidence indicates that pathogen defense signaling is not a linear single-response event, but a complex network involving multiple effectors and defense signals (Pieterse et al., 2009). Therefore, to further elucidate the molecular machinery underpinning BR-mediated susceptibility to P. graminicola, we focused on exploring the interaction of BR with other plant defense regulators. Given the paramount importance of SA in mediating foliar plant immunity (Robert-Seilaniantz et al., 2011), and the recent identification of BR-SA cross talk in the context of abiotic stress signaling (Divi et al., 2010), we first assessed the involvement of SA-modulated immune responses. Interestingly, we found that application of BL negates resistance conferred by the synthetic SA analog benzothiadiazole (BTH) (Fig. 3A), which is suggestive of negative cross talk in the direction of BR damping SA action. To test this hypothesis, wild-type Nipponbare and SA-deficient NahG transgenics (Yang et al., 2004) were routinely treated with 1 μm BRZ and tested for expression of induced resistance. As shown in Figure 3B, NahG plants were significantly more sensitive to pathogen attack than corresponding wild-type seedlings, demonstrating the importance of SA biosynthesis in basal resistance to P. graminicola. SA accumulation, however, did not appear to be a prerequisite for BRZ-induced resistance, as BRZ application was equally effective in wild-type Nipponbare and NahG plants, causing an approximate 30% reduction in basal disease susceptibility in both genotypes.
Figure 3.
BRs antagonize SA-mediated root immunity against P. graminicola. A, Exogenous BL (1 μm) attenuates BTH-induced resistance. For chemical induction of resistance, seeds were briefly dipped into a BTH solution (0.5 mm) and subsequently sown on GB5 medium containing 50 mg L21 BTH. BL treatment, pathogen inoculation, and disease evaluation was performed exactly as described in legend to Figure 1. Different letters indicate statistically significant differences (Mann-Whitney; n = 12; α = 0.05). Representative pictures were taken 7 dpi. Minus sign (−) = mock control, plus sign (+) = infected. B, Effect of BRZ pretreatment (1 μm) on Pythium susceptibility in wild-type Nipponbare and SA-deficient NahG plants. Statistically significant differences between treatments are labeled with different letters (Mann-Whitney; n = 24; α = 0.05). Numbers below graph represent disease index values. C, qRT-PCR analysis of the SA regulatory genes OsWRKY45 and OsNPR1 in control and BL-treated Nipponbare roots inoculated with P. graminicola. Transcript levels were normalized using actin as an internal reference and for each time point expressed relative to mock-inoculated controls. Data are means ± sd of two technical and two biological replicates from a representative experiment, each biological replicate representing a pooled sample from at least six individual plants. D, BRZ-induced resistance is compromised in OsWRKY45 and OsNPR1 RNAi lines. All genotypes were pretreated with 1 μm BRZ and subsequently inoculated with P. graminicola PB912 132. Bars labeled with different letters are significantly different (Mann-Whitney; n = 24; α = 0.05). All experiments were repeated at least twice with similar results. [See online article for color version of this figure.]
To further probe the nature of the SA-BR signal interaction, we next monitored the expression of the SA regulatory genes OsNPR1 and OsWRKY45 at various times after inoculation. Both genes encode master regulatory proteins that control distinct branches of the SA signaling cascade in rice (Shimono et al., 2007; Yuan et al., 2007). Following a rapid strong up-regulation upon pathogen infection, transcription of OsNPR1 and OsWRKY45 gradually decreased throughout the course of infection, an effect that was greatly accelerated by exogenous BL (Fig. 3C). Intriguingly, temporal expression of both genes seemed to be inversely correlated with that of the BR markers OsBLE2, OsCPD1, and OsRAVL1 (Fig. 2), especially within the first 3 dpi. In a similar vein, BL application also prevented the full age-dependent expression of OsNPR1 in noninoculated samples (red arrows on Supplemental Fig. S4). Along with the negative impact of exogenous BL on the BTH-inducible resistance (Fig. 3A), these results further support negative interactions between SA and BR during P. graminicola infection. To assess the functional relevance of such putative BR-SA antagonism, we then quantified the level of BRZ-induced resistance in OsWRKY45 and OsNPR1 RNAi lines (Fig. 3D). Interestingly, we found that plants silenced for OsWRKY45 are more susceptible to P. graminicola than the wild-type background and also fail to develop resistance when treated with BRZ. In contrast, no significant differences in disease susceptibility could be observed between nontreated wild type (cv Taipei) and OsNPR1 RNAi plants. However, similar to the OsWRKY45 transgenics, OsNPR1-silenced plants were unable to mount resistance when induced by BRZ, suggesting that BR antagonizes SA-mediated defenses downstream of SA accumulation, but upstream of OsNPR1 and OsWRKY45.
Repression of GA-Mediated Defenses Is a Crucial Facet of BR-Provoked Susceptibility to P. graminicola
GAs are a large family of tetracyclic diterpenoid hormones that control nearly every aspect of plant growth and development. Recent work on wild-type tissues of various plants has provided multiple physiological and molecular links to support extensive interplay between the BR and GA signaling pathways (Zhang et al., 2009). Considering these findings and given the emerging role of GA as a bona fide immunity hormone (Robert-Seilaniantz et al., 2011), we sought to extend our analysis of the disease-promoting effect of BRs by scrutinizing potential BR-GA cross talk. As shown in Supplemental Figure S5A, treatment of wild-type Nipponbare with increasing concentrations of GA3 enhanced resistance to P. graminicola in a concentration-dependent manner. Conversely, depletion of endogenous GA levels using the GA biosynthesis inhibitor uniconazole (Izumi et al., 1984) promoted disease susceptibility. Although care should be taken when interpreting these results since the specificity of uniconazole is not entirely clear (Bidadi et al., 2010), these data strongly suggest that GA acts as a positive player in rice immunity to P. graminicola (Supplemental Fig. S5B). Interestingly, supplying plants with both GA3 and BL recapitulated the BL-conferred disease susceptibility phenotype (Supplemental Fig. S5A), whereas cotreatment of uniconazole with BL did not confer an additive effect on the level of susceptibility (Supplemental Fig. S6A). Essentially identical results were obtained at the physiological level, with BL and uniconazole inhibiting primary root length to a similar extent (Supplemental Fig. S7A). Moreover, roots of the GA-deficient mutant d35 were less sensitive to exogenous BL than were wild-type roots (Supplemental Fig. S7C). Together, these data infer (1) that endogenous GA levels represent an important reservoir needed to mount a full innate immune response to P. graminicola, (2) that BR inhibits GA responses in roots at least in part by interfering with GA metabolism, and (3) that BR-induced susceptibility may develop coincidently with decreases in endogenous GA content. Supporting these conclusions, BRZ failed to stimulate root elongation (Supplemental Fig. S7A) and increase disease resistance (Supplemental Fig. S6B) when combined with 10 μm uniconazole.
The DELLA Protein and GA Repressor SLR1 Is a Key Player in Resistance to P. graminicola
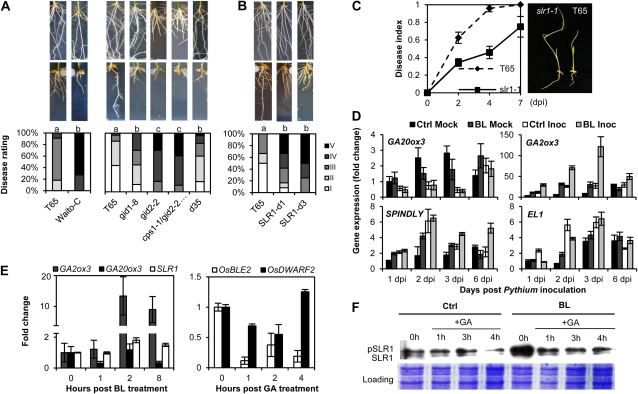
The observation that BR-induced susceptibility to P. graminicola is steered at least in part through negative cross talk with GA-controlled responses prompted us to further interrogate the role of GA in the rice-P. graminicola interaction. In contrast to its well-documented role as a plant growth regulator, GA has only recently been implicated in plant immunity. Current concepts suggest that at least in Arabidopsis, GA modulates plant disease resistance by inducing the degradation of DELLAs, a class of nuclear growth-repressing proteins that act as central suppressors of GA signaling (Navarro et al., 2008). Contrary to the five DELLA proteins present in Arabidopsis, rice has only a single DELLA gene, designated SLENDER RICE1 (SLR1; Ikeda et al., 2001). To discern the role of SLR1 and its position within the signal transduction path controlling disease and resistance against P. graminicola, several GA-deficient and/or insensitive rice mutants, all of which are known to overaccumulate SLR1 (Ueguchi-Tanaka et al., 2008), were tested for their susceptibility to P. graminicola. As shown in Figure 4A, all mutants tested, either impaired in GA biosynthesis (Waito-C, d35), insensitive to GA (gid1-8, gid2-2), or both (cps1-1/gid2-2), showed increased susceptibility compared with wild-type T65 plants. Enhanced susceptibility to P. graminicola was also observed in two independent SLR1 gain-of-function mutants (Asano et al., 2009; Fig. 4B), whereas slr1-1, a loss-of-function allele displaying a constitutive GA response phenotype (Ikeda et al., 2001), exhibited reduced necrosis and increased resistance to pathogen infection compared to inoculated T65 (Fig. 4C). In keeping with these findings, qRT-PCR analyses revealed SLR1 to be transiently up-regulated in response to both pathogen inoculation and BL treatment (Supplemental Fig. S8), further implicating SLR1 as a core participant in rice responses to P. graminicola.
Figure 4.
BR-induced susceptibility to P. graminicola involves repression of effectual GA-mediated defenses. A, Bioassays with several GA-deficient and -insensitive mutants reveal a positive role of GA in rice immunity to P. graminicola. All genotypes were inoculated as described in legend to Figure 1. Different letters indicate statistically significant differences (Mann-Whitney; n ≥ 12; α = 0.05). B, Hypersusceptibility of two independent SLR1 gain-of-function mutants. Bars with different letters are significantly different (Mann-Whitney; n = 12; α = 0.05). Photographs of noninoculated controls are shown in the top section, representative disease symptoms are depicted in the bottom section. C, Growth of P. graminicola PB912 132 on wild-type T65 and the constitutive GA mutant slr1-1. Data represent means ± se from three independent biological replicates (n = 15). D, Effect of BL pretreatment on the transcription of GA-associated genes in roots of cv Nipponbare inoculated with P. graminicola. BL treatment, pathogen inoculation, and qRT-PCR analysis were performed exactly as described in Figures 1 and 2. E, Cross-talk experiments demonstrating reciprocal negative interactions between BRs and GA in root tissue of cv Nipponbare. Six-day-old seedlings grown on GB5 medium were treated with either 1 μm BL or 50 μm GA3 for the indicated times and subjected to qRT-PCR analysis exactly as described in the legend to Figure 2. F, Immunoblot analysis of SLR1 protein accumulation in roots of 7-d-old Nipponbare plants grown in the presence or absence of 1 μm BL. SLR1 degradation was induced by submerging roots in 500 μm GA3 for the indicated times and protein levels were analyzed by western blotting using a SLR1-specific antibody. A duplicate protein gel was stained with Coomassie Blue (CBB) as loading control. [See online article for color version of this figure.]
Multilevel Interactions Mediate BR-GA Antagonism in P. graminicola-Inoculated Roots
Theoretically, cross talk between hormone pathways may occur at the level of biosynthesis regulation, signal transduction, and/or gene expression. To gain further insight into the molecular mechanisms of above-mentioned BR-GA interplay, we measured the transcript levels of multiple GA biosynthetic and regulatory genes in roots of inoculated Nipponbare grown in the presence or absence of 1 μm BL. As shown in Figure 4D, Pythium inoculation strongly induced the GA-degrading enzyme GIBBERELLIN 2-b-DIOXYGENASE3 (OsGA2ox3) in both control and BL-treated samples, whereas the GA-biosynthesis enzyme GIBBERELLIN 20-OXIDASE3 (OsGA20ox3) was severely down-regulated following inoculation. Similar results were obtained in our microarray analysis where perception of P. graminicola, despite being associated with activation of multiple genes involved in biosynthesis of inactive GA precursors (i.e. diterpenoids and GA12), led to a general repression of GA 20-oxidases but activation of GA 2-oxidases (Supplemental Table S2). In wild-type rice, expression of GA-biosynthetic genes is feedback inhibited by GA (Itoh et al., 2002); however, on the basis of the strong inhibitory effect of BL on GA-induced Pythium resistance as well as the likely role of GA biosynthesis in promoting immunity, we interpret these data to suggest that P. graminicola recruits the BR pathway, at least in part, to actively suppress GA biosynthesis in inoculated tissues. In support of this assumption, we found exogenously administered BL to inhibit GA20ox3 and induce GA2ox3 within 8 h of incubation (Fig. 4E). Intriguingly, treating roots with 50 μm GA3 likewise resulted in a fast and strong down-regulation of the BR response and BR biosynthesis genes OsBLE2 and OsDWARF2, respectively, indicating that BR and GA cause cross-inhibitory effects on the reciprocal hormone biosynthesis pathways to interact in a mutually antagonistic manner.
Besides interfering with GA metabolism, BR also may inhibit GA action through transcriptional activation of GA repressor genes. Over the last few years, several rice proteins with an inhibitory function in GA signaling have been characterized. These include the O-linked GlcNAc transferase SPINDLY (Shimada et al., 2006), and the casein kinase EARLY FLOWERING1 (EL1; Dai and Xue, 2010). Although expression of SPINDLY was transiently up-regulated by BL alone, the most dramatic changes in SPINDLY kinetics were seen following pathogen inoculation, with mRNA levels peaking at 2 dpi at approximately 6 times the levels found in noninoculated controls (Fig. 4D). A similar, albeit slightly more pronounced trend was noticed in BL-pretreated roots where pathogen-induced expression of SPINDLY was evident as late as 6 dpi. Interestingly, transcript accumulation of EL1 resembled the profile observed for SPINDLY, suggesting that P. graminicola triggers the expression of GA repressor proteins, possibly in a BL-dependent manner, as yet another mechanism to antagonize GA.
Given the central role of DELLA proteins in modulating hormonal cross talk in stress and developmental processes (Robert-Seilaniantz et al., 2011), and the reported ability of SPINDLY and EL1 to fine tune the suppressive function of rice SLR1 (Shimada et al., 2006; Dai and Xue, 2010), we finally asked whether BR-GA antagonism also is manifested at the level of SLR1 protein stability. To this end, we examined the level of SLR1 (using a monoclonal anti-SLR1 antibody) in wild-type Nipponbare roots grown for 7 d in the presence or absence of 1 μm BL. Consistent with previous reports (Sasaki et al., 2003), SLR1 migrated as two bands in SDS-PAGE gels, representing the phosphorylated (top band) and nonphosphorylated (bottom band) form of the protein, respectively. Levels of immunologically detectable SLR1 were obviously higher in BL-treated plants than in nontreated controls before the onset of GA3 treatment (Fig. 4F). Following GA3 treatment, however, no clear-cut differences in SLR1 levels were apparent between treatments, except for a slight increase in BL-treated roots at 4 h post GA application. Therefore, our findings argue that BL does not intervene with GA-mediated SLR1 protein turnover per se, but rather indirectly promotes SLR1 stabilization by inactivating GA through the combined repression of GA synthesis and transcriptional activation of GA repressor genes.
DISCUSSION
Originally discovered in the pollen of Brassica napus (Grove et al., 1979), BRs encompass a family of over 40 structurally and functionally related steroid compounds. Apart from their well-established function in orchestrating growth and developmental processes, there is extensive literature demonstrating the remarkable ability of BRs to boost plant responses to a suite of biotic and abiotic stresses. However, despite this broad functional repertoire, very little is known about the mechanisms underlying BR-mediated stress responses. In an effort to further our fundamental understanding of the immune-regulatory role of BR, we have studied its impact, dynamics, and interrelationship with other hormones during the interaction between rice and the soilborne oomycete P. graminicola. Challenging the prevailing view that BRs positively regulate plant pathogen responses, we show that BRs suppress root immunity to P. graminicola, resulting in increased pathogen proliferation and substantially enhanced disease susceptibility. Moreover, our results support a scenario whereby P. graminicola coopts the plant BR machinery as a decoy strategy to tap into the immune signaling circuitry and interfere with effectual SA- and GA-controlled defenses.
To date, genetic studies aimed at elucidating the role and action mechanisms of BR in plant stress adaptation have been confounded by the often strong and pleiotropic phenotypes of BR biosynthesis and signaling mutants, including extreme dwarfism, sterility, dark-green and epinastic leaves, and delayed development (Bishop, 2003). Moreover, current techniques for BR measurements require copious amounts of plant tissue and, despite allowing for quantification of specific intermediates in the BR pathway, often fail to capture stress-induced changes in the levels of the biologically active BRs, castasterone and BL (Shimada et al., 2001; Wu et al., 2008). As a result, most reports currently available are based on exogenous hormone applications. Under our experimental conditions, exogenous BL markedly promoted susceptibility to P. graminicola, whereas treatment with BRZ, a highly specific inhibitor of BR biosynthesis, induced substantial levels of resistance (Fig. 1B). Coupled with the extensive transcriptional reprogramming of various BR biosynthetic, signaling, and response genes in pathogen-inoculated roots and the overrepresentation of BR-responsive cis-elements in the promoters of P. graminicola-dependent rice genes (Fig. 2; Table I; Supplemental Table S1), these findings strongly argue that P. graminicola hijacks the rice BR biosynthesis and signaling machinery to cause disease, thus exploiting BRs as virulence factors. In compliance with this concept, many other microbial pathogens have lately been shown to disarm the plant’s weaponry by manipulating host hormone signaling (De Vleesschauwer et al., 2010; Robert-Seilaniantz et al., 2011), highlighting the central importance of hormone homeostasis in molding pathological outcomes.
As for many other hormones, BR cellular homeostasis is achieved mainly by end product feedback regulation with activation of BR signaling suppressing BR synthesis (He et al., 2005). The finding that in pathogen-inoculated roots, in spite of the strong activation of several BR-responsive genes, feedback control did not set in, but on the contrary OsDWARF2 and OsCPD1 were induced (Fig. 2), infers that P. graminicola concomitantly affects BR biosynthesis and ensuing signaling. One possible mechanism involves the B3 DNA-binding domain protein RAVL1. Recently identified as a transcriptional activator of both BR biosynthesis and BR signaling genes (Il Je et al., 2010), expression of RAVL1 was nonresponsive to exogenous BR, but strongly induced following P. graminicola inoculation (Fig. 2). However, other than attenuating negative feedback inhibition, P. graminicola also seems to impinge on downstream signal processing as revealed by the pathogen-specific suppression of the key BR repressor gene OsMADS55 (Fig. 2). Taken together, these findings strongly argue that P. graminicola targets multiple regulatory modules of the BR pathway to disturb BR cellular homeostasis and ensure prolonged activation of the pathway throughout the course of infection.
At least two different, not mutually exclusive, scenarios can be hypothesized to explain the negative impact of BRs on P. graminicola resistance. First, endogenous BRs may directly benefit the growth and/or virulence of the pathogen. Similar to oomycetes of the genus Phytophthora, Pythium species lack sterol biosynthetic pathways and thus need to assimilate sterols from their environment or host to support growth and initiate sexual reproduction (Hendrix, 1964). In view of this, it is tempting to speculate that P. graminicola usurps the plant’s BR machinery to secure access to sterols during the infection process and initiate production of oospores. However attractive, such a concept is hard to reconcile with the inability of exogenous BL to promote hyphal growth (data not shown) and the fairly similar rates of pathogen colonization in control and BRZ-treated roots (Fig. 1E). Moreover, despite their strong potential to enhance pathogen growth in culture and in sharp contrast with previous observations in the Arabidopsis-P. syringae pathosystem (Griebel and Zeier, 2010), we failed to observe any disease-promoting effect of exogenously administered plant sterols, including sitosterol and stigmasterol (data not shown). Therefore, rather than directly promoting pathogen fitness, host-produced BRs may enhance susceptibility to P. graminicola by interfering with specific plant defense signaling pathways. In compliance with this concept, there is ample evidence demonstrating extensive interplay between BR and other small-molecule hormones (Zhang et al., 2009). For example, BR modulates the biosynthesis of ET (Hansen et al., 2009) and intimately interacts with cytokinins and auxin in regulating various developmental processes (Goda et al., 2004; Vert et al., 2008; Peleg et al., 2011; Vercruyssen et al., 2011). Moreover, several microarray experiments and more recent biochemical approaches point to cross talk between BR and ABA and JA signaling pathways, respectively (Goda et al., 2002; Nemhauser et al., 2006; Zhang et al., 2009).
Recently, however, BRs were also found to interact with GA (Zhang et al., 2009; Sun et al., 2010). Although most examples of BR-GA signal connections relate to physiological processes, several lines of evidence indicate that BR/GA antagonism also fulfills a pivotal role during the BR-mediated susceptibility to P. graminicola. First, disease development was more severe on several GA-deficient and/or -insensitive mutants (Fig. 4, A and B), implying a positive role of GA in resistance to P. graminicola. Second, disruption of endogenous GA content using the GA biosynthesis inhibitor, uniconazole, yielded levels of susceptibility similar to those observed in BL-fed plants, whereas coapplication of BL and uniconazole did not cause an additive effect. Third, uniconazole treatment negated the resistance-inducing effect of BRZ, indicating a crucial requirement of de novo GA biosynthesis in the BRZ-triggered resistance. Fourth, BL treatment positively regulated the abundance of the DELLA protein and central GA repressor SLR1, a phenomenon accompanied by BL suppression of the GA biosynthetic gene GA20ox3 and induction of GA2ox3 oxidase involved in GA deactivation (Fig. 4, E and F). In conjunction with the central importance of SLR1 in P. graminicola resistance (Fig. 4, B and C; Supplemental Fig. S8), these findings support a scenario whereby BR triggers susceptibility to P. graminicola at least in part by counteracting pathogen-induced GA synthesis, leading to indirect stabilization of the DELLA protein SLR1. Supporting this hypothesis, various other phytohormones, including ABA, cytokinin, auxin, and ET have previously been shown to affect the GA-induced destabilization of DELLA proteins. Thus, auxin promotes GA-induced proteolysis of DELLAs, whereas stress-induced ABA, cytokinin, and ET enhance DELLA stabilization and delay its degradation by GA (Fu and Harberd, 2003; Vriezen et al., 2004; Brenner et al., 2005; Achard et al., 2007). Moreover, recent data have brought DELLA proteins to the fore as pivotal regulators of plant-microbe interactions. In Arabidopsis, DELLAs positively regulate resistance to necrotrophs but impede immunity to biotrophs (Navarro et al., 2008). Accordingly, we found pathogen-induced SLR1 expression to occur predominantly within the first 2 dpi (Supplemental Fig. S8), a time frame corresponding to the assumed biotrophic phase of the P. graminicola infection cycle (Fig. 1E).
Intriguingly, the positive effect of BRs on DELLA protein stability could also provide a mechanistic explanation for the observation that BL induces resistance to rice blast and bacterial blight (Nakashita et al., 2003), both of which diseases are known to be favored by high endogenous GA levels (Yang et al., 2008). In a similar vein, BR-modulated DELLA abundance may offer a mechanistic framework for how BRs trigger resistance to abiotic stresses. In some elegant work using Arabidopsis mutants lacking four of the five DELLA proteins, Achard et al. (2006) showed a positive correlation between DELLA protein levels and tolerance to various abiotic stresses. These authors also demonstrated that DELLA proteins elevate the expression of antioxidant enzymes and thereby limit oxidative stress-induced cell death (Achard et al., 2008), two effects which, at least in cucumber (Cucumis sativus), also have been attributed to elevated BR activity (Xia et al., 2009b). In line with our results and given the central role of reactive oxygen species (ROS) as initiating agents in myriad plant stress responses (Miller et al., 2008), BR-conferred abiotic stress tolerance could therefore be explained by an indirect effect based on BR induction of DELLA-mediated ROS detoxification.
Besides the negative effect of BR on GA-controlled and SLR1-dependent defenses, our data implicate a crucial role of BR-SA signal interactions in determining rice-P. graminicola outcomes. Using a combination of exogenous hormone treatments, qPCR-based expression experiments, and bioassays with SA-deficient and -insensitive transgenics, we found that BR interferes with SA-dependent defenses downstream of SA biosynthesis, but upstream of the master SA regulators OsWRKY45 and OsNPR1 (Fig. 3). Interestingly, apparently contradictory results demonstrate that BL application enhances tolerance to salt and thermo stress, two traits that are associated with increased SA signaling and that require functional NPR1 (Divi et al., 2010). Furthermore, in both Arabidopsis and cucumber, BL treatment induces the expression of the classical SA marker gene PR1 (Xia et al., 2009b; Divi et al., 2010), while the BR response regulator BES1 was recently found to physically bind and activate the key defense regulator AtMYB30 (Li et al., 2009). Nevertheless, recent findings suggest that BR may also antagonize plant defense signaling via its transcriptional regulator BZR1. Upon activation by BR, BZR1 binds and represses the promoters of various defense-associated genes, including the flagellin receptor FLS2 and the major R gene SNC1 (Sun et al., 2010). This apparently ambivalent BR response is also evident in rice. Indeed, whereas our data clearly uncover BR as a negative regulator of SA-mediated immunity against P. graminicola, Nakashita et al. (2003) previously demonstrated BL application to enhance resistance against M. oryzae and the bacterium Xanthomonas oryae pv oryzae, presumably in an SA-independent manner. Hence, a complex picture is emerging in which steroid hormones, like the other defense hormones SA, JA/ET, and ABA, function as global multifaceted regulators of plant innate immunity, with apparently divergent outcomes. Given the widespread role of BRs in various aspects of growth and development, this notion also implies that BRs are positioned at the interface of hormone, developmental, and stress signaling, and thus may serve as important regulators of the often-reported trade-off between enhanced disease resistance and plant growth (Purrington, 2000; Bolton, 2009).
A key question arising from our findings, therefore, is whether the immune-suppressive effect of pathogen-induced BR is unique for the rice-P. graminicola pathosystem, or, alternatively, reflects a conserved virulence mechanism based on the manipulation of in planta BR signaling and/or biosynthesis. In this respect, it is noteworthy that transgenic Arabidopsis expressing AvrPto, a bacterial effector that targets the BR coreceptor BAK1, not only displays compromised immunity but also exhibits phenotypes similar to these of BR-insensitive mutants (Shan et al., 2008). Significantly, AvrPto expression does not affect the plant response to other growth-promoting hormones, raising the possibility that AvrPto specifically binds BAK1 to modify BR signaling and attenuate host defenses (Shan et al., 2008). Similarly, it was recently reported that enhanced BR signaling resulting from either a gain-of-function mutation in BAK1 (Jaillais et al., 2011), ectopic expression of BRI1 (Belkhadir et al., 2012), or application of BL (Albrecht et al., 2012) impedes innate immunity to P. syringae conditioned by the conserved microbial signature flg22. These findings are particularly interesting in light of recent reports demonstrating that the obligate biotrophic oomycete Albugo laibachii is equipped with a near-complete BR biosynthesis pathway and thus may produce BR itself (Kemen et al., 2011). Also, several fungal pathogens synthesize toxins that closely resemble steroid hormones such as zearalenone (Robert-Seilaniantz et al., 2007). By analogy with coronatine-mediated disease susceptibility (Cui et al., 2005; Melotto et al., 2006), one may envision that pathogenic microbes employ such BR mimicry as a virulence strategy to break into the plant’s signaling infrastructure and suppress host defense responses. Considering the vital role of steroid hormones in coordinating and integrating myriad cellular, developmental, and physiological processes (Sun et al., 2010; Yu et al., 2011), manipulating host BR signaling and hijacking BR hormone cross-talk mechanisms likely represents a powerful virulence strategy controlling the outcome of many plant-pathogen interactions.
CONCLUSION
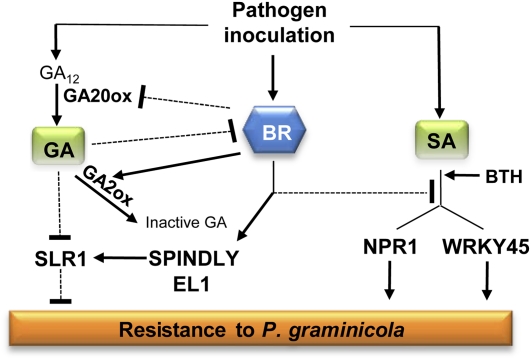
In summary, we have shown that both exogenously administered and endogenous BRs negatively influence root immunity to the rice pathogen P. graminicola. Moreover, our results favor a scenario whereby P. graminicola hijacks the rice BR machinery as a decoy to shut down effective SA- and GA-mediated immune responses (Fig. 5). While challenging the common assumption that BRs act as positive players in the plant’s defense signaling circuitry, the findings presented in this study highlight the importance of BRs in modulating immunity during plant growth and uncover pathogen-mediated manipulation of plant steroid homeostasis as a core virulence strategy.
Figure 5.
Model illustrating how BR, SA, and GA and defense-related cross talk among this triad of stress hormones molds rice immunity to P. graminicola. Perception of P. graminicola leads to pronounced increases in endogenous BR levels and resultant activation of BR signal transduction. This pathogen-triggered BR action, which likely results from combined induction of de novo BR biosynthesis, attenuation of negative feedback regulation, and potentiated derepression of BR signaling, plays a dual role in promoting susceptibility. On the one hand, BR antagonizes effective SA-mediated defenses by interfering downstream of SA accumulation, but upstream of OsNPR1 and OsWRKY45. On the other hand, BR dampens effectual immune responses directed by GA. Operating at both the level of biosynthesis regulation and signal transduction, this BR-GA antagonism leads to indirect stabilization of the DELLA protein SLR1. Sharp, black lines represent positive effects; blunted, dotted lines depict antagonistic interactions. [See online article for color version of this figure.]
MATERIALS AND METHODS
Plant Materials and Growth Conditions
Rice (Oryza sativa) lines used in this work included the japonica cultivar Nipponbare, the corresponding NahG (Yang et al., 2004) and OsWRKY45 RNAi transgenics (Shimono et al., 2007), the BR-deficient mutant alleles brd1-1 and brd1-2 (Mori et al., 2002), as well as an OsNPR1-silenced line (Yuan et al., 2007) and its wild-type Taipei (cv japonica). The GA-deficient and/or insensitive mutants d35 (ent-kaurene oxidase mutant; Itoh et al., 2004), Waito-C (GA3ox2 mutant; Itoh et al., 2001), gid1-8 (GA receptor GID1 mutant; Ueguchi-Tanaka et al., 2007), gid2-2 (F-box protein GID2 mutant; Sasaki et al., 2003), cps1-1/gid2-2 (ent-copalyl diphosphate synthase mutant crossed with gid2-2; Ueguchi-Tanaka et al., 2008), and the SLR1 loss- or gain-of-function alleles slr1-1 (Ikeda et al., 2001), SLR1-d1, and SLR1-d3 (Asano et al., 2009), respectively, all are in the background of japonica cultivar T65, as are the BR-insensitive and BR-deficient mutant lines d61-1, d61-2, d2-1, and d2-2 (Yamamuro et al., 2000; Hong et al., 2003). For seed multiplication, plants were propagated in the greenhouse (30°C ± 4°C, 16-h photoperiod) and fertilized with 0.5% ammonium sulfate every 2 weeks until flowering.
Pythium graminicola Bioassays
Pythium graminicola strain PB912 132, isolated from a diseased aerobic rice field in Los Baños, The Philippines (Van Buyten et al., 2012), was cultivated at 28°C on potato dextrose agar (PDA; Difco Laboratories). Rice seeds were surface sterilized by agitation in 2% sodium hypochlorite for 20 min, rinsed three times with sterile demineralized water, plated in square petri dishes (12 × 12 cm) on standard strength Gamborg B5/1% plant agar medium, and subsequently grown in a growth chamber at 28°C (day)/26°C (night) under 12-h photoperiod. Three days after imbibition, i.e. when primary roots were approximately 1 cm in length, germinated seedlings were inoculated by carefully placing a 0.6-cm-diameter agar plug taken from the margin of 7-d-old PDA cultures in between each two plants. Control samples were mock inoculated with 0.6-cm-diameter PDA plugs. Disease symptoms were scored 7 dpi and disease rates were expressed on the basis of diseased root area and reduction in root length using a 1-to-5 disease severity scale. To account for differences in root growth caused by genetic mutations, exogenous hormones or chemical inhibitor compounds per se, all ratings were expressed relative to the respective noninfected controls: class I, root length more than 60% of the respective mock treatment, very little necrosis covering less than 10% of total root area; II, root length more than 60% of the respective mock treatment, more than 10% of total root area necrotic; III, root length between 20% and 60% of the respective mock treatment; IV, root length <20% of the respective mock treatment, less than 75% of root area necrotic; V, root length <20% of the respective mock treatment, more than 75% of root area affected. Disease index values were calculated according to the following formula: [((1 × a) + (2 × b) + (3 × c) + (4 × d) + (5 × e))/(a + b + c + d + e)] × 100/5, where a, b, c, d, and e are the number of roots with scores I, II, III, IV, and V, respectively.
Chemical Treatments
BTH (BION 50 WG, Syngenta) and GA3 (Sigma-Aldrich) were dissolved in water at the indicated concentrations. Stock solutions of BL (Sigma-Aldrich) and uniconazole (Wako) were in ethanol, whereas BRZ (Wako) was dissolved in dimethyl sulfoxide. Equivalent volumes of both solvents were added to separate control treatments. All chemicals were added to autoclaved media after cooling to approximately 60°C.
Microscopy of Root Infection
Root infection was monitored at various times after inoculation with P. graminicola PB912 132 using a combination of bright-field and epifluorescence microscopy. Root samples were fixed in a 50 mm sodium phosphate buffer (pH 7.2) containing 4% paraformaldehyde and 1% glutaraldehyde, dehydrated in a graded series of ethanol, and infiltrated with Technovit 7100 solution. The infiltrated root samples were embedded in plastic cubes filled with Technovit 7100 histo-embedding medium (Heraeus Kulzer). A Leica RM2265 motorized rotary microtome (Leica Microsystems) was used to obtain 5-μm cross sections of the embedded tissue. Pythium structures were visualized by staining tissue sections in 0.1% (w/v) trypan blue in 10% (v/v) acetic acid for 5 min. The oxidation of phenolic compounds, which is related with induced necrosis, was visualized by staining sections in 0.1% fluorescent brightener 28 (Calcofluor White M2R) for 1 min. After rinsing and drying, the sections were mounted with neutral mounting medium (DPX; Klinipath). Images were digitally acquired with an Olympus BX51 microscope equipped with an Olympus ColorView III camera and further processed with the Olympus analysis cell^F software (Olympus Soft Imaging Solutions) and ImageJ 1.44p.
Western-Blot Analysis
Extraction of total proteins and western blotting were performed as described by Shimada et al. (2006) with minor modifications. Total protein samples were extracted from 7-d-old rice seedlings grown on Gamborg 5 agar plates with 2× extraction buffer (20 mm Tris-HCl, pH 7.5, 150 mm NaCL, 0.5% Tween 20, 1 mm EDTA, 1 mm phenylmethylsulfonyl fluoride). Following quantification using a Bradford assay (Bio-Rad), protein extracts were mixed with an equal volume of 2× sample buffer (150 mm Tris-HCl, pH 6.8, 2% w/v SDS, 10% w/v glycerol, 0.01% w/v bromophenol blue, and 0.1 m w/v dithiothreitol), boiled for 5 min, separated on 7.5% SDS-PAGE gels, and transferred to a Hybond ECL membrane (Amersham Pharmacia Biotech). Polyclonal anti-SLR1 antibody and goat anti-rabbit HRP-conjugated secondary antibody (Dako) were used to quantify SLR1. Peroxidase activity was detected using ECL plus western-blotting substrate (Pierce biotechnology) according to the manufacturer’s instructions.
RNA Extraction and Quantitative RT-PCR
Total RNA was extracted from frozen root tissue using the spectrum plant total RNA kit (Sigma-Aldrich) and subsequently Turbo DNase treated according to the provided protocol (Ambion). First-strand cDNA was synthesized from 2 μg of total RNA using Multiscribe reverse transcriptase (Applied Biosystems) and random primers following the manufacturer’s instructions. Quantitative PCR amplifications were conducted in optical 96-well plates with the Mx3005P real-time PCR detection system (Stratagene), using Sybr Green master mix (Fermentas) to monitor dsDNA synthesis. The expression of each gene was assayed in duplicate in a total volume of 25 μL including a passive reference dye (ROX) according to the manufacturer’s instructions (Fermentas). The thermal profile used consisted of an initial denaturation step at 95°C for 10 min, followed by 40 cycles of 95°C for 15s, 59°C for 30s, and 72°C for 30s. To verify amplification of one specific target cDNA, a melting-curve analysis was included according to the thermal profile suggested by the manufacturer (Stratagene). The amount of plant RNA in each sample was normalized using OsACTIN1 (LOC_Os03g50885) as internal control and samples collected from control plants at 1 dpi were selected as calibrator. The data were analyzed using Stratagene’s Mx3005P software. Nucleotide sequences of all primers used are listed in Supplemental Table S3.
Microarray Analysis and Data Processing
Rice plants (cultivar Nipponbare) were grown and infected with P. graminicola strain PB912 132 as described before. Samples from control mock-infected and Pythium-inoculated plants were taken 1, 2, and 4 dpi, each sample representing a pool from at least 20 individual plants. Three biological replicates were performed, giving a total of 18 RNA samples (two treatments × three time points × three replicates). For microarray analysis, we used a previously described two-dye method allowing direct comparison between two samples on the same microarray (Satoh et al., 2010). In brief, cyanine 3- or cyanine 5-labeled cRNA samples were synthesized from 850 ng of total RNA using a low-input RNA labeling kit (Agilent Technologies) and hybridized to custom-made 60-mer Agilent arrays according to the manufacturer’s protocols (Agilent Technologies). Following washing, slide image files were generated using a DNA microarray scanner (G2505B; Agilent Technologies) and signal intensities were extracted and normalized within each array using Feature Extraction version 9.5 (Agilent Technologies). Signal intensities among all arrays were normalized according to the quantile method for standardization (global scaling) using EXPANDER ver 4.1. Significance analysis was performed using a fixed linear model (ANOVA) implemented in R/MAANOVA. Differentially expressed genes were defined as genes with a log2-based signal ratio >0.585 or <−0.585 (i.e. 1.5-fold) and a false discovery rate of less than 0.05. All microarray data generated in this study are available in the Gene Expression Omnibus (NCBI-GEO; http://www.ncbi.nlm.nih.gov/geo) database under the reference GSE32582. For motif search and cis-element identification, 1-kb promoter regions of all differentially expressed genes were extracted from the Rice Genome Annotation (http://rice.plantbiology.msu.edu) and Orygenes (http://orygenesdb.cirad.fr) databases and analyzed using the RSAT (http://rsat.ulb.ac.be/rsat/), Osiris (http://www.bioinformatics2.wsu.edu/Osiris/), and PLACE programs (http://www.dna.affrc.go.jp/PLACE/).
Supplemental Data
The following materials are available in the online version of this article.
Supplemental Figure S1. Effect of BL pretreatment on resistance to P. graminicola in the BR-deficient mutant d2-2.
Supplemental Figure S2. Morphology and dynamics of P. graminicola colonization of rice roots.
Supplemental Figure S3. Expression of BR-responsive (OsBLE2, OsXTH1) and BR-biosynthetic (OsCPD1, OsDWARF2) genes in wild-type T65 and signal-defective d61-1 seedlings inoculated with P. graminicola.
Supplemental Figure S4. Effect of BL pretreatment on transcript accumulation of the SA marker genes OsNPR1 and OsWRKY45 in roots of cv Nipponbare inoculated with P. graminicola.
Supplemental Figure S5. Repression of GA biosynthesis is an integral part of BR-mediated susceptibility to P. graminicola.
Supplemental Figure S6. Uniconazole treatment mimics BL-induced susceptibility but attenuates BRZ-triggered resistance.
Supplemental Figure S7. Physiological readouts of GA-BL interactions in rice leaf and root tissue.
Supplemental Figure S8. Effect of BL pretreatment on transcript accumulation of the DELLA gene SLR1 in roots of cv Nipponbare inoculated with P. graminicola.
Supplemental Table S1. BR-associated genes differentially expressed after P. graminicola infection.
Supplemental Table S2. GA metabolism genes significantly induced or repressed following inoculation of rice roots with P. graminicola.
Supplemental Table S3. Primer sequences used in quantitative RT-PCR.
Supplementary Material
Acknowledgments
We thank Ilse Delaere for technical assistance; Miyako Ueguchi-Tanaka, Makoto Matsuoko, Motoyuki Ashikari, Hiroshi Takatsuji, Zuhua He, and Yinong Yang for providing BR, GA, and SA mutant and transgenic rice lines; and Hanne Van Gorp for excellent discussions during the preparation of the manuscript.
References
- Achard P, Baghour M, Chapple A, Hedden P, Van Der Straeten D, Genschik P, Moritz T, Harberd NP. (2007) The plant stress hormone ethylene controls floral transition via DELLA-dependent regulation of floral meristem-identity genes. Proc Natl Acad Sci USA 104: 6484–6489 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Achard P, Cheng H, De Grauwe L, Decat J, Schoutteten H, Moritz T, Van Der Straeten D, Peng JR, Harberd NP. (2006) Integration of plant responses to environmentally activated phytohormonal signals. Science 311: 91–94 [DOI] [PubMed] [Google Scholar]
- Achard P, Renou J-P, Berthomé R, Harberd NP, Genschik P. (2008) Plant DELLAs restrain growth and promote survival of adversity by reducing the levels of reactive oxygen species. Curr Biol 18: 656–660 [DOI] [PubMed] [Google Scholar]
- Albrecht C, Boutrot F, Segonzac C, Schwessinger B, Gimenez-Ibanez S, Chinchilla D, Rathjen JP, de Vries SC, Zipfel C. (2012) Brassinosteroids inhibit pathogen-associated molecular pattern-triggered immune signaling independent of the receptor kinase BAK1. Proc Natl Acad Sci USA 109: 303–308 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Asami T, Min YK, Nagata N, Yamagishi K, Takatsuto S, Fujioka S, Murofushi N, Yamaguchi I, Yoshida S. (2000) Characterization of brassinazole, a triazole-type brassinosteroid biosynthesis inhibitor. Plant Physiol 123: 93–100 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Asano K, Hirano K, Ueguchi-Tanaka M, Angeles-Shim RB, Komura T, Satoh H, Kitano H, Matsuoka M, Ashikari M. (2009) Isolation and characterization of dominant dwarf mutants, Slr1-d, in rice. Mol Genet Genomics 281: 223–231 [DOI] [PubMed] [Google Scholar]
- Bajguz A, Hayat S. (2009) Effects of brassinosteroids on the plant responses to environmental stresses. Plant Physiol Biochem 47: 1–8 [DOI] [PubMed] [Google Scholar]
- Belkhadir Y, Jaillais Y, Epple P, Balsemão-Pires E, Dangl JL, Chory J. (2012) Brassinosteroids modulate the efficiency of plant immune responses to microbe-associated molecular patterns. Proc Natl Acad Sci USA 109: 297–302 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bidadi H, Yamaguchi S, Asahina M, Satoh S. (2010) Effects of shoot-applied gibberellin/gibberellin-biosynthesis inhibitors on root growth and expression of gibberellin biosynthesis genes in Arabidopsis thaliana. Plant Root 4: 4–11 [Google Scholar]
- Bishop GJ. (2003) Brassinosteroid mutants of crops. J Plant Growth Regul 22: 325–335 [DOI] [PubMed] [Google Scholar]
- Bolton MD. (2009) Primary metabolism and plant defense—fuel for the fire. Mol Plant Microbe Interact 22: 487–497 [DOI] [PubMed] [Google Scholar]
- Brenner WG, Romanov GA, Köllmer I, Bürkle L, Schmülling T. (2005) Immediate-early and delayed cytokinin response genes of Arabidopsis thaliana identified by genome-wide expression profiling reveal novel cytokinin-sensitive processes and suggest cytokinin action through transcriptional cascades. Plant J 44: 314–333 [DOI] [PubMed] [Google Scholar]
- Brooks DM, Bender CL, Kunkel BN. (2005) The Pseudomonas syringae phytotoxin coronatine promotes virulence by overcoming salicylic acid-dependent defences in Arabidopsis thaliana. Mol Plant Pathol 6: 629–639 [DOI] [PubMed] [Google Scholar]
- Chun SC, Schneider RW. (1998) Sites of infection by pythium species in rice seedlings and effects of plant age and water depth on disease development. Phytopathology 88: 1255–1261 [DOI] [PubMed] [Google Scholar]
- Clouse SD, Sasse JM. (1998) Brassinosteroids: essential regulators of plant growth and development. Annu Rev Plant Physiol Plant Mol Biol 49: 427–451 [DOI] [PubMed] [Google Scholar]
- Cui J, Bahrami AK, Pringle EG, Hernandez-Guzman G, Bender CL, Pierce NE, Ausubel FM. (2005) Pseudomonas syringae manipulates systemic plant defenses against pathogens and herbivores. Proc Natl Acad Sci USA 102: 1791–1796 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dai C, Xue H-W. (2010) Rice early flowering1, a CKI, phosphorylates DELLA protein SLR1 to negatively regulate gibberellin signalling. EMBO J 29: 1916–1927 [DOI] [PMC free article] [PubMed] [Google Scholar]
- De Vleesschauwer D, Yang YN, Cruz CV, Höfte M. (2010) Abscisic acid-induced resistance against the brown spot pathogen Cochliobolus miyabeanus in rice involves MAP kinase-mediated repression of ethylene signaling. Plant Physiol 152: 2036–2052 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dhaubhadel S, Browning KS, Gallie DR, Krishna P. (2002) Brassinosteroid functions to protect the translational machinery and heat-shock protein synthesis following thermal stress. Plant J 29: 681–691 [DOI] [PubMed] [Google Scholar]
- Divi UK, Krishna P. (2009) Brassinosteroid: a biotechnological target for enhancing crop yield and stress tolerance. New Biotechnol 26: 131–136 [DOI] [PubMed] [Google Scholar]
- Divi UK, Rahman T, Krishna P. (2010) Brassinosteroid-mediated stress tolerance in Arabidopsis shows interactions with abscisic acid, ethylene and salicylic acid pathways. BMC Plant Biol 10: 151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Duan K, Li L, Hu P, Xu S-P, Xu Z-H, Xue H-W. (2006) A brassinolide-suppressed rice MADS-box transcription factor, OsMDP1, has a negative regulatory role in BR signaling. Plant J 47: 519–531 [DOI] [PubMed] [Google Scholar]
- Fu XD, Harberd NP. (2003) Auxin promotes Arabidopsis root growth by modulating gibberellin response. Nature 421: 740–743 [DOI] [PubMed] [Google Scholar]
- Goda H, Sawa S, Asami T, Fujioka S, Shimada Y, Yoshida S. (2004) Comprehensive comparison brassinosteroid-regulated of auxin-regulated and brassinosteroid-regulated genes in Arabidopsis. Plant Physiol 134: 1555–1573 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goda H, Shimada Y, Asami T, Fujioka S, Yoshida S. (2002) Microarray analysis of brassinosteroid-regulated genes in Arabidopsis. Plant Physiol 130: 1319–1334 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grant MR, Jones JDG. (2009) Hormone (dis)harmony moulds plant health and disease. Science 324: 750–752 [DOI] [PubMed] [Google Scholar]
- Griebel T, Zeier J. (2010) A role for beta-sitosterol to stigmasterol conversion in plant-pathogen interactions. Plant J 63: 254–268 [DOI] [PubMed] [Google Scholar]
- Grove MD, Spencer GF, Rohwedder WK, Mandava N, Worley JF, Warthen JD, Steffens GL, Flippenanderson JL, Cook JC. (1979) Brassinolide, a plant growth-promoting steroid isolated from Brassica napus pollen. Nature 281: 216–217 [Google Scholar]
- Hansen M, Chae HS, Kieber JJ. (2009) Regulation of ACS protein stability by cytokinin and brassinosteroid. Plant J 57: 606–614 [DOI] [PMC free article] [PubMed] [Google Scholar]
- He JX, Gendron JM, Sun Y, Gampala SSL, Gendron N, Sun CQ, Wang ZY. (2005) BZR1 is a transcriptional repressor with dual roles in brassinosteroid homeostasis and growth responses. Science 307: 1634–1638 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hendrix JW. (1964) Sterol induction of reproduction and stimulation of growth of Pythium and Phytophthora. Science 144: 1028–1029 [DOI] [PubMed] [Google Scholar]
- Hong Z, Ueguchi-Tanaka M, Umemura K, Uozu S, Fujioka S, Takatsuto S, Yoshida S, Ashikari M, Kitano H, Matsuoka M. (2003) A rice brassinosteroid-deficient mutant, ebisu dwarf (d2), is caused by a loss of function of a new member of cytochrome P450. Plant Cell 15: 2900–2910 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ikeda A, Ueguchi-Tanaka M, Sonoda Y, Kitano H, Koshioka M, Futsuhara Y, Matsuoka M, Yamaguchi J. (2001) slender rice, a constitutive gibberellin response mutant, is caused by a null mutation of the SLR1 gene, an ortholog of the height-regulating gene GAI/RGA/RHT/D8. Plant Cell 13: 999–1010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Itoh H, Tatsumi T, Sakamoto T, Otomo K, Toyomasu T, Kitano H, Ashikari M, Ichihara S, Matsuoka M. (2004) A rice semi-dwarf gene, Tan-Ginbozu (D35), encodes the gibberellin biosynthesis enzyme, ent-kaurene oxidase. Plant Mol Biol 54: 533–547 [DOI] [PubMed] [Google Scholar]
- Itoh H, Ueguchi-Tanaka M, Sato Y, Ashikari M, Matsuoka M. (2002) The gibberellin signaling pathway is regulated by the appearance and disappearance of SLENDER RICE1 in nuclei. Plant Cell 14: 57–70 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Itoh H, Ueguchi-Tanaka M, Sentoku N, Kitano H, Matsuoka M, Kobayashi M. (2001) Cloning and functional analysis of two gibberellin 3 beta-hydroxylase genes that are differently expressed during the growth of rice. Proc Natl Acad Sci USA 98: 8909–8914 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Izumi K, Yamaguchi I, Wada A, Oshio H, Takahashi N. (1984) Effects of a new plant-growth retardant (E)-1-(4-chlorophenyl)-4,4-dimethyl-2-(1,2,4-triazol-1-Yl)-1-penten-3-Ol (S-3307) on the growth and gibberellin content of rice plants. Plant Cell Physiol 25: 611–617 [Google Scholar]
- Jaillais Y, Belkhadir Y, Balsemão-Pires E, Dangl JL, Chory J. (2011) Extracellular leucine-rich repeats as a platform for receptor/coreceptor complex formation. Proc Natl Acad Sci USA 108: 8503–8507 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Je BI, Piao HL, Park SJ, Park SH, Kim CM, Xuan YH, Park SH, Huang J, Do Choi Y, An G, et al. (2010) RAV-Like1 maintains brassinosteroid homeostasis via the coordinated activation of BRI1 and biosynthetic genes in rice. Plant Cell 22: 1777–1791 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jung K-H, An G, Ronald PC. (2008) Towards a better bowl of rice: assigning function to tens of thousands of rice genes. Nat Rev Genet 9: 91–101 [DOI] [PubMed] [Google Scholar]
- Kagale S, Divi UK, Krochko JE, Keller WA, Krishna P. (2007) Brassinosteroid confers tolerance in Arabidopsis thaliana and Brassica napus to a range of abiotic stresses. Planta 225: 353–364 [DOI] [PubMed] [Google Scholar]
- Kankanala P, Czymmek K, Valent B. (2007) Roles for rice membrane dynamics and plasmodesmata during biotrophic invasion by the blast fungus. Plant Cell 19: 706–724 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kemen E, Gardiner A, Schultz-Larsen T, Kemen AC, Balmuth AL, Robert-Seilaniantz A, Bailey K, Holub E, Studholme DJ, Maclean D, et al. (2011) Gene gain and loss during evolution of obligate parasitism in the white rust pathogen of Arabidopsis thaliana. PLoS Biol 9: e1001094. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim T-W, Guan S, Sun Y, Deng Z, Tang W, Shang J-X, Sun Y, Burlingame AL, Wang Z-Y. (2009) Brassinosteroid signal transduction from cell-surface receptor kinases to nuclear transcription factors. Nat Cell Biol 11: 1254–1260 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim T-W, Wang Z-Y. (2010) Brassinosteroid signal transduction from receptor kinases to transcription factors. Annu Rev Plant Biol 61: 681–704 [DOI] [PubMed] [Google Scholar]
- Lee S, Choi SC, An G. (2008) Rice SVP-group MADS-box proteins, OsMADS22 and OsMADS55, are negative regulators of brassinosteroid responses. Plant J 54: 93–105 [DOI] [PubMed] [Google Scholar]
- Li J, Wen JQ, Lease KA, Doke JT, Tax FE, Walker JC. (2002) BAK1, an Arabidopsis LRR receptor-like protein kinase, interacts with BRI1 and modulates brassinosteroid signaling. Cell 110: 213–222 [DOI] [PubMed] [Google Scholar]
- Li L, Yu X, Thompson A, Guo M, Yoshida S, Asami T, Chory J, Yin Y. (2009) Arabidopsis MYB30 is a direct target of BES1 and cooperates with BES1 to regulate brassinosteroid-induced gene expression. Plant J 58: 275–286 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Melotto M, Underwood W, Koczan J, Nomura K, He SY. (2006) Plant stomata function in innate immunity against bacterial invasion. Cell 126: 969–980 [DOI] [PubMed] [Google Scholar]
- Miller G, Shulaev V, Mittler R. (2008) Reactive oxygen signaling and abiotic stress. Physiol Plant 133: 481–489 [DOI] [PubMed] [Google Scholar]
- Mori M, Nomura T, Ooka H, Ishizaka M, Yokota T, Sugimoto K, Okabe K, Kajiwara H, Satoh K, Yamamoto K, et al. (2002) Isolation and characterization of a rice dwarf mutant with a defect in brassinosteroid biosynthesis. Plant Physiol 130: 1152–1161 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakamura A, Fujioka S, Sunohara H, Kamiya N, Hong Z, Inukai Y, Miura K, Takatsuto S, Yoshida S, Ueguchi-Tanaka M, et al. (2006) The role of OsBRI1 and its homologous genes, OsBRL1 and OsBRL3, in rice. Plant Physiol 140: 580–590 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakashita H, Yasuda M, Nitta T, Asami T, Fujioka S, Arai Y, Sekimata K, Takatsuto S, Yamaguchi I, Yoshida S. (2003) Brassinosteroid functions in a broad range of disease resistance in tobacco and rice. Plant J 33: 887–898 [DOI] [PubMed] [Google Scholar]
- Navarro L, Bari R, Achard P, Lisón P, Nemri A, Harberd NP, Jones JDG. (2008) DELLAs control plant immune responses by modulating the balance of jasmonic acid and salicylic acid signaling. Curr Biol 18: 650–655 [DOI] [PubMed] [Google Scholar]
- Nemhauser JL, Hong F, Chory J. (2006) Different plant hormones regulate similar processes through largely nonoverlapping transcriptional responses. Cell 126: 467–475 [DOI] [PubMed] [Google Scholar]
- Peleg Z, Reguera M, Tumimbang E, Walia H, Blumwald E. (2011) Cytokinin-mediated source/sink modifications improve drought tolerance and increase grain yield in rice under water-stress. Plant Biotechnol J 9: 747–758 [DOI] [PubMed] [Google Scholar]
- Pieterse CMJ, Leon-Reyes A, Van der Ent S, Van Wees SCM. (2009) Networking by small-molecule hormones in plant immunity. Nat Chem Biol 5: 308–316 [DOI] [PubMed] [Google Scholar]
- Purrington CB. (2000) Costs of resistance. Curr Opin Plant Biol 3: 305–308 [DOI] [PubMed] [Google Scholar]
- Robert-Seilaniantz A, Grant MR, Jones JDG. (2011) Hormone crosstalk in plant disease and defense: more than just jasmonate-salicylate antagonism. Annu Rev Phytopathol 49: 26.21–26.27 [DOI] [PubMed] [Google Scholar]
- Robert-Seilaniantz A, Navarro L, Bari R, Jones JD. (2007) Pathological hormone imbalances. Curr Opin Plant Biol 10: 372–379 [DOI] [PubMed] [Google Scholar]
- Sasaki A, Itoh H, Gomi K, Ueguchi-Tanaka M, Ishiyama K, Kobayashi M, Jeong DH, An G, Kitano H, Ashikari M, et al. (2003) Accumulation of phosphorylated repressor for gibberellin signaling in an F-box mutant. Science 299: 1896–1898 [DOI] [PubMed] [Google Scholar]
- Satoh K, Kondoh H, Sasaya T, Shimizu T, Choi IR, Omura T, Kikuchi S. (2010) Selective modification of rice (Oryza sativa) gene expression by rice stripe virus infection. J Gen Virol 91: 294–305 [DOI] [PubMed] [Google Scholar]
- Shan L, He P, Li J, Heese A, Peck SC, Nürnberger T, Martin GB, Sheen J. (2008) Bacterial effectors target the common signaling partner BAK1 to disrupt multiple MAMP receptor-signaling complexes and impede plant immunity. Cell Host Microbe 4: 17–27 [DOI] [PMC free article] [PubMed] [Google Scholar]
- She J, Han Z, Kim T-W, Wang J, Cheng W, Chang J, Shi S, Wang J, Yang M, Wang Z-Y, et al. (2011) Structural insight into brassinosteroid perception by BRI1. Nature 474: 472–476 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shimada A, Ueguchi-Tanaka M, Sakamoto T, Fujioka S, Takatsuto S, Yoshida S, Sazuka T, Ashikari M, Matsuoka M. (2006) The rice SPINDLY gene functions as a negative regulator of gibberellin signaling by controlling the suppressive function of the DELLA protein, SLR1, and modulating brassinosteroid synthesis. Plant J 48: 390–402 [DOI] [PubMed] [Google Scholar]
- Shimada Y, Fujioka S, Miyauchi N, Kushiro M, Takatsuto S, Nomura T, Yokota T, Kamiya Y, Bishop GJ, Yoshida S. (2001) Brassinosteroid-6-oxidases from Arabidopsis and tomato catalyze multiple C-6 oxidations in brassinosteroid biosynthesis. Plant Physiol 126: 770–779 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shimono M, Sugano S, Nakayama A, Jiang C-J, Ono K, Toki S, Takatsuji H. (2007) Rice WRKY45 plays a crucial role in benzothiadiazole-inducible blast resistance. Plant Cell 19: 2064–2076 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun Y, Fan X-Y, Cao D-M, Tang W, He K, Zhu J-Y, He J-X, Bai M-Y, Zhu S, Oh E, et al. (2010) Integration of brassinosteroid signal transduction with the transcription network for plant growth regulation in Arabidopsis. Dev Cell 19: 765–777 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tang W, Kim T-W, Oses-Prieto JA, Sun Y, Deng Z, Zhu S, Wang R, Burlingame AL, Wang Z-Y. (2008) BSKs mediate signal transduction from the receptor kinase BRI1 in Arabidopsis. Science 321: 557–560 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ueguchi-Tanaka M, Hirano K, Hasegawa Y, Kitano H, Matsuoka M. (2008) Release of the repressive activity of rice DELLA protein SLR1 by gibberellin does not require SLR1 degradation in the gid2 mutant. Plant Cell 20: 2437–2446 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ueguchi-Tanaka M, Nakajima M, Katoh E, Ohmiya H, Asano K, Saji S, Hongyu X, Ashikari M, Kitano H, Yamaguchi I, et al. (2007) Molecular interactions of a soluble gibberellin receptor, GID1, with a rice DELLA protein, SLR1, and gibberellin. Plant Cell 19: 2140–2155 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Van Buyten E, Banaay C, Vera-Cruz C, Höfte M. (March 15, 2012) Identity and variability of Pythium species associated with yield decline in aerobic rice cultivation in the Philippines. Plant Pathol http://dx.doi.org/10.1111/j.1365-3059.2012.02607.x [Google Scholar]
- Vercruyssen L, Gonzalez N, Werner T, Schmülling T, Inzé D. (2011) Combining enhanced root and shoot growth reveals cross talk between pathways that control plant organ size in Arabidopsis. Plant Physiol 155: 1339–1352 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Verhage A, van Wees SCM, Pieterse CMJ. (2010) Plant immunity: it’s the hormones talking, but what do they say? Plant Physiol 154: 536–540 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vert G, Chory J. (2006) Downstream nuclear events in brassinosteroid signalling. Nature 441: 96–100 [DOI] [PubMed] [Google Scholar]
- Vert G, Walcher CL, Chory J, Nemhauser JL. (2008) Integration of auxin and brassinosteroid pathways by Auxin Response Factor 2. Proc Natl Acad Sci USA 105: 9829–9834 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vriezen WH, Achard P, Harberd NP, Van Der Straeten D. (2004) Ethylene-mediated enhancement of apical hook formation in etiolated Arabidopsis thaliana seedlings is gibberellin dependent. Plant J 37: 505–516 [DOI] [PubMed] [Google Scholar]
- Wang ZY, Nakano T, Gendron J, He JX, Chen M, Vafeados D, Yang YL, Fujioka S, Yoshida S, Asami T, et al. (2002) Nuclear-localized BZR1 mediates brassinosteroid-induced growth and feedback suppression of brassinosteroid biosynthesis. Dev Cell 2: 505–513 [DOI] [PubMed] [Google Scholar]
- Wu CY, Trieu A, Radhakrishnan P, Kwok SF, Harris S, Zhang K, Wang J, Wan J, Zhai H, Takatsuto S, et al. (2008) Brassinosteroids regulate grain filling in rice. Plant Cell 20: 2130–2145 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xia X-J, Huang L-F, Zhou Y-H, Mao W-H, Shi K, Wu J-X, Asami T, Chen Z, Yu J-Q. (2009a) Brassinosteroids promote photosynthesis and growth by enhancing activation of Rubisco and expression of photosynthetic genes in Cucumis sativus. Planta 230: 1185–1196 [DOI] [PubMed] [Google Scholar]
- Xia X-J, Wang Y-J, Zhou Y-H, Tao Y, Mao W-H, Shi K, Asami T, Chen Z, Yu J-Q. (2009b) Reactive oxygen species are involved in brassinosteroid-induced stress tolerance in cucumber. Plant Physiol 150: 801–814 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamamuro C, Ihara Y, Wu X, Noguchi T, Fujioka S, Takatsuto S, Ashikari M, Kitano H, Matsuoka M. (2000) Loss of function of a rice brassinosteroid insensitive1 homolog prevents internode elongation and bending of the lamina joint. Plant Cell 12: 1591–1606 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang D-L, Li Q, Deng Y-W, Lou Y-G, Wang M-Y, Zhou G-X, Zhang Y-Y, He Z-H. (2008) Altered disease development in the eui mutants and Eui overexpressors indicates that gibberellins negatively regulate rice basal disease resistance. Mol Plant 1: 528–537 [DOI] [PubMed] [Google Scholar]
- Yang GX, Matsuoka M, Iwasaki Y, Komatsu S. (2003) A novel brassinolide-enhanced gene identified by cDNA microarray is involved in the growth of rice. Plant Mol Biol 52: 843–854 [DOI] [PubMed] [Google Scholar]
- Yang YN, Qi M, Mei CS. (2004) Endogenous salicylic acid protects rice plants from oxidative damage caused by aging as well as biotic and abiotic stress. Plant J 40: 909–919 [DOI] [PubMed] [Google Scholar]
- Yin YH, Vafeados D, Tao Y, Yoshida S, Asami T, Chory J. (2005) A new class of transcription factors mediates brassinosteroid-regulated gene expression in Arabidopsis. Cell 120: 249–259 [DOI] [PubMed] [Google Scholar]
- Yu X, Li L, Zola J, Aluru M, Ye H, Foudree A, Guo H, Anderson S, Aluru S, Liu P, et al. (2011) A brassinosteroid transcriptional network revealed by genome-wide identification of BESI target genes in Arabidopsis thaliana. Plant J 65: 634–646 [DOI] [PubMed] [Google Scholar]
- Yuan Y, Zhong S, Li Q, Zhu Z, Lou Y, Wang L, Wang J, Wang M, Li Q, Yang D, et al. (2007) Functional analysis of rice NPR1-like genes reveals that OsNPR1/NH1 is the rice orthologue conferring disease resistance with enhanced herbivore susceptibility. Plant Biotechnol J 5: 313–324 [DOI] [PubMed] [Google Scholar]
- Yun HS, Bae YH, Lee YJ, Chang SC, Kim S-K, Li J, Nam KH. (2009) Analysis of phosphorylation of the BRI1/BAK1 complex in Arabidopsis reveals amino acid residues critical for receptor formation and activation of BR signaling. Mol Cells 27: 183–190 [DOI] [PubMed] [Google Scholar]
- Zhang SWY, Wei Y, Lu Y, Wang X. (2009) Mechanisms of brassinosteroids interacting with multiple hormones. Plant Signal Behav 4: 1117–1120 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.