Abstract
Flax (Linum usitatissimum) stems contain cells showing contrasting cell wall structure: lignified in inner stem xylem tissue and hypolignified in outer stem bast fibers. We hypothesized that stem hypolignification should be associated with extensive phenolic accumulation and used metabolomics and transcriptomics to characterize these two tissues. 1H nuclear magnetic resonance clearly distinguished inner and outer stem tissues and identified different primary and secondary metabolites, including coniferin and p-coumaryl alcohol glucoside. Ultrahigh-performance liquid chromatography-Fourier transform ion cyclotron resonance-mass spectrometry aromatic profiling (lignomics) identified 81 phenolic compounds, of which 65 were identified, to our knowledge, for the first time in flax and 11 for the first time in higher plants. Both aglycone forms and glycosides of monolignols, lignin oligomers, and (neo)lignans were identified in both inner and outer stem tissues, with a preponderance of glycosides in the hypolignified outer stem, indicating the existence of a complex monolignol metabolism. The presence of coniferin-containing secondary metabolites suggested that coniferyl alcohol, in addition to being used in lignin and (neo)lignan formation, was also utilized in a third, partially uncharacterized metabolic pathway. Hypolignification of bast fibers in outer stem tissues was correlated with the low transcript abundance of monolignol biosynthetic genes, laccase genes, and certain peroxidase genes, suggesting that flax hypolignification is transcriptionally regulated. Transcripts of the key lignan genes Pinoresinol-Lariciresinol Reductase and Phenylcoumaran Benzylic Ether Reductase were also highly abundant in flax inner stem tissues. Expression profiling allowed the identification of NAC (NAM, ATAF1/2, CUC2) and MYB transcription factors that are likely involved in regulating both monolignol production and polymerization as well as (neo)lignan production.
Flax (Linum usitatissimum) has been grown for many thousands of years, and recent studies have found traces of flax fibers in 30,000-year-old Paleolithic caves in Georgia (Kvavadze et al., 2009). This species is cultivated for seeds (linseed) and fibers. The seeds contain high amounts of unsaturated fatty acids such as α-linolenic acid (C18:3) as well as biologically active lignans that are used in the chemical and nutraceutical industries (Toure and Xu, 2010; Singh et al., 2011). The extremely long primary fibers (bast fibers) are located in the outer tissues of the stem between the phloem and the epidermis and have an average length of around 7 cm. They are used to produce textiles (linen) and to reinforce composite materials (Summerscales et al., 2010). The secondary cell walls of flax bast fibers are unusual in that they are rich in cellulose but contain only low amounts of lignin (2%) as compared with most other plant secondary cell walls (Day et al., 2005b). In contrast, the secondary walls of flax xylem cells located in the inner stem tissues show a typical lignification profile containing around 25% lignin. Although present in only low amounts in flax bast fibers, lignin has a negative impact on fiber quality, since the presence of this phenolic polymer hinders fiber separation during the retting process. Conversely, the presence of lignin can facilitate fiber-fiber and fiber-matrix interactions in composite materials (Ramires et al., 2010). Lignin, therefore, occupies a key role in determining fiber quality in flax, and a better understanding of its biosynthesis and regulation in this species is necessary to tailor textile and composite performances.
Lignin is a complex three-dimensional phenolic polymer naturally present in the walls of certain specialized cells (xylem, sclerenchyma), where it plays an important role in plant mechanical support and defense. The lignin polymer is assembled in the cell wall by the oxidative polymerization of hydroxycinnamyl alcohols (monolignols) synthesized via the phenylpropanoid pathway. The three major monolignols p-coumaryl alcohol, coniferyl alcohol, and sinapyl alcohol give rise to the so-called H-units (p-hydroxyphenyl), G-units (guaiacyl), and S-units (syringyl) of the lignin polymer, respectively (Boerjan et al., 2003). The relative proportion of the different monolignols varies between different plant groups, species, and tissues. Lignin from gymnosperms and other lower vascular plants is rich in G-units, contains low amounts of H-units, and, except for a few exceptions, contains no S-units (Weng and Chapple, 2010). In contrast, dicot lignin contains G- and S-units as well as low amounts of H-units. Monocot lignin also contains higher amounts of H-units. Over the last two decades, different research programs have targeted individual genes of the phenylpropanoid pathway and clearly demonstrated that up-/down-regulation of these genes can modify lignin content and structure (Vanholme et al., 2008). In flax, we have recently shown that transcripts of monolignol biosynthetic genes are significantly more abundant in inner stem tissues when compared with outer stem tissues, suggesting that hypolignification in the flax stem is also regulated by the differential expression of lignin genes (Fénart et al., 2010).
The perturbation of lignin biosynthesis in mutants and transgenics is often associated with the accumulation of various phenolic storage and detoxification products (Leplé et al., 2007; Fornalé et al., 2010; Vanholme et al., 2010). We hypothesized that natural flax hypolignification might similarly be associated with an accumulation of a wide range of phenolic compounds. Interestingly, this species is one of the richest sources of biologically active lignans that, like lignin, are derived from the phenylpropanoid pathway (Westcott and Muir, 2003). Lignans are dimeric structures containing two monolignols linked together by 8-8′ carbon bonds. The dimerization of coniferyl alcohol results in the formation of pinoresinol, which is then converted into lariciresinol and secoisolariciresinol via the action of the enzyme pinoresinol-lariciresinol reductase (PLR; Davin and Lewis, 2003). In flax seeds, secoisolariciresinol is then glycosylated and accumulates to high levels as secoisolariciresinol diglucoside (SDG; Hano et al., 2006b). Coniferyl alcohol-derived units can also be linked by other (non 8-8′) carbon-carbon bonds, and the term neolignan is used (Moss, 2000). Low-Mr monolignol-coupling products containing carbon-carbon bonds, as well as ether bonds, are also formed during the oxidative step of lignin polymerization and are collectively referred to as oligolignols (Morreel et al., 2004, 2010a). Lignans, neolignans, and other monolignol-coupling products, therefore, are all oligolignols. These compounds can all be glycosylated, so there exists a wide range of potential products that can be formed from monolignols not incorporated into the lignin polymer (Vanholme et al., 2010). In order to test our hypothesis that flax hypolignification is associated with the accumulation of phenolic compounds, we decided to characterize monolignols and monolignol-derived phenolics in flax stems using a previously developed lignomics approach (Morreel et al., 2004, 2010b). This approach has allowed the identification of 36 and 55 different phenolic compounds in tobacco (Nicotiana tabacum) xylem tissue (Dauwe et al., 2007) and in Arabidopsis (Arabidopsis thaliana) stems (Vanholme et al., 2010), respectively.
In order to complete our knowledge about how monolignol biosynthesis, oligolignol accumulation, and hypolignification in flax are regulated at the transcriptional level, we used Nimblegen high-density microarrays to compare phenylpropanoid gene expression profiles in inner and outer stem tissues. Recent studies have demonstrated that up-/down-regulation of genes encoding different NAC or MYB transcription factors (TFs) modulates the expression of monolignol biosynthesis genes and modifies both lignification and secondary cell wall formation (Zhong et al., 2010; Zhang et al., 2011). Therefore, we focused our attention on genes encoding TFs that were differentially expressed in inner and outer stem tissues in order to identify potential regulators of flax hypolignification.
In this paper, we report the results of combined metabolomic profiling, lignomics, and transcriptomics of flax inner and outer stem tissues. In addition to providing novel information on mono(oligo)lignol metabolism in flax, our results also suggest that a wide range of phenolics accumulate under natural hypolignification conditions as well as in low-lignin conditions induced by the engineering of target genes.
RESULTS
Flax Xylem and Bast Fiber Cell Walls Show Highly Contrasting Lignification Profiles
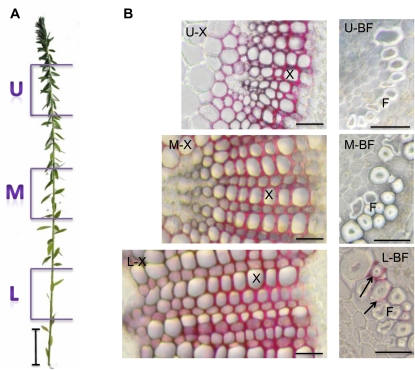
Examination of phloroglucinol-HCl-stained cross-sections made at different heights in the flax stem (Fig. 1) showed that both inner and outer stem tissues follow a developmental gradient along the length of the stem from the upper, young tissues to the lower, more mature tissues (Fig. 1). In stem inner tissues, this developmental gradient is associated with the progressive formation of xylem tissue and extensive lignification of cell walls, as indicated by the red color. In outer tissues, the developmental gradient is associated with progressive thickening of the secondary cell wall of bast fibers and by lignin deposition in the middle lamella/primary walls of certain of these fibers in the lower region.
Figure 1.
Two-month-old flax plant (A) and stem cross-sections (B) from upper (U), middle (M), and lower (L) regions of the stem. U-X and U-BF = microphotographs of xylem and bast fibers from the upper region, M-X and M-BF = microphotographs of xylem and bast fibers from the middle region, and L-X and L-BF = microphotographs of xylem and bast fibers from the lower region. Cross-sections are stained with phloroglucinol-HCl, and lignified cell walls are colored red. F, Bast fiber; X, xylem cell. Arrows show lignin deposits in bast fiber middle lamella/primary cell wall. Bars = 5 cm (A) and 20 μm (B). [See online article for color version of this figure.]
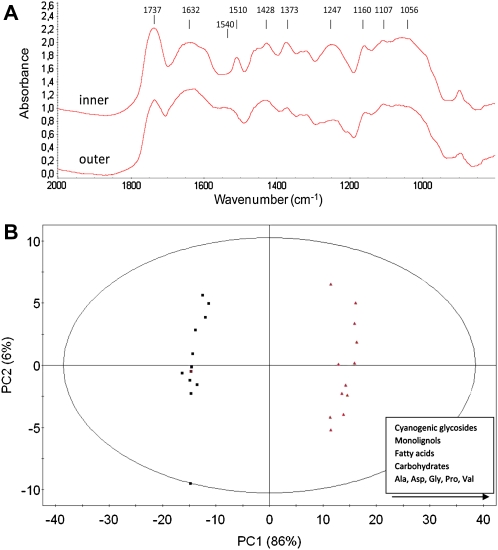
Fourier transform infrared (FTIR) spectroscopy (Fig. 2A) of cell walls from flax inner and outer stem tissues confirmed these observations. The inner, but not the outer, tissues are characterized by a major absorption peak at 1,510 cm−1, indicating the presence of high amounts of lignin. The observation that peaks at 1,428, 1,373, and 1,247 cm−1 characteristic of nonspecific lignin functional groups are more intense in inner tissues further supports this observation (Faix, 1991). No differences between the two tissue types are observed in the 1,000 to 1,200 cm−1 range characteristic of both cellulose and hemicelluloses (Marechal and Chanzy, 2000). The outer tissue peak at 1,540 cm−1 could be assigned to amide groups from proteins (Hano et al., 2006a). The predominance of proteins in these samples is potentially due to the incomplete removal of soluble fractions from cells surrounding bast fibers in outer stem tissues. The peak at approximately 1,737 cm−1 in outer and inner stem tissues is probably associated with pectin- or xylan-associated ester functions (Kacurakova et al., 2000).
Figure 2.
A, FTIR spectroscopy of flax stem inner and outer tissues. Peaks corresponding to lignin (1,510 cm−1), pectin/xylan (1,750 cm−1), and probable proteins (1,550 and 1,630 cm−1) are indicated by arrows. Cellulose and hemicellulose peaks are situated in the 1,000- to 1,200-cm−1 range. B, 1H NMR metabolic profiling. PCA of flax inner stem tissues (black squares) and outer stem tissues (red triangles) is shown. [See online article for color version of this figure.]
Inner and Outer Stem Tissues Are Metabolically Distinct
In order to more fully characterize flax inner and outer stem tissues, we performed metabolic profiling using 1H NMR. Principal component analysis (PCA) of the 1H NMR data provided an initial overview of the flax stem inner and outer tissue metabolome and allowed these two tissues to be clearly differentiated (Fig. 2B). The major metabolites contributing to the PCA-based clustering of both tissues were subsequently identified and are listed in Table I. Primary metabolites included Suc, α-/β-Glc, lactate, fumarate (tricarboxylic acid cycle), formic acid (C1 metabolism), and the amino acids Ile, Val, Thr, Ala, Pro, Gln, Asp, Gly, and Trp. Molecules involved in lipid metabolism, namely choline and the unsaturated fatty acids linoleic acid (C18:2) and α-linolenic acid (C18:3), which are also present in flax seeds (linseed), were identified as well. Other compounds present included the cyanogenic glycosides lotaustralin and linamarin identified previously in flax organs (Niedźwiedź-Siegień, 1998) and the alkaloid trigonelline (nicotinic acid betaine), an osmolyte previously shown to be implicated in the response to drought stress (Charlton et al., 2008). The glucosylated forms of p-coumaryl alcohol and coniferyl alcohol (coniferin) were also identified in flax stem tissues. The semiquantitative analysis revealed that the different metabolites accumulated in higher amounts in outer stem tissues. For example, the monolignol glucosides accumulated up to five times more, cyanogenic glucosides more than three times more, and primary metabolites around two times more except for Gln and formic acid (Table I). Overall, these data suggest that flax outer tissues are metabolically more active than inner stem tissues on a per gram dry weight basis. This most likely reflects the fact that inner stem tissues contain a higher proportion of dead cells in the xylem.
Table I. 1Principal compounds specifically identified in flax inner and outer stem tissues by 1H-NMR.
nd, Not determined.
| Metabolite | Chemical Shift and (Coupling Constants) | Intensity Ratio (Outer Tissue-Inner Tissue) |
| ppm | ||
| Ile | 0.95 (t 7.5 Hz) | nd |
| 1.02 (d 7 Hz) | ||
| Linoleic acid | 0.96 (t 7.6 Hz) | 3.6 |
| Val | 1.00 (d 7 Hz) | 1.7 |
| 1.05 (d 7.1 Hz) | ||
| Linolenic acid | 1.01 (t 7.6 Hz) | 2.1 |
| Lotaustralin | 1.07 (t 7.6 Hz) | 3.4 |
| 1.60 (s) | ||
| 1.64 (s) | ||
| 1.89 (m) | ||
| 1.98 (m) | ||
| Lactate | 1.33 (d 5.4 Hz) | 1.8 |
| Thr | 1.33 (d 5.4 Hz) | |
| 3.51 (d 5.1 Hz) | ||
| 4.22 (m) | ||
| Ala | 1.48 (d 7.3 Hz) | 2.1 |
| 3.72 (q 7.3 Hz) | ||
| Linamarin | 1.69 (s) | 3.6 |
| 1.69 (s) | ||
| 3.25 (dd 7.8–9.3 Hz) | ||
| 3.39 (t 9.4 Hz) | ||
| 3.49 (t 9.1 Hz) | ||
| 3.87 (dd 2–12 Hz) | ||
| 4.72 (d 7.7 Hz) | ||
| Pro | 2.00 (m) | 2.1 |
| 2.09 (m) | ||
| 2.34 (m) | ||
| 3.41 (m) | ||
| 4.07 (dd 6.5–8.7 Hz) | ||
| Gln | 2.13 (m) | 1 |
| 2.45 (m) | ||
| 3.71 (t 6.2 Hz) | ||
| Asp | 2.64 (dd 9.4–17.4 Hz) | 2 |
| 2.81 (dd 3.6–17.4 Hz) | ||
| 3.83 (dd 3.6–9.4 Hz) | ||
| Choline | 3.21 (s) | 1.8 |
| Suc | 3.43 (t 9.6 Hz) | 2.3 |
| 3.50 (dd 3.9–9.9 Hz) | ||
| 3.65 (s) | ||
| 4.02 (t 8.4 Hz) | ||
| 4.17 (d 8.6 Hz) | ||
| 5.4 (d 3.8 Hz) | ||
| Gly | 3.50 (s) | 1.7 |
| Coniferin | 3.90 (s) | 5.1 |
| 4.24 (dd 1.2–6 Hz) | ||
| 5.01 (m) | ||
| 6.32 (td 5.8–16 Hz) | ||
| 6.57 (d 16 Hz) | ||
| 7.03 (dd 2–8.3 Hz) | ||
| 7.12 (d 8.4 Hz) | ||
| 7.14 (d 2 Hz) | ||
| Trigonelline | 4.45 (s) | 2.4 |
| 8.10 (m) | ||
| 8.86 (m) | ||
| 9.14 (s) | ||
| β-Glc | 4.58 (d 8 Hz) | 2.7 |
| p-Coumaryl alcohol glucoside | 5.00 (d 7.7 Hz) | 3.5 |
| 6.29 (td 5.8–16 Hz) | ||
| 6.58 (d 16 Hz) | ||
| 7.07 (d 8.7 Hz) | ||
| 7.42 (d 8.7 Hz) | ||
| α-Glc | 5.18 (d 3.7 Hz) | |
| Fumaric acid | 6.55 (s) | 2 |
| Trp | 7.13 (m) | nd |
| 7.21 (m) | ||
| 7.28 (s) | ||
| 7.46 (d 8 Hz) | ||
| 7.71 (d 7.8 Hz) | ||
| Formic acid | 8.46 (s) | 1 |
Flax Stem Tissues Are Rich in Oligolignols
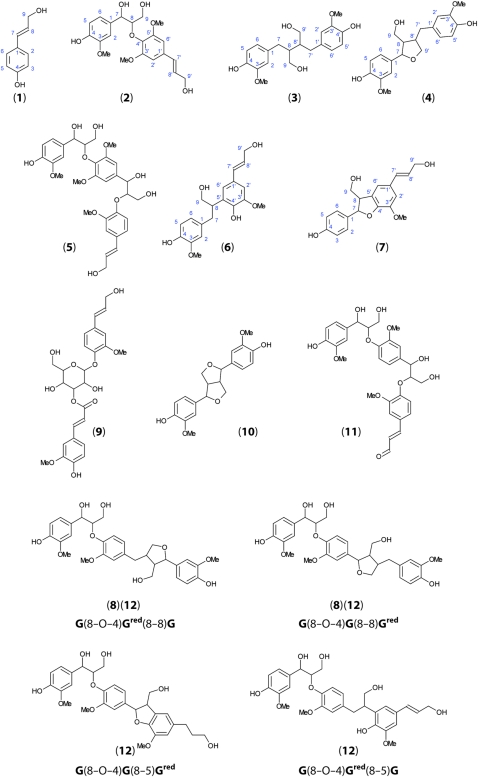
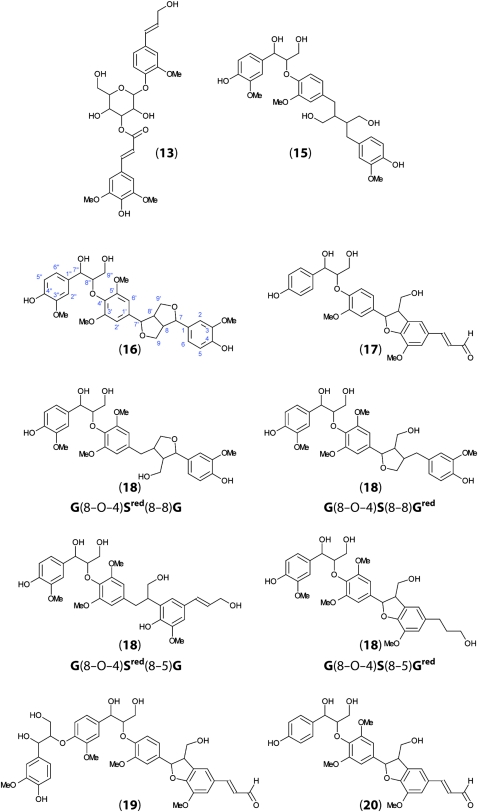
To characterize monolignol-coupling products from flax stem tissues, we used a lignomics approach (Morreel et al., 2004, 2010b). Methanol extracts from stem outer and inner tissues were analyzed by ultrahigh-performance liquid chromatography-Fourier transform ion cyclotron resonance-mass spectrometry (UHPLC-FT-ICR-MS), and elucidation of the MS2 spectra was based on the lignin oligomer/(neo)lignan sequencing approach and the fragmentation rules of the different linkage types as described previously (Morreel et al., 2010a, 2010b). Altogether, we identified 81 different phenolic compounds in flax stem tissues (Table II), of which 65 are identified, to our knowledge, for the first time in flax and 11 for the first time in plants. Twenty-eight compounds (20 when excluding isomers) had negative ionization mode electrospray-MS2 spectra that had not been described before, to our knowledge (Table II). The probable identities and molecular structures of these compounds, based on MS2 fragmentation data, are given in Figure 3 and Supplemental Data S1.
Figure 3.
Proposed molecular structures of novel flax oligolignols identified by UHPLC-FT-IR-MS. Numbers refer to MS data presented in Table II. The identification of compounds based upon MS2 and MS3 fragmentation patterns is described in Supplemental Data S1. [See online article for color version of this figure.]
Table II. Mass spectral data of flax phenolics.
Phenolic profiling was performed with reverse-phase UHPLC coupled to atmospheric pressure chemical ionization-ion trap-FT-ICR-MS. Accurate m/z values were obtained with FT-ICR-MS, whereas the MS2 and MS3 (in parentheses after the corresponding MS2 in italics) collision-induced dissociation spectra were recorded via ion-trap-MS. The relative abundance of each peak in inner and outer tissues is based on the selected ion current. Shorthand names of oligolignols/(neo)lignans are based on Morreel et al. (2004). p-Hydroxyphenyl, guaiacyl, and syringyl units as well as units derived from 5-hydroxyconiferyl alcohol, ferulic acid, and coniferaldehyde are designated H, G, S, 5H, FA, and G′, respectively. The linkage type is indicated in parentheses. The term “red” indicates reduced unit or adjacent linkage (Morreel et al., 2010a); e.g. Gred(8-8)G or G(8-8)Gred, Gred(8-8)Gred, Gred(8-5)G, and G(8-5)Gred refer to lariciresinol, secoisolariciresinol, isodihydrodehydrodiconiferyl alcohol, and dihydrodehydrodiconiferyl alcohol, respectively. A forward slash indicates that two units or two linkage positions are equally possible at this position in the shorthand name. In this case, the various structures are shown in Figure 3. hex, Hexose or hexoside; tR, retention time; a, intensity based on acetate adduct ion; nd, not detected. ?, MS2 spectrum matches the standard but not the retention time; ??, MS2 spectrum does not fully match that of the suggested compound based on the literature reference. “Compound” refers to the name of the compound: roman text indicates compounds identified for the first time in flax, boldface italic text indicates compounds previously identified in flax. Ref number after a compound name refers to a bibliographic reference for MS data (Supplemental Data S1), F Ref number after a compound name refers to a previous bibliographic reference (Supplemental Data S1) identifying the compound in flax, and S number after a compound name refers to the molecular structures presented in Figure 3. No bibliographic references for the MS data of S numbered compounds were available, and their MS-based structural elucidation is described in Supplemental Data S1. Asterisks indicate compounds described, to our knowledge, for the first time in plants.
| tR | Relative Abundance | Ratio Outer-Inner | m/z | Formula | Δppm | m/z MS2 (MS3) | Compound (Ref, F Ref, S, *) | |
| Inner | Outer | |||||||
| min | ||||||||
| 4.16 | nd | 4.5 × 104 | Outer | 367.10391 | C17H19O9 | 1.24 | 191 (MS3: 173, 127, 85) | Feruloyl quinic acid (Ref 1) |
| 4.50 | 2.4 × 103 | 8.5 × 104 | 36.9 | 357.11952 | C16H21O9 | 1.16 | 195 (MS3: 177, 151, 136, 123, 119) | Dihydroferulic acid hex (Ref 2) |
| 4.87 | 2.3 × 106 a | 1.0 × 107 a | 4.3 | 311.11488 | C15H19O7 | 4.03 | 149 (MS3: 131) | p-Coumaryl alcohol hex (S 1) |
| 5.61 | nd | 7.0 × 104 | Outer | 551.17932 | C26H31O13 | 4.18 | 389, 341, 193 (MS3: 341, 193, 181, 151) | G(8-O-4)FA hex (Ref 3) |
| 5.83 | 1.1 × 104 | 5.4 × 105 | 49.1 | 551.17898 | C26H31O13 | 3.57 | 389, 341, 193 (MS3: 341, 193) | G(8-O-4)FA hex (Ref 3) |
| 5.86 | 3.2 × 106 a | 1.2 × 107 a | 3.8 | 341.12541 | C16H21O8 | 3.57 | 179 (MS3: 161, 146) | Coniferin (Ref 2, F Ref 1) |
| 5.92 | 2.2 × 104 | 8.4 × 105 | 38.2 | 551.17745 | C26H31O13 | 0.79 | 389, 341, 193 (MS3: 341, 193) | G(8-O-4)FA hex (Ref 3) |
| 6.71 | 7.7 × 103 | 4.6 × 105 | 59.7 | 551.17807 | C26H31O13 | 1.91 | 389, 341, 193 (MS3: 341, 193) | G(8-O-4)FA hex (Ref 3) |
| 6.98 | 5.0 × 103 a | 1.8 × 104 a | 3.6 | 431.15684a | C19H27O11a | 2.21 | 371, 209 (MS3: 194, 191) | Syringin (Ref 2) |
| 7.01 | 1.3 × 104 | 8.9 × 104 | 6.8 | 537.19884 | C26H33O12 | 2.03 | 375, 345, 327 (MS3: 345, 327) | G(8-O-4)G hex?? (Ref 2, F Ref 2) |
| 7.37 | 5.6 × 102 | 5.6 × 104 | 100 | 355.10375 | C16H19O9 | 0.83 | 295, 235, 217, 193, 175 (MS3: 178, 149, 134) | Feruloyl hex (Ref 1, F Ref 3) |
| 7.59 | 1.5 × 103 a | 1.7 × 104 a | 11.3 | 401.14582a | C18H25O10a | 1.25 | 341, 179 (MS3: 179) | Coniferyl alcohol hex? (Ref 2, F Ref 1) |
| 7.88 | nd | 1.4 × 104 | Outer | 385.11444 | C17H21O10 | 1.09 | 325, 265, 247, 223, 205 (MS3: 208, 179, 164) | Sinapoyl hex (Ref 1) |
| 8.42 | nd | 3.4 × 104 | Outer | 533.16680 | C26H29O12 | 0.66 | 371, 353, 341 (MS3: 353, 341, 327, 191) | G(8-5)FA hex (Ref 2) |
| 8.52 | 7.8 × 102 | 1.5 × 104 | 19.2 | 389.12484 | C20H21O8 | 1.67 | 341, 193, 181, 165 (MS3: 178, 149, 134) | G(8-O-4)FA (Ref 3) |
| 8.76 | 5.6 × 104 a | 5.8 × 105 a | 10.4 | 537.19854 | C26H33O12 | 1.47 | 375 (MS3: 327, 195, 165) | G(t8-O-4)G hex (Ref 2, F Ref 2) |
| 9.37 | nd | 2.0 × 104 | Outer | 355.10379 | C16H19O9 | 0.94 | 327, 235, 217, 193, 175 (MS3: 178, 149, 134) | Feruloyl hex (Ref 1, F Ref 3) |
| 9.37 | nd | 1.6 × 104 | Outer | 551.17696 | C26H31O13 | −0.10 | 491, 389, 193 (MS3: 341, 193) | G(8-O-4)feruloyl hex (Ref 3) |
| 9.66 | 4.8 × 103 | 1.3 × 104 | 2.7 | 179.07169 | C10H11O3 | 1.80 | 164, 161, 146 (MS3: 146) | Coniferyl alcohol (Ref 1, F Ref 4) |
| 10.91 | 1.9 × 103 | 8.9 × 104 | 46.8 | 551.17778 | C26H31O13 | 1.39 | 461, 389, 341, 193 (MS3: 341, 193) | G(8-O-4)feruloyl hex (Ref 3) |
| 10.97 | 8.5 × 103 a | 1.8 × 105 | 21.2 | 627.23060a | C29H39O15a | 1.84 | 567, 405 (MS3: 357, 209, 195, 165) | G(8-O-4)S hex (S 2) |
| 11.17 | 1.1 × 104 | 1.8 × 105 | 16.4 | 537.19789 | C26H33O12 | 0.26 | 507, 489, 375, 357, 301, 271, 195 | G(e8-O-4)G hex (Ref 2, F Ref 2) |
| 11.40 | 2.1 × 104 | 3.1 × 105 | 14.8 | 523.21976 | C26H35O11 | 2.44 | 361 (MS3: 346, 313, 179, 165) | Secoisolariciresinol hex (S 3) |
| 11.55 | 8.3 × 103 | 1.1 × 105 | 13.4 | 521.20407 | C26H33O11 | 2.37 | 359, 329 (MS3: 299, 178) | Lariciresinol hex (S 4) |
| 12.00 | 2.8 × 103 | 2.3 × 104 | 8.2 | 601.23007 | C31H37O12 | 1.70 | 553, 421, 405, 403, 373, 357, 225, 195 | G(8-O-4)S(8-O-4)G (S 5)* |
| 12.18 | 7.8 × 105 a | 4.5 × 106 a | 5.8 | 579.21004a | C28H35O13a | 2.98 | 519, 357, 339, 327 (MS3: 324) | G(8-5)G hex (Ref 2, F Ref 5, 6) |
| 12.32 | 1.1 × 103 a | 1.8 × 104 a | 16.4 | 595.20322a | C28H35O14a | −0.02 | 535, 373, 193 (MS3: 355, 193,179) | G(8-O-4)5H hex (Ref 4) |
| 12.43 | 5.7 × 104 | 4.3 × 104 | 0.8 | 375.14573 | C20H23O7 | 2.14 | 327, 195, 165 | G(t8-O-4)G (Ref 5, F Ref 2) |
| 12.51 | 1.3 × 104 | 2.8 × 105 | 21.5 | 521.20381 | C26H33O11 | 1.87 | 359, 329 | Lariciresinol hex (S 4) |
| 12.58 | 2.1 × 104 a | 5.6 × 105 a | 26.7 | 581.22511a | C28H37O13a | 1.97 | 521, 359, 341, 329 (MS3: 326) | IDDDC hex (S 6) |
| 12.69 | 3.6 × 104 | 3.1 × 104 | 0.9 | 375.14569 | C20H23O7 | 2.04 | G(8-O-4)G (Ref 5, F Ref 2) | |
| 12.73 | 2.7 × 103 a | 3.2 × 104 | 11.9 | 549.19884a | C27H33O12a | 1.98 | 489, 327, 309, 297 | H(8-5)G hex (S 7)* |
| 13.41 | 2.6 × 104 | nd | Inner | 523.21918 | C26H35O11 | 1.33 | 361 (MS3: 346, 343, 313, 179, 165) | Secoisolariciresinol hex (S 3) |
| 13.58 | 4.4 × 103 | 5.5 × 104 | 12.5 | 717.27850 | C36H45O15 | 2.94 | 699, 669, 555, 525, 507 (MS3: 537, 525, 507, 477, 359, 329, 195) | G(8-O-4)Gred/G(8-8)G/Gred hex (S 8)* |
| 13.62 | 2.0 × 103 a | 3.9 × 104 | 19.5 | 579.20903a | C28H35O13a | 1.23 | 519, 339, 327 | G(8-5)G hex (Ref 2, F Ref 5, 6) |
| 13.62 | nd | 1.3 × 105 | Outer | 517.17154 | C26H29O11 | 0.01 | 337, 193 (MS3: 277, 217, 193, 175) | Feruloyl coniferin (S 9)* |
| 13.81 | 3.8 × 103 | 2.5 × 104 | 6.6 | 571.22002 | C30H35O11 | 2.69 | 523, 391, 375, 343, 327, 195 | G(8-O-4)G(8-O-4)G (Ref 5) |
| 13.89 | 1.5 × 103 | 8.9 × 104 | 59.3 | 521.20347 | C26H33O11 | 1.22 | 503, 359, 341, 329, 179, 161 | IDDDC hex (S 6) |
| 13.90 | 1.1 × 104 | 7.9 × 104 | 7.2 | 553.20917 | C30H33O10 | 2.26 | 505, 339 (MS3: 487, 475, 457) | G(t8-O-4)G(8-5)G (Ref 5) |
| 14.08 | 6.6 × 103 | 1.1 × 105 | 16.7 | 519.18875 | C26H31O11 | 3.01 | 501, 357 (MS3: 342, 327, 311, 151, 136) | Pinoresinol hex (S 10) |
| 14.21 | 4.9 × 103 a | 2.4 × 103 a | 0.5 | 433.15153a | C22H25O9a | 2.60 | 373 (MS3: 355, 325, 195, 177) | G(8-O-4)G′ (Ref 5) |
| 14.31 | 4.0 × 103 a | 2.1 × 104 a | 5.3 | 433.15157a | C22H25O9a | 2.69 | G(8-O-4)G′ (Ref 5) | |
| 14.50 | 7.6 × 102 | 2.0 × 104 | 26.3 | 521.20325 | C26H33O11 | 0.80 | 503, 491, 359, 341 (MS3: 329, 314), | IDDDC hex (S 6) |
| 14.72 | 1.0 × 103 | 1.5 × 104 | 15 | 405.15636 | C21H25O8 | 2.14 | 357, 225 (MS3: 342) | S(t8-O-4)G (Ref 6) |
| 15.00 | 4.9 × 103 | 6.0 × 104 | 12.2 | 375.14579 | C20H23O7 | 2.30 | 327, 195, 165 (MS3: 312) | G(t8-O-4)G (Ref 5, F Ref 2) |
| 15.06 | 3.4 × 103 | 3.5 × 103 | 1.1 | 569.20468 | C30H33O11 | 3.24 | 521, 391, 177, 162 | G(8-O-4)G(8-O-4)G′ (S 11)* |
| 15.24 | 2.6 × 103 | 4.5 × 103 | 1.7 | 373.13006 | C20H21O7 | 2.10 | 355, 343, 325, 249, 231, 219, 203, 193, 179 | G(8-O-4)5H ? (Ref 6) |
| 15.28 | 4.2 × 103 | 3.9 × 103 | 0.9 | 601.23041 | C31H37O12 | 2.26 | 551, 421, 373, 225, 195 | G(8-O-4)S(8-O-4)G (S 5)* |
| 15.71 | 3.0 × 104 | 1.8 × 105 | 6 | 583.21962 | C31H35O11 | 1.95 | 535, 369, 357 (MS3: 505) | G(t8-O-4)S(8-5)G (Ref 5) |
| 15.80 | 1.7 × 103 | 8.5 × 103 | 5 | 571.21852 | C30H35O11 | 0.06 | 523, 391, 343, 195 | G(8-O-4)G(8-O-4)G (Ref 5) |
| 15.91 | 2.4 × 103 | 1.4 × 104 | 5.8 | 571.21980 | C30H35O11 | 2.30 | 523, 449, 301, 271, 195 | G(8-O-4)G(8-O-4)G (Ref 5) |
| 15.99 | 3.1 × 103 | 5.6 × 103 | 1.8 | 601.23037 | C31H37O12 | 2.20 | 553, 421, 225, 195 | G(8-O-4)S(8-O-4)G (S 5)* |
| 16.06 | 1.2 × 103 | 7.0 × 103 | 5.8 | 747.28685 | C37H47O16 | −0.15 | 585, 537 (MS3: 537, 359, 345, 343) | S(t8-O-4)Gred/G(8-8/5)G/Gred hex (S 12)* |
| 16.13 | 1.9 × 103 | 1.7 × 104 | 8.9 | 571.21975 | C30H35O11 | 2.21 | 523, 497, 449, 301, 271 | G(8-O-4)G(8-O-4)G (Ref 5) |
| 16.42 | 2.9 × 103 | 2.4 × 104 | 8.3 | 571.21976 | C30H35O11 | 2.23 | 523, 391, 343, 301, 271, 195 | G(8-O-4)G(8-O-4)G (Ref 5) |
| 16.64 | 1.4 × 103 | 1.3 × 105 | 92.8 | 547.18218 | C27H31O12 | 0.15 | 367 (MS3: 307, 265, 247, 223, 205, 190, 179, 164) | Sinapoyl coniferin (S 13)* |
| 16.65 | 1.6 × 104 a | 1.2 × 104 a | 0.75 | 419.17204a | C22H27O8a | 2.14 | 359, 329 (MS3: 329) | Lariciresinol (Ref 8, F Ref 7) |
| 16.75 | 2.9 × 103 | 9.2 × 104 | 31.7 | 714.25241 | C34H40O14N3 | 1.07 | 696, 533, 517 (MS3: 337, 193, 175), 337 | Nitrogen-containing derivate of feruloyl coniferin (S 14)* |
| 16.87 | 9.5 × 104 | 3.0 × 104 | 0.3 | 357.13516 | C20H21O6 | 2.23 | 339, 327, 221, 203, 191 (MS3: 324, 203, 123) | DDC (Ref 5, F Ref 2) |
| 17.13 | 1.1 × 103 | 1.6 × 103 | 1.5 | 557.24022 | C30H37O10 | 1.79 | 539, 509, 415, 361 | G(e8-O-4)Gred(8-8)Gred (S 15) |
| 17.18 | 2.9 × 103 | 2.2 × 103 | 0.8 | 373.13008 | C20H21O7 | 2.15 | 355, 343, 193, 179 (MS3: 164, 161, 146) | G(8-O-4)5H (Ref 6) |
| 17.37 | 1.5 × 104 | 4.8 × 103 | 0.3 | 555.22464 | C30H35O10 | 1.93 | 525, 507, 477, 329 | G(e8-O-4)Gred(8-8)G (Ref 7) |
| 17.74 | 9.4 × 103 | 4.5 × 103 | 0.5 | 553.20920 | C30H33O10 | 2.31 | 535, 523, 505, 339, 327, 195, 165 (MS3: 324) | G(t8-O-4)G(8-5)G (Ref 7) |
| 17.78 | 2.1 × 103 | 1.1 × 104 | 5.2 | 745.27291 | C37H45O16 | 2.15 | 583 (MS3: 535, 387, 195), 535, 387) | G(t8-O-4)S(8-8)G hex (S 16) |
| 17.90 | 4.7 × 103 | 2.1 × 104 | 4.5 | 745.27362 | C37H45O16 | 3.10 | 583, 535, 387 | G(8-O-4)S(8-8)G hex (S 16) |
| 18.49 | 1.0 × 103 | nd | Inner | 551.19358 | C30H31O10 | 2.38 | 503, 373 | G(8-5)G(8-O-4)G′ (Ref 5) |
| 18.59 | 9.5 × 102 | 4.1 × 103 | 4.3 | 745.27205 | C37H45O16 | 0.99 | 583, 535, 387 | G(8-O-4)S(8-8)G hex (S 16) |
| 19.09 | 1.5 × 103 | 7.1 × 103 | 4.7 | 553.20831 | C30H33O10 | 0.70 | 535, 523, 505, 339, 327, 195 | G(t8-O-4)G(8-5)G (Ref 5) |
| 19.12 | 1.7 × 103 | 1.6 × 103 | 0.9 | 521.18310 | C29H29O9 | 2.67 | 473, 337, 325, 190 | H(8-O-4)G(8-5)G′ (S 17)* |
| 19.17 | 4.7 × 102 | 3.0 × 103 | 6.4 | 583.21915 | C31H35O11 | 1.14 | 535, 339, 327, 225 | S(t8-O-4)G(8-5)G (Ref 5) |
| 19.39 | 2.4 × 103 | 1.4 × 104 | 5.8 | 553.20907 | C30H33O10 | 2.08 | 523, 505, 357, 339, 303 | G(t8-O-4)G(8-5)G (Ref 5) |
| 19.59 | 5.7 × 104 | 1.8 × 104 | 0.3 | 583.21852 | C31H35O11 | 0.06 | 535, 369, 357 | G(t8-O-4)S(8-5)G (Ref 5) |
| 19.74 | 2.3 × 104 | 7.1 × 103 | 0.3 | 551.19348 | C30H31O10 | 2.19 | 503, 485, 337, 325, 218 | G(t8-O-4)G(8-5)G′ (Ref 5) |
| 19.89 | 2.8 × 103 | 3.1 × 103 | 1.1 | 357.13497 | C20H21O6 | 1.70 | 342, 339, 327, 311, 151, 136 (MS3: 136) | Pinoresinol (Ref 5, F Ref 7,8) |
| 20.11 | 5.9 × 103 | 2.1 × 103 | 0.4 | 585.23525 | C31H37O11 | 1.90 | 537, 359, 195 | G(t8-O-4)Sred/S(8-8/5)G/Gred (S 18) |
| 20.20 | 2.6 × 103 | 1.2 × 103 | 0.5 | 747.26721 | C40H43O14 | 1.85 | 699, 551, 503, 337 | G(t8-O-4)G(8-O-4)G(8-5)G′ (S 19)* |
| 20.30 | 1.2 × 104 | 4.6 × 103 | 0.4 | 583.21863 | C31H35O11 | 0.25 | 535, 369, 357 | G(e8-O-4)S(8-5)G (Ref 5) |
| 20.92 | 2.2 × 103 | 8.1 × 102 | 0.4 | 551.19354 | C30H31O10 | 2.30 | 533, 503, 367, 355, 218 | H(t8-O-4)S(8-5)G′ (S 20)* |
| 21.11 | 9.9 × 102 | 2.6 × 103 | 2.6 | 613.22927 | C32H37O12 | 0.36 | 565, 369, 357 | S(t8-O-4)S(8-5)G (Ref 5) |
| 21.70 | 1.9 × 103 | 4.2 × 103 | 2.2 | 583.21974 | C31H35O11 | 2.15 | 535, 369, 357 | G(t8-O-4)S(8-5)G (Ref 5) |
| 21.80 | 3.2 × 104 | 7.5 × 103 | 0.2 | 581.20430 | C31H33O11 | 2.52 | 551, 533, 515, 367, 355, 218 | G(t8-O-4)S(8-5)G′ (Ref 5) |
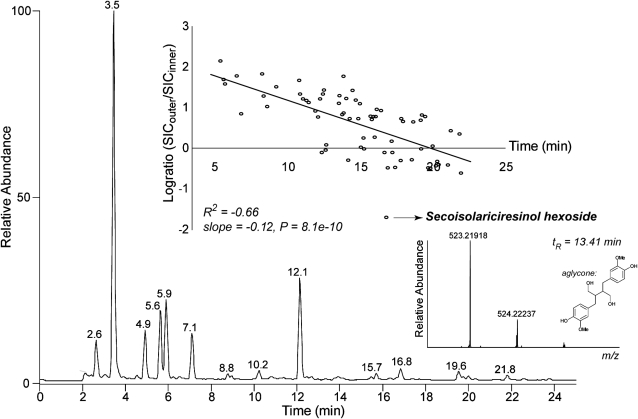
Approximately half of the peaks (40) represented lignin oligomers, which are predominantly present in the second half of the chromatogram, whereas the remainder of the peaks represented mainly glycosylated (neo)lignans (28 compounds). The oligolignols were essentially composed of G-units and to a lesser extent of S- and FA- (for ferulic acid-derived) units, and only a few contained an H-unit. When the ratio of oligolignol abundance in outer versus inner tissues was computed, a negative correlation with the retention time was observed (Fig. 4). This confirms the observation (Table II) that the more hydrophilic glycosylated (neo)lignans are more abundant in the outer tissues, whereas the less polar lignin oligomers are more abundant in inner tissues. However, in contrast with this general observation, one of the two secoisolariciresinol hexoside isomers was present in inner tissues yet below the detection limit in the outer tissues. In both tissues, p-coumaryl alcohol hexoside and coniferin were abundantly present (Table II), whereas only trace levels were observed for syringin. Of the three monolignol glucosides, only the aglycone of coniferin (i.e. coniferyl alcohol) was detected. Remarkably, secondary metabolites comprising a coniferin moiety (i.e. feruloyl coniferin, sinapoyl coniferin, and a nitrogen-containing derivate of feruloyl coniferin) were also traced in the profiles.
Figure 4.
Phenolic profile of flax inner tissue. The profile was obtained via reverse-phase UHPLC analysis of ethanol extracts. Mass spectral detection was performed with atmospheric pressure chemical ionization-FT-ICR-MS equipped with an ion-trap mass spectrometer in which collision-induced dissociation was performed. Phenolic profiles of inner and outer tissues were very similar. Based on the selected ion current (SIC), the logarithmically transformed ratio (log ratio) of the abundance of each peak was computed. A plot of the log ratio versus the retention time indicated a significant correlation. One outlier is secoisolariciresinol hexoside (only present in inner tissue). An enlarged view of the full FT-ICR-MS scan at their retention time is shown.
Monolignol Genes Are Differentially Expressed in Inner and Outer Stem Tissues
High-density 385K Nimblegen flax microarrays representing 40,000 flax unigenes were used for transcriptomics (Fénart et al., 2010). The accuracy of gene expression levels was confirmed by quantitative reverse transcription-PCR of 10 different genes showing low, medium, and high expression levels as determined by the microarrays (data not shown). Our results (Supplemental Data S2) show that a total of 775 genes are differentially expressed (log2 ratio > 1.5) between the two tissues. Transcripts of 486 genes are significantly more abundant in inner tissues as compared with outer tissues and those of 289 genes are significantly more abundant in outer versus inner stem tissues. The functional classification (Gene Ontology [GO] biological processes) of these genes is given in Supplemental Data S3.
In agreement with the contrasting lignification profiles of inner and outer stem tissues, transcripts corresponding to 27 flax unigenes encoding the major lignin biosynthetic enzymes were significantly more abundant in inner tissues when compared with outer tissues (Fig. 5; Supplemental Data S2, category secondary metabolic process). No transcripts corresponding to lignin unigenes were significantly more abundant in outer tissues. Transcripts for eight laccase unigenes encoding enzymes potentially involved in the oxidative polymerization of monolignols were also significantly more abundant in inner stem tissues. Transcripts corresponding to three peroxidase unigenes were more abundant in stem inner tissues, and transcripts for 15 peroxidase unigenes were less abundant (Supplemental Data S2, category other metabolic process). Analyses of subcellular localization by TargetP (http://www.cbs.dtu.dk/services/TargetP; data not shown) indicated that all differentially regulated peroxidase unigenes encode class III peroxidases targeted to the cell wall. The most highly expressed unigene (log2 ratio = 4.32) in inner tissues encodes a PLR catalyzing the synthesis of the lignans lariciresinol and secoisolariciresinol. Transcripts for two other unigenes encoding a PLR were also significantly abundant in inner tissues, as were those for two unigenes encoding phenylcoumaran benzylic ether reductase (PCBER) catalyzing the biosynthesis of the neolignan isodihydrodehydrodiconiferyl alcohol (IDDDC; Gang et al., 1999; Vander Mijnsbrugge et al., 2000).
Figure 5.
Schematic view of lignin and lignan biosynthetic and polymerization genes up- and down-regulated in flax inner and outer stem tissues. CAD, Cinnamyl alcohol dehydrogenase; CCoAOMT, caffeoyl-CoA O-methyltransferase; CCR, cinnamoyl-CoA reductase; C3H, p-coumaroyl shikimate 3′-hydroxylase; C4H, cinnamate 4-hydroxylase; 4CL, 4-hydroxycinnamoyl-CoA ligase; COMT, caffeic acid O-methyltransferase; F5H, ferulic acid/coniferaldehyde/coniferyl alcohol 5-hydroxylase; HCT, hydroxycinnamoyl-CoA:shikimate hydroxycinnamoyl transferase; LAC, laccase; PAL, Phe ammonia lyase; PCBER, phenylcoumaran benzylic ether reducatse; PLR, pinoresinol lariciresinol reductase; POX, peroxidase. Heat map red squares show genes up-regulated in inner stem tissues as compared with outer stem tissues, and green squares show genes up-regulated in outer stem tissues as compared with inner stem tissues (log2 ratio > 1.5 and < −1.5). [See online article for color version of this figure.]
Laccase/Peroxidase Genes Are Differentially Expressed in Upper, Middle, and Lower Stems
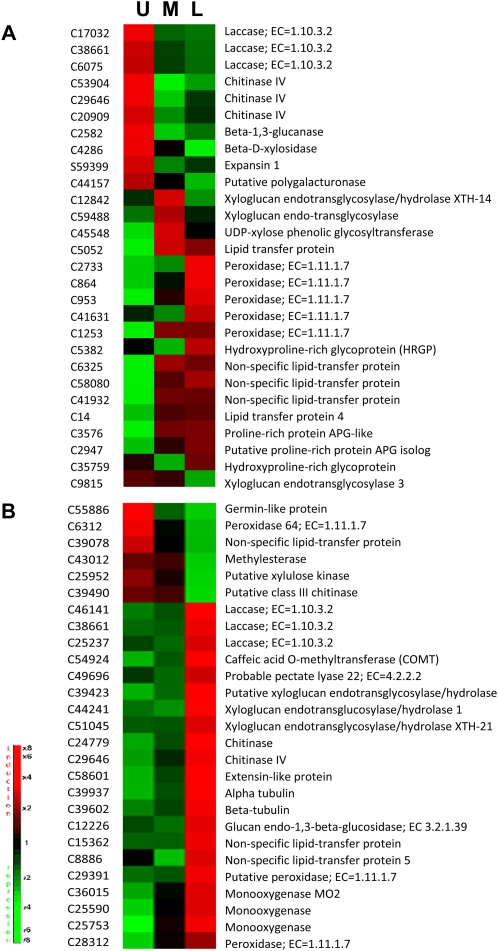
Flax stem tissues and cell walls show a developmental gradient along the length of the stem from the upper, young tissues to the lower, more mature tissues (Fig. 1). In stem inner tissues, this developmental gradient is associated with the progressive formation of xylem tissue, secondary cell wall formation, and extensive lignification. In outer tissues, the developmental gradient is associated with bast fiber elongation in the upper tissues and with the progressive formation and reorganization of the secondary cell wall in the middle and lower stem tissues (Gorshkova and Morvan, 2006). To obtain information about developmentally related gene expression in flax stems, microarrays were used to characterize gene expression at three different levels (upper, middle, and lower) in both inner and outer stem tissues. A total of 123 and 144 unigenes were differentially expressed between different stem levels in stem inner and outer tissues, respectively (Supplemental Data S4 and S5). The functional classification (GO biological processes) of these genes is given in Supplemental Data S6.
In the inner stem tissues, 28 unigenes potentially involved in lignin- and cell wall-related processes were differentially expressed between the upper, middle, and lower regions (Fig. 6A). Transcripts corresponding to monolignol biosynthetic genes were not differentially accumulated between the different stem regions. In contrast, genes encoding the two major classes of cell wall-related oxidoreduction enzymes (laccases and peroxidases) show distinct developmental regulation along the stem; transcripts of three laccase unigenes were significantly more abundant in upper stem tissues and less abundant in middle and lower stem tissues. Two of these unigenes (C17032 and C26075) were also more significantly expressed in whole inner stem tissues compared with whole outer stem tissues (Fig. 5; Supplemental Data S2). Transcripts for five peroxidase unigenes were either less abundant or showed no significant differences in upper and middle stem samples but were more abundant in lower stem tissues. All of these peroxidase unigenes were more significantly expressed in whole stem outer tissues compared with whole stem inner tissues (Fig. 5; Supplemental Data S2).
Figure 6.
Profile expression of differentially expressed cell wall-related genes (−1.5 < log2 ratio > 1.5) in inner (A) and outer (B) stem tissues during stem development. Log2 ratio was calculated as gene expression for each developmental level of inner/outer tissues (U, upper; M, middle; L, lower) versus entire inner/outer tissues. [See online article for color version of this figure.]
In the outer stem tissues, 27 unigenes encoding proteins potentially involved in lignin- and cell wall-related processes were differentially expressed between the upper, middle, and lower regions (Fig. 6B). In contrast to the profile observed for inner tissues, transcripts for three laccase unigenes were more abundant in the lower region. One of these unigenes (C38661) corresponded to one of the three laccase genes more expressed in stem upper tissues. Transcripts of one lignin unigene (COMT), one peroxidase unigene, and six unigenes encoding proteins involved in cell wall-remodeling events (pectate lyase, glucan endo-1,3-β-glucosidase, and xyloglucan endotransglucosylase/hydrolases) and the cytoskeleton (α- and β-tubulin) were also more abundant in lower stem samples. The peroxidase unigene (C23812) was also more highly expressed in whole stem outer tissues compared with whole stem inner tissues (Fig. 5; Supplemental Data S2). In contrast, one peroxidase gene (C6312) was more highly expressed in the upper region of the stem. Interestingly, this gene was also more highly expressed in whole stem inner tissues compared with whole stem outer tissues (Fig. 5; Supplemental Data S2).
Expression Profiling Identifies TFs Potentially Involved in Regulating Monolignol Metabolism and Cell Wall Formation in Flax
In order to investigate the transcriptional regulation of oligolignol metabolism and secondary cell wall formation in flax, we compared the expression profiles of genes encoding flax TFs with oligolignol- and cell wall-related genes. First, we selected all TF genes (613 unigenes) in the flax Genolin data set (Fénart et al., 2010; http://urgi.versailles.inra.fr/Species/Flax/Download-sequences; Table III; Supplemental Data S7). TF unigenes were then grouped into two clusters: (1) genes up-regulated in stem inner tissues when compared with stem outer tissues, and (2) genes down-regulated in stem inner tissues when compared with stem outer tissues. Sixty-three TF unigenes were significantly up-regulated in stem inner tissues, and 18 TF unigenes were up-regulated in stem outer tissues (Fig. 7A). Approximately 50% of the up-regulated genes in both tissues belonged to major TF classes known to be involved in regulating secondary cell wall formation and lignin/phenolic metabolism (Zhong et al., 2010).
Figure 7.
Putative regulators of cell wall-related genes. A, Cluster representation of TFs and cell wall-related genes differentially expressed in inner (I) and outer (O) tissues (1, up-regulated genes; 2, down-regulated genes) and with specific profiles by level (U, upper; M, middle; L, lower) in each tissue. B, Similar profile expression (K-means clustering) of TFs and cell wall-related genes in different stem levels. B1, C680-4CL, C3640-4CL, C519-CCoAOMT, C395-CCoAOMT, C11786-glucosyltransferase, C21188-glucosyltransferase, C6820-glucosyltransferase, C1284-glucosyltransferase, C6568-TF-like, C20655-NAM; B2, C5820-C3H, C6449-glucosyltransferase, C19744-Myb13, C50209-Myb; B3, C1423-LTP, C9104-bZIP35; B4, C28312-peroxidase, C11887-HB52; B5, C3323-laccase, C4333-laccase11, C13388-NAC; B6, C188-chitinase, C39490-class III chitinase, C43012-methylesterase, C31970-YAB5, C10018-GLK1; B7, C59528-PAL, C247-PAL, C32427-pectate lyase, C13388 NAC; B8, C3700-ketoacyl-CoA synthase, C8414-3-ketoacyl-CoA synthase 1, C29546-C3700-ketoacyl-CoA synthase, C10964-WRKY33. C, Similar profile expression (K-means clustering) of TFs arbitrarily considered as repressors and cell wall-related genes in different stem levels. C1, C10591-endoglucanase 2, C3396-fascicline-like AGP11, C5556-fascicline-like AGP11, C46914-hydroxy-Pro-rich glycoprotein, C37539-laccase, C835-PAL, C3205-UDP-d-glucuronate carboxy-lyase, C33158-xylan-1,4-β-xylosidase, C1122-Myb59, C30843-Myb59; C2, C6449-glycosyltransferase, C1465-UDP-Glc-6-dehydrogenase, C15-LTP, C16481-GRAS32; C3, C1190-chitinase-like, C34084-chitinase-like, C9696-Myb4; C4, C4294-endo-1,4-β glucanase, C38661-laccase, C3505-laccase, C3107-laccase, C36731-HSFA4C, C53556-TF-like. TFs from B1 to B3 and C1 and C2 profiles belong to A1 up-regulated genes in inner tissues, and those from B4 to B8 and C3 and C4 profiles belong to A2 up-regulated genes in outer tissues. [See online article for color version of this figure.]
Table III. Number of flax unigenes in major TF classes.
| TF Family | No. of Unigenes | No. of Unigenes Up-Regulated in Stem Inner Tissues | No. of Unigenes Up-Regulated in Stem Outer Tissues | No. of Unigenes Not Differentially Regulated |
| Myb | 62 | 8 | 3 | 51 |
| ERF | 13 | 0 | 0 | 13 |
| bZip | 35 | 4 | 0 | 31 |
| MADS | 9 | 0 | 0 | 9 |
| bHLH | 25 | 1 | 1 | 23 |
| HD-ZIP | 18 | 8 | 0 | 10 |
| WRKY | 35 | 3 | 1 | 31 |
| LIM | 13 | 4 | 0 | 9 |
| NAC | 18 | 2 | 2 | 14 |
| DRE | 5 | 0 | 0 | 5 |
| ATH | 3 | 1 | 0 | 2 |
| GRAS | 7 | 1 | 0 | 6 |
| BEL | 3 | 0 | 0 | 3 |
| Others | 367 | 31 | 11 | 325 |
| Total | 613 | 63 | 18 | 532 |
We then clustered the 613 TF genes with 273 cell wall- and oligolignol-related genes (CWGs) that were differentially expressed between either inner and outer stem tissues or different levels (upper, middle, and lower) within stem inner or outer tissues (Fig. 7A; Supplemental Data S7). K-means cluster analyses allowed us to identify TF genes and CWGs showing similar expression profiles in upper, middle, and lower regions from either stem inner or stem outer tissues (Fig. 7B; Table IV). Altogether, we identified eight major profiles (Fig. 7, profiles B1–B8) showing coexpression of TF genes and oligolignol- or cell wall-related genes. In certain profiles (B1, B2, B5, and B7), NAC and MYB TF genes were coexpressed with monolignol biosynthesis genes and laccase genes potentially involved in oxidative polymerization. These profiles all showed an increase in lignin gene expression along the stem length (upper to lower) in outer tissues, in agreement with the developmentally related lignification process in these tissues. In stem inner tissues, lignin gene expression decreased progressively (profile B1), or increased in the middle stem region before decreasing in the lower stem region (profile B2), or else did not change significantly (profiles B5 and B7). Certain TF genes coregulated with monolignol genes were also coexpressed with other CWGs, including several glucosyltransferases and a pectate lyase. Other TF genes were coexpressed with CWGs but not with lignin genes. We did not detect any coexpression of TF gene expression and genes specifically involved in lignan biosynthesis such as PLR and PCBER.
Table IV. TFs and cell wall genes (lignin, other) showing similar expression profiles (K-means clustering) in flax stem inner and outer tissues.
| Profile | TF | Cell Wall Gene (Lignin) | CWG (Other) |
| B1 | C6568-TF-like, C20655-NAM | C680-4CL, C3640-4CL, C519-CCoAOMT, C395-CCoAOMT | C11786-glucosyltransferase, C21188-glucosyltransferase, C6820-glucosyltransferase, C1284-glucosyltransferase |
| B2 | C19744-Myb13, C50209-Myb | C5820-C3H | C6449-glucosyltransferase |
| B3 | C9104-bZIP35 | C1423-LTP | |
| B4 | C11887-HB52 | C28312-peroxidase | |
| B5 | C13388-NAC | C3323-laccase, C4333-laccase11 | |
| B6 | C31970-YAB5, C10018-GLK1 | C188-chitinase, C39490-class III chitinase, C43012-methylesterase | |
| B7 | C13388 NAC | C59528-PAL, C247-PAL | C32427-pectate lyase |
| B8 | C10964-WRKY33 | C3700-ketoacyl-CoA synthase, C8414-3-ketoacyl-CoA synthase 1, C29546- C3700-ketoacyl-CoA synthase | |
| C1 | C1122-Myb59, C30843-Myb59 | C37539-laccase, C835-PAL | C10591-endoglucanase 2, C3396-fascicline-like AGP11, C5556-fascicline-like AGP11, C46914-Hyp-rich glycoprotein, C3205-UDP-d-glucuronate carboxy-lyase, C33158-xylan-1,4-β-xylosidase |
| C2 | C16481-GRAS32 | C6449-glycosyltransferase, C1465-UDP-Glc 6-dehydrogenase, C15-LTP | |
| C3 | C9696-Myb4 | C1190-chitinase-like, C34084-chitinase-like | |
| C4 | C36731-HSFA4C, C53556-TF-like | C38661-laccase, C3505-laccase, C3107-laccase, | C4294-endo-1,4-β glucanase |
K-means cluster analysis allows the identification of coregulated genes showing similar expression profiles but does not allow the direct detection of genes that show opposite expression patterns (e.g. repressor TF genes and their target genes). In order to identify potential repressor TFs in flax tissues, we arbitrarily inversed the signs of our lignin/CWG expression data (i.e. negative log2 ratios were considered as positive values) and did K-means cluster analysis with our TF expression data. We hypothesized that this approach would allow us to detect potential associations between an increase in TF expression and a decrease in oligolignol/CWG expression. This analysis allowed us to identify four major profiles (Fig. 7C; Table IV) and to identify a MYB TF gene negatively correlated with the expression of a laccase gene and a PAL gene (profile C1) together with two other TF genes that were negatively coregulated with three laccase unigenes (profile C4). These TF genes were also negatively coregulated with other CWGs, including endoglucanases, a β-xylosidase, and two FLA unigenes. Another MYB TF gene was negatively coexpressed with chitinase-like genes but not with lignin genes. Similarly, a GRAS TF gene was negatively coregulated with genes encoding a glycosyltransferase, a UDP-Glc-6-dehydrogenase, and a lipid transfer protein (LTP). Altogether, expression profiling enabled us to identify flax TFs potentially involved in a transcriptional regulatory network controlling lignification, (neo)lignan formation, and secondary cell wall development.
DISCUSSION
Monolignols Are Used in Different Metabolisms in Flax Stems
Induced hypolignification by the engineering of different target genes in model plant species is often associated with the accumulation of different low-Mr soluble phenolic compounds. We hypothesized that the natural hypolignification of bast fibers in flax outer stem tissues might also be characterized by the presence of a wide range of soluble phenolics. In order to test our hypothesis, we used combined metabolome profiling and lignomics of inner and outer stem tissues.
A number of primary and secondary metabolites were identified, and PCA of the 1H NMR data showed a clear discrimination between inner and outer tissues. Generally, outer stem tissues contained higher amounts of both primary and secondary metabolites than inner stem tissues per gram of dry weight, most probably because of the higher proportion of living cells in outer stem tissues. Many of these metabolites were previously identified in flax, and their presence in outer stem tissues is in general agreement with the physiology of this tissue. 1H NMR metabolomic profiling also identified the glucosylated forms of p-coumaryl alcohol and coniferyl alcohol (coniferin) but not the corresponding aglycones in both outer and inner stem tissues, suggesting that the free monolignol is rapidly glycosylated or channeled into other metabolic pathways.
Lignomics using UHPLC-FT-ICR-MS provided further extremely detailed information on flax stem phenolics. Altogether, we identified 81 different aromatic compounds, including, to our knowledge, 65 compounds previously unidentified in flax and 11 new compounds previously unidentified in plants. Flax seeds have long been known to be rich in lignans such as lariciresinol and secoisolariciresinol, and a number of phenolics have been previously identified in this species (Beejmohun et al., 2007; Struijs et al., 2009). However, to our knowledge, this is the first time a comprehensive phenolic profiling has been performed in flax. Lignomics of poplar (Populus spp.) xylem recently identified 134 oligolignols ranging from dilignols to hexalignols (Morreel et al., 2010a); therefore, we were not surprised to find oligolignols in flax inner stem tissues consisting of mainly xylem tissue. Remarkably, almost all of these oligolignols were also present in outer stem tissues, in agreement with our initial hypothesis suggesting that hypolignification in the flax stem would be associated with the accumulation of phenolics. Generally, glycosylated (neo)lignans were more abundant in the hypolignified outer tissues.
Of the three main monolignols, only coniferyl alcohol was detected in both inner and outer stem tissues. The presence of this monolignol in stems is not unexpected, since flax lignin is rich in G-lignin (Day et al., 2005b; del Rio et al., 2011). Of interest is the observation that coniferyl alcohol was nearly three times more abundant in outer stem tissues as compared with inner stem tissues, and this despite the fact that the majority of lignin genes are significantly underexpressed in outer stem tissues. Since laccase (and certain peroxidase) gene transcripts are significantly more abundant in stem inner tissues, one possible explanation is that the pool of free alcohol units is continually depleted to drive inner stem (xylem) lignification. Glycosylated forms of all three monolignols were detected in both inner and outer stem tissues and were approximately three to four times more abundant in the latter. Coniferin and the p-coumaryl alcohol glucoside were abundant in both inner and outer tissues, whereas only trace amounts of syringin were detected. Monolignol glycosylation has been proposed to facilitate monolignol storage in the vacuole and/or their transport toward the apoplast and incorporation into lignin following deglycosylation and activation by laccases and/or peroxidases (Weng and Chapple, 2010). However, recent results in Arabidopsis have shown that glycosylation is necessary for monolignol transport to the vacuole and subsequent storage, whereas the aglycone alcohols can be actively transported across the plasma membrane (Miao and Liu, 2010).
Closer examination of the flax stem oligolignol pool suggested that monolignols were utilized in three different but related metabolisms (Fig. 8). First, monolignols can be incorporated into the lignin polymer. The structure of low-Mr oligolignols is believed to reflect the radical coupling conditions present during lignin formation and therefore to provide information about lignin structure that is complementary to other widespread chemical or physical techniques such as thioacidolysis and NMR (Lapierre et al., 1985; Lu and Ralph, 2003; Morreel et al., 2004, 2010a, 2010b). Recently, a comprehensive study has provided detailed information on the structure of flax milled wood lignin from both stem inner tissues (shives) and bast fibers (del Rio et al., 2011). That study underlined the fact that flax lignin is rich in G-units and also contains high amounts of H-units, as reported previously in flax bast fibers (Day et al., 2005b). Analyses by pyrolysis-gas chromatography-MS indicated that H-units represented 5% and 13% of shive lignin and fiber lignin, respectively. Both two-dimensional NMR studies and thioacidolysis confirmed the presence of G- and H-units and indicated that flax lignin is highly condensed (40%–50%) and contains high proportions of β-β (resinol; 9%) and β-5 (phenylcoumaran; 14%) linkages (del Rio et al., 2011). (Note that two different nomenclatures are used to describe the propane carbon atoms involved in monolignol linkages in the lignin polymer and in oligolignols. In oligolignols, the numbers 7, 8, and 9 are used, whereas in the lignin polymer, α, β, and γ are used to describe the same carbon atoms. A β-β lignin linkage, therefore, corresponds to an 8-8 oligolignol linkage.) These results are in close agreement with our observations showing that the great majority of oligolignols contain G-units and also H- and S-units. In addition, the frequency of different intermonomeric linkages in our oligolignol results is very similar to that observed by del Rio et al. (2011), with 8-O-4 linkages representing 62% of the total bonds and 8-8 and 8-5 bonds representing 16% and 22%, respectively. The observed increase in the percentage of condensed bonds can be related to the high proportion of H-units. Examination of the oligolignols showed that the H-units were present as phenolic end groups in the flax oligolignols, as predicted by the higher oxidation potential of p-coumaryl alcohol as compared with those of coniferyl and sinapyl alcohol (Syrjanen and Brunow, 1998). In this case, the 8-O-4 coupling of p-coumaryl alcohol to a lignin oligomer blocks further chain elongation with incoming coniferyl or sinapyl alcohol radicals, resulting in a relative increase of 8-5 and 8-8 linkages. Similar increases have also been observed in NMR analyses of the H-rich lignins from HCT down-regulated pine (Pinus spp.) and alfalfa (Medicago sativa; Shadle et al., 2007; Wagner et al., 2007) and C3H down-regulated alfalfa (Ralph et al., 2006). We did not see any evidence in our transcriptomic data to suggest that the flax HCT/C3H genes were underexpressed in comparison with other lignin biosynthesis genes; however, in the absence of suitable flax mutants/transgenics for comparison, it is difficult draw a conclusion on the likely explanation for the high H-unit content of flax lignin. Nevertheless, the high proportion of G- and H-units, as well as the high percentage of condensed bonds, render the flax lignin of both xylem and bast fibers particularly difficult to degrade and have an important impact on the industrial processing of this species (Day et al., 2005b; del Rio et al., 2011).
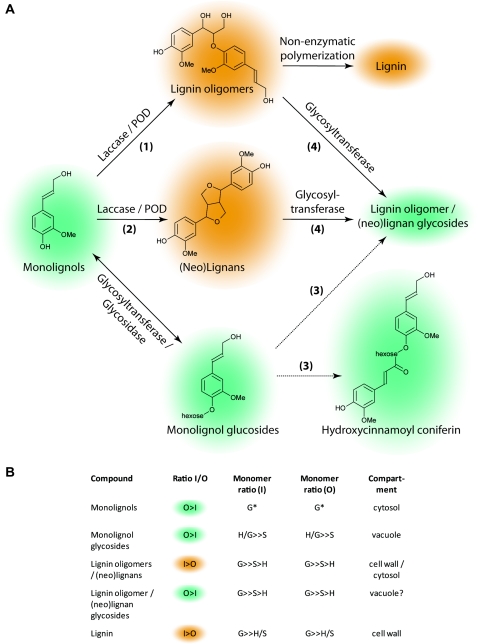
Figure 8.
Monolignol metabolism in flax stems. A, Different possible metabolic pathways of monolignols in flax stems (coniferyl alcohol is used as an example). Monolignols may be converted into phenoxy radicals in the cell wall by laccases and/or peroxidases and then undergo nonenzymatic polymerization to form lignin oligomers and lignin (pathway 1). Monolignols may also be converted into phenoxy radicals by laccases and/or peroxidases before being dimerized to form (neo)lignans (pathway 2). Monolignols can also be glycosylated by glycosyltransferases and then further complexed with other monolignols via an as yet uncharacterized pathway (pathway 3). Lignin oligomers and (neo)lignans may also be glycosylated (pathway 4). Orange/green shading in A and B (column 2) indicate the relative ratios of different monolignol compounds in flax stem inner and outer tissues (orange shading = compounds present in higher amounts in inner stem tissues, and green shading = compounds present in higher amounts in outer stem tissues). POD, Peroxidase. B, Relative ratios of H-, G-, and S-unit compounds in stem inner tissues (column 3) and outer tissues (column 4). The supposed subcellular localization of the different compounds is shown in column 5. G* indicates that p-coumaryl alcohol and sinapyl alcohol were not detected in either inner or outer stem tissues. [See online article for color version of this figure.]
Second, monolignols can also be dimerized to form lignans. We detected a range of lignans and neolignans, including pinoresinol, lariciresinol, secoisolariciresinol, dehydrodiconiferyl alcohol (DDC), and IDDDC, as well as their glycosylated forms in both inner and outer stem tissues of flax. These results clearly indicate that coniferyl alcohol is utilized in (neo)lignan biosynthesis as well as in G-lignin formation. Although the PLR gene encoding the enzyme catalyzing the formation of lariciresinol is more highly expressed (log2 ratio = 3.6–4.3) in inner stem tissues, pinoresinol and lariciresinol levels are similar in both inner and outer stem tissues. In inner stem tissues, coniferyl alcohol could be rapidly incorporated into the lignin polymer and therefore may become limiting for subsequent lignan formation. The observation that both coniferyl alcohol and coniferin are more abundant in outer stem tissues supports this argument. Previous gene expression analyses have demonstrated that a PLR gene is expressed in different flax seed tissues (Hano et al., 2006b; Venglat et al., 2011). Although promoter-GUS analyses and semiquantitative PCR indicated that the LuPLR1 gene cloned was not expressed in flax stem tissues (Hano et al., 2006b), the recent cloning of a second PLR gene (LuPLR2; Hemmati et al., 2010) from leaves and stems of flax plants, together with our results, suggest that PLR genes in flax form a small multigene family with organ-/tissue-specific expression patterns. Two other highly abundant neolignan unigene transcripts in flax inner stem tissues correspond to the enzyme PCBER, catalyzing the reduction of DDC into IDDDC and identified previously as the most abundant protein in poplar xylem (Gang et al., 1999; Vander Mijnsbrugge et al., 2000). The corresponding gene is also expressed in aspen stems (Populus spp.; Sterky et al., 2004; Prassinos et al., 2005). Although PCBER transcripts were more highly abundant in inner stem tissues, IDDDC was not detected in either inner or outer stem tissues, and this despite the presence of DDC in both tissues. In contrast, IDDDC hexosides were detected in both stem tissues, suggesting that the neolignan is rapidly glycosylated.
Biologically active (neo)lignans are widely present in tracheophytes (Weng and Chapple, 2010) and in the woody tissues of different tree species (Davin and Lewis, 2003; Holmborn et al., 2003), where they are believed to protect the vascular tissue from pathogens (Kwon et al., 2001). In flax, lignans accumulate to high quantities in seeds, with SDG being the major lignan accumulating in flax hulls (Singh et al., 2011). Anhydrosecoisolariciresinol, the anhydrous form of secoisolariciresinol, has also previously been shown to accumulate in flax capsules, seeds, and roots but not in leaves and stems (Charlet et al., 2002). To our knowledge, this is the first report of the presence of such a large range of (neo)lignans in flax stems. As indicated above, lignans are known to accumulate in the woody tissues of trees; therefore, their presence in flax xylem tissues is not unexpected. The accumulation of (neo)lignans in outer stem tissues is in agreement with our initial hypothesis suggesting a link between hypolignification and the accumulation of phenolic compounds. According to this idea, (neo)lignans in flax represent monolignol detoxification/storage forms. Subsequent glycosylation would also increase their water solubility, thereby facilitating transport to the flowers and potential participation in SDG accumulation in seeds.
Third, the detection of feruloyl and sinapoyl coniferin, and a nitrogen-containing derivative of feruloyl coniferin, points to the use of monolignols in other, partially unknown secondary pathways. Interestingly, these compounds were uniquely present (or present in higher amounts) in the hypolignified stem outer tissues; therefore, it is possible that the coniferin-containing compounds in flax outer stem tissues also represent storage or detoxification forms of coniferyl alcohol.
Altogether, our results provide novel detailed information concerning the fate of monolignols in flax stem tissues and underline the fact that further research is necessary to improve our understanding of the factors controlling monolignol partitioning between these different pathways as well as the relation between them. Further work is also necessary to identify the subcellular compartments in which the different monolignol-derived compounds and their glycosides are synthesized and stored (Fig. 8). While it is generally accepted that monolignols are converted into phenoxy radicals by laccases and/or class III peroxidases and then polymerized to form lignin oligomers and lignin in the cell wall, much less is known about the subcellular localization of (neo)lignans. A recent study (Attoumbré et al., 2010) localized the flax lignan secoisolariciresinol to the cell wall in the outer integument of seeds, suggesting that (neo)lignans may also be localized in this compartment. In contrast, coniferin and the other monolignol glycosides are supposedly stored in the vacuole (Whetten and Sederoff, 1995). The compartments in which (neo)lignans are glycosylated and stored have not yet been identified. If monolignols are dimerized in the cell wall under the action of laccases/peroxidases, are they then transported back into the cytosol prior to glycosylation and subsequent transfer to the vacuole? The study of Attoumbré et al. (2010) showed that secoisolariciresinol was also weakly detected in cytoplasmic inclusions, suggesting that the lignan is also present in this compartment. Since the antibodies used in this study also showed a weak cross-reactivity with the glycosylated form (SDG), it is possible that glycosylated (neo)lignans may be present in both the cell wall and cytosol compartments. Recent work (Miao and Liu, 2010) has shown that plasma membrane ATP-binding cassette-like (ABC) transporters are capable of transporting aglycone monolignols but not the corresponding glucosides, whereas vacuolar ATP-binding cassette-like transporters can transport the glucosides but not the aglycone forms. These results clearly indicate that glycosylation plays a central role in monolignol transport and storage, and it would obviously be interesting to see whether a similar mechanism exists for monolignol dimer transport. Since the ABC transporter superfamily is one of the largest transporter protein families in plants and different members are involved in the transport of a wide range of different molecules, including phenolics (Yazaki, 2006; Kaneda et al., 2011), it is not impossible that ABC transporters may also be involved in transporting monolignol dimers. Our results (Supplemental Data S2) indicated that flax transcripts corresponding to two ABC transporters (orthologs of AtABCG11 and AtABCG40) are significantly less abundant in flax outer stem tissues. Functional characterization of these transporters could help to clarify this point.
Hypolignification in Flax Stems Involves Differential Expression of Monolignol Genes
The hypolignification of flax fibers can theoretically be controlled at a number of points (e.g. transcriptional regulation of monolignol biosynthesis genes, precursor transport into the cell wall, and oxidative polymerization; Weng and Chapple, 2010). We have previously shown that transcripts for flax monolignol biosynthetic genes are more abundant in inner stem tissues as compared with outer stem tissues (Fénart et al., 2010). Our results here showing that transcripts for all (27 of 27) flax lignin biosynthesis unigenes are less abundant in stem outer tissues as compared with inner tissues confirm this earlier investigation and suggest that fiber hypolignification is regulated, at least partially, via the down-regulation of monolignol biosynthetic genes. These results are in agreement with similar studies in hemp (Cannabis sativa), which also possesses less heavily lignified outer stem fibers (De Pauw et al., 2007; van den Broeck et al., 2008). In addition, our results suggest that hypolignification is also controlled at the monolignol polymerization step. Oxidation of monolignols to form lignin phenoxy radicals is catalyzed by laccases and/or peroxidases. Although previous studies in flax have suggested that peroxidases are involved in stem lignification (McDougall, 1992), our results showing that transcripts for eight laccase unigenes and three peroxidase genes are more abundant in stem inner tissues suggest that laccases are also involved in this process. In support of this hypothesis, it is interesting that the five most abundant laccase transcripts in flax inner stem tissues are orthologs of the IRX12/AtLAC4 and AtLAC17 genes that have previously been shown to be involved in lignin formation in Arabidopsis (Brown et al., 2005; Berthet et al., 2011). The observation that three laccase gene transcripts are significantly more abundant in the upper region of inner stem tissues, whereas five peroxidase gene transcripts are more abundant in the lower region, could suggest that laccases are involved in the early stages of lignin formation followed by peroxidases, as hypothesized previously (Sterjiades et al., 1993). This idea is also supported by our results showing that three laccase transcripts and a transcript corresponding to the lignin gene COMT are significantly more abundant in the lower region of stem outer tissues, where lignification of bast fibers is first observed.
Whereas laccase unigenes were more abundant in flax stem inner tissues when compared with outer stem tissues, the opposite was largely true with regard to peroxidase genes, and 15 of 18 peroxidase unigene transcripts were more abundant in outer stem tissues. All these flax peroxidases are class III peroxidases targeted to the cell wall. Class III peroxidases belong to large multigene families (73 in Arabidopsis and 138 in rice [Oryza sativa]; Passardi et al., 2004) and catalyze diverse oxidoreductions between hydrogen peroxide (H2O2) and various reductants. Peroxidases, like laccases, have been implicated in monolignol coupling during lignin synthesis as well as in other cell wall reticulation processes. Peroxidases are also capable of generating hydroxyl radicals from H2O2 via a nonenzymatic Fenton reaction and have been implicated in cell wall loosening and cell growth (Chen and Schopfer, 1999; Passardi et al., 2004); therefore, it is possible that the observed up-regulation of peroxidase genes in flax stem outer tissues is related to fiber elongation and not to cell wall reticulation and/or lignification. Our observation that one peroxidase unigene is significantly expressed in the upper region of outer stem tissues supports this idea, since bast fiber cell walls are not lignified at this stage. Interestingly, class III peroxidases have been shown to be involved in cell wall loosening and the massive cell expansion associated with cotton fiber elongation (Qin et al., 2008).
Both laccases and peroxidases have been implicated in the oxidation of monolignols necessary for (neo)lignan formation (Davin and Lewis, 2003); therefore, it is probable that some of the corresponding proteins are also involved in the formation of these phenolics. Whether the observed differences in the abundance of (neo)lignan versus lignin oligomers in flax stem inner and outer tissues can be linked to the marked differences in laccase/peroxidase gene expression profiles requires further investigation. In addition, it is clear that the identification and functional characterization of genes encoding the various glycosyltransferases and glycosidases acting on monolignols represent a major challenge to fully understanding flax oligolignol metabolism.
In conclusion, our results show that the natural hypolignification of flax outer stem tissues involves the differential expression of lignin biosynthesis genes and laccase/peroxidase genes potentially involved in the oxidative polymerization of monolignols. However, a complete explanation of flax bast fiber hypolignification and lignin structure should also take into account other factors, such as the cellulose-rich environment of the cell wall and the availability of H2O2, since these have been shown to affect lignification (Ros Barcelo, 1998; Touzel et al., 2003). Although we did not see any differential accumulation of transcripts coding for H2O2-generating NADPH oxidases, we did observe a differential accumulation of transcripts (Supplemental Data S2) corresponding to six flax germin-like proteins potentially involved in H2O2 generation via the oxidation of oxalic acid (Davidson et al., 2009). Three flax unigenes were more highly expressed in stem inner tissues, while another three unigenes were more highly expressed in outer stem tissues. Functional characterization of these germin-like proteins should help to evaluate whether H2O2 supply plays a role in regulating lignification in flax.
Expression Profiling Identifies TFs Potentially Involved in Controlling Lignification, Phenolic Metabolism, and Cell Wall Formation
Comparison of the expression levels in flax stems enabled us to identify TF genes showing significant differential expression between inner and outer stem tissues. A number of these genes were orthologs of TFs known to be involved in the control of vascular tissue differentiation and secondary cell wall formation in other species. For example, transcripts for eight unigenes encoding HD-ZIP III TFs involved in controlling the balance between xylem and phloem tissue formation in both Arabidopsis and poplar were significantly more abundant in flax inner stem tissues (Prigge et al., 2005; Ko et al., 2006). Similarly, transcripts for four bZIP genes were more abundant in stem inner tissues. Members of this TF family are expressed during stem development and secondary vascular tissue formation in Arabidopsis, Eucalyptus, and poplar (Paux et al., 2004; Ehting et al., 2005; Dharmawardhana et al., 2010). Other differentially regulated TF genes included two bHLH, four WRKY, and four LIM genes. Members of these TF families have also been previously implicated in vascular tissue development and/or the control of secondary cell wall formation (Demura and Fukuda, 2007).
The identification of AC elements in the promoters of several flax lignin biosynthesis genes (data not shown) suggested a role for the involvement of MYB TFs in the transcriptional regulation of lignin biosynthesis in this species. In support of this hypothesis, we observed that transcripts for the orthologs of AtMYB20 and AtMYB42 were more abundant in stem inner tissues. In Arabidopsis, these genes form part of a secondary cell wall transcriptional regulatory network and are downstream of so-called master switch TFs such as NAC and other MYB factors (Zhong et al., 2010). In contrast, no orthologs corresponding to the MYB factors AtMYB58 or AtMYB63 directly implicated in the regulation of lignin biosynthesis (Zhou et al., 2009) were differentially regulated. We also identified a MYB TF transcript that was significantly more abundant in stem outer tissues and belongs to subgroup 4, which includes repressors of the lignin biosynthetic pathway such as AtMYB4, EgMYB1, and ZmMYB31/42 (Jin et al., 2000; Fornalé et al., 2006, 2010; Legay et al., 2010). Although functional analyses are obviously necessary, the expression of a potential lignin repressor in outer tissues is consistent with the hypolignified nature of this tissue. AC elements are also present in the promoter of the flax LuPLR1 gene (Hano et al., 2006b), suggesting that transcriptional regulation of lignan biosynthesis might also involve MYB factors. In support of this idea, a recent study has shown that MYB TF genes are expressed in flax seed tissues (Venglat et al., 2011). In this latter study, different MYB TF genes were expressed in flax stem tissues and seeds, although six MYB TF genes were also expressed in both tissues. We also observed that the flax orthologs of several genes encoding NAC factors, such as SND1, SND2, XND1, VNI2, and KNAT7, previously shown to be involved in the control of secondary cell wall formation and/or xylem vessel differentiation and forming part of a transcriptional control network, were also differentially regulated between inner and outer stem tissues (Zhao et al., 2008; Yamaguchi et al., 2010; Zhang et al., 2011; Zhao and Dixon, 2011).
Transcriptome profiling at different levels in flax stem inner and outer tissues allowed us to more precisely identify TFs that were coregulated with different lignin- and cell wall-related genes. Once again, these results underlined the likely central role of NAC and MYB TFs in regulating lignin biosynthesis in flax. Monolignol biosynthetic genes 4CL and CCoAOMT were coregulated with the flax ortholog of SND2, and the monolignol gene C3H was coregulated with the flax ortholog of AtMYB20. Both of these TFs form part of a transcriptional regulatory network controlling lignification and secondary cell wall formation in Arabidopsis (Zhong et al., 2010). Another NAC factor (ANAC002/ATAF1) was coregulated with two PAL unigenes and the orthologs of AtLAC4 and AtLAC11, suggesting that the regulation of both monolignol production and oxidative polymerization in flax involve NAC TFs. AtLAC4 has been shown to play a central role together with AtLAC17 in controlling interfascicular fiber lignification in Arabidopsis (Berthet et al., 2011) and to be a direct target of AtMYB58 (Zhou et al., 2009). Taken together, these results suggest that hypolignification of flax fibers is controlled via a complex regulatory transcriptional network, with different NAC and MYB TFs playing central roles in controlling the expression of genes responsible for producing the lignin monomers and the oxidative polymerization step.
The presence of (neo)lignans in stem tissues suggests that these TFs might also be involved in regulating the production of these dimeric structures. Although we did not detect any coexpression of TF gene expression and genes specifically involved in lignan biosynthesis such as PLR and PCBER, the observation that flax NAC genes are coregulated with monolignol biosynthetic genes, together with results (Venglat et al., 2011) showing that MYB TFs are expressed in developing seeds, could suggest that a similar transcriptional regulatory network plays a role in controlling (neo)lignan production in the flax stem.
CONCLUSION
A combined metabolomic profiling, lignomics, and transcriptomics approach provides, to our knowledge for the first time, a detailed picture of monolignol gene expression and metabolism in flax stems. In addition to providing novel information about the transcriptional regulation of lignification in flax plants, our study suggests that the natural hypolignification of outer stem tissues is associated with the accumulation of a wide range of phenolics. Recently, a number of tools have been developed for flax, including high-density microarrays, cDNA libraries, physical and genetic maps, molecular markers, and mutant populations, which should allow an integrated approach to investigate different processes in this biologically interesting species (Cloutier et al., 2010; Fénart et al., 2010; Ragupathy et al., 2011; Venglat et al., 2011).
MATERIALS AND METHODS
Plant Material
Nine-week-old greenhouse-grown flax (Linum usitatissimum) plants were used for all experiments. For total stem transcriptomics, metabolomics, and lignomics, stem outer tissues were separated from inner tissues by peeling off the bark as described previously (Day et al., 2005a). Inner and outer stem tissues from 20 plants were immediately frozen in liquid nitrogen, pooled, and stored at −80°C prior to RNA extraction. For upper, middle, and lower stem transcriptomics, stems from five plants were divided into upper, middle, and lower regions. Outer tissues were separated from inner tissues by peeling, frozen in liquid nitrogen, pooled, and stored at −80°C prior to RNA extraction.
Microscopy
Freehand cross-sections were made of flax stems using a fresh razor blade, stained with phloroglucinol-HCl, and examined using a Nikon Eclipse TS100 microscope to visualize lignified cell walls. Photographs were made using a Nikon D5000 camera.
FTIR
Six independent inner and outer stem tissue samples from the pooled material were repeatedly 80% ethanol extracted to remove most of the extractive components, resulting in cell wall-enriched fractions. Diffuse reflectance infrared Fourier transform spectroscopy was performed using NICOLET 6700 (Thermo Electron). FTIR spectra of cell walls were recorded in the mid range (4,000–450 cm−1) at 4 cm−1 resolution. All spectra were baseline corrected and then normalized at 1,550 cm−1. Average spectra obtained from the six biological replicates of inner and outer tissues are shown in the window 2,000 to 800 cm−1 as described previously (Hano et al., 2006a).
1H NMR Metabolomics
Extraction Procedure
Six independent inner and outer stem tissues samples from the pooled material were reduced to a homogeneous powder using a grinder. Fifty milligrams was transferred to a 2-mL microtube, and 1.5 mL of extraction buffer (750 μL of methanol-d4 and 750 μL of KH2PO4 buffer in D2O containing 1% [w/v] trimethyl silyl propionic acid sodium salt [TMSP-d4], pH 6.0) was added to each sample. The tube was shaken at room temperature for 1 min with a vortex before ultrasonication (25 min) and centrifugation at 20,000g at room temperature for 30 min. pH was adjusted to 6. Six hundred microliters of the supernatant was then transferred into a 5-mm NMR tube.
NMR Spectra Measurements
All spectra were recorded at 300 K on a Bruker Avance III 600 spectrometer operating at 600.17 MHz for 1H and at 150.91 MHz for 13C that was equipped with a TXI 5-mm z-gradient probe. Shim control is performed automatically by gradient shimming and final line shape optimization (Topshim 1D procedure). Each spectrum consisted of 128 scans of 64 K data points with a spectral width of 6,602 Hz and a water suppression pulse sequence with a relaxation delay of 2 s. The resulting 1H spectra were manually phased, baseline corrected, and calibrated to TMSP at 0.0 ppm, all using Topspin (version 2.1; Bruker). Free induction decay was multiplied by an exponential weighing function corresponding to a line broadening of 0.3 Hz prior to Fourier transformation. Two-dimensional J-resolved and 1H NMR spectra were acquired using 32 scans per 128 increments that were collected into 16 K data points, using spectral widths of 6,602 Hz in F2 and 50 Hz in F1. J-resolved spectra were tilted by 45° and symmetrized about F1. Coherence order selective gradient heteronuclear single quantum coherence spectra were recorded for a data matrix of 256 × 4,096 points covering 30,185 × 6,602 Hz with 128 scans for each increment. INEPT transfer delays were optimized for a heteronuclear coupling of 145 Hz, and a relaxation delay of 1.5 s was applied. Data were linear predicted in F1 to 512 × 4,096 using 32 coefficients and then zero filled to 2,048 × 4,096 points prior to echo-anti echo-type two-dimensional Fourier transformation, and a sine bell-shaped window function shifted by π/2 in both dimensions was applied.
Data and Statistical Analyses
The 1H NMR spectra were automatically reduced to ASCII files using MestReNova (version 5.2.5; Mestrelab Research). Spectral intensities were scaled to TMSP and reduced to integrated regions or “buckets” of equal width (0.04 ppm) corresponding to the region δ 10.4 ppm to δ −0.6 ppm. The region between δ 4.85 ppm and δ 4.75 ppm was removed from the analysis because of the residual signal of water. The residual proton signals corresponding to methanol-d4 (δ 3.33 ppm to δ 3.25 ppm) and the proton signals corresponding to TMSP-d4 (at δ 0.0 ppm) were also removed. The generated ASCII file was imported into Microsoft Excel for the addition of labels. PCA was performed with SIMCA-P software (version 11.0; Umetrics). Each NMR signal identified by PCA as discriminant was integrated. The resulting area was used to test the significance of the differences observed between inner and outer stem tissues by Student’s t test performed with Microsoft Excel software. It was considered significant at P < 0.01.
Oligolignol Profiling by UHPLC-FT-ICR-MS
An inner and outer stem tissue sample from the pooled material was homogenized in liquid nitrogen and extracted with 25 mL of ethanol:water (4:1, v/v). Following five consecutive extractions, the extracts were combined and centrifuged, and the supernatants were evaporated using a Buchnii rotavapor. After redissolving the pellet in methanol (2:1, v/w), 200 μL was evaporated in a SpeedVac and extracted with 800 μL of water:cyclohexane (1:1, v/v). Phenolic profiling was performed using 10 μL of the water phase. Extracts were analyzed with an Accela UHPLC system (Thermo Electron) consisting of an Accela autosampler coupled to an Accela pump and further hyphenated to a LTQ FT Ultra (Thermo Electron) MS unit consisting of a linear ion-trap mass spectrometer connected with a FT-ICR-MS device. The separation was performed on a reverse-phase Acquity UPLC BEH C18 column (150 mm × 2.1 mm, 1.7 μm; Waters) with aqueous 0.1% acetic acid and acetonitrile:water (99:1, v/v; acidified with 0.1% acetic acid) as solvents A and B. At a flow of 300 μL min−1 and a column temperature of 80°C, the following gradient was applied: 0 min at 5% B, 30 min at 45% B, 35 min at 100% B. The autosampler temperature was 10°C. Analytes were negatively ionized with an atmospheric pressure chemical ionization source using the following parameter values: source current, 5 μA; capillary temperature, 200°C; vaporizer temperature, 350°C; sheath gas 39 (arb), aux gas 5 (arb), sweep gas 7 (arb). Full FT-ICR-MS spectra between 120 and 1,000 mass-to-charge ratio (m/z) were recorded (1.2–1.7 s scan−1) at a resolution of 100,000. In parallel, three data-dependent MSn spectra were recorded on the ion-trap MS apparatus using the preliminary low-resolution data obtained during the first 0.1 s of the previous full FT-ICR-MS scan: a MS2 scan of the most abundant m/z ion of the full FT-ICR-MS scan, followed by two MS3 scans of the most abundant first product ions. MSn scans were obtained with 35% collision energy. Chemical formulae of the compounds of interest were obtained with the Qual Browser in Xcalibur version 2.0 SR2.
Nimblegen Microarray Transcriptomics
RNA Extraction
Total RNA was isolated from pooled flax inner and outer stems using the NucleoSpin RNA Plant kit (Macherey-Nagel) following the manufacturer’s guidelines. To obtain a sufficient amount of RNA for microarray analysis (10 μg), a minimum of three extractions with up to 150 mg of fresh tissue were necessary for each sample. To eliminate DNA contamination, on-column treatment was done using the RNase-free DNase included in the kit. RNA integrity and concentration were evaluated with RNA StdSens Chips using the Experion automated electrophoresis system (Bio-Rad). For each sample, the three RNA extracts were pooled and final concentrations were adjusted to 1 μg μL−1. Two biological replicates were performed for each sample.
Double-stranded cDNA (ds-cDNA) was synthesized from 10 μg of total RNA using an Invitrogen SuperScript ds-cDNA synthesis kit in the presence of 250 ng of random hexamer primers. ds-cDNA was cleaned and labeled in accordance with the Nimblegen Gene Expression Analysis protocol (Nimblegen Systems). Briefly, ds-cDNA was incubated with 4 μg of RNase A (Promega) at 37°C for 10 min and cleaned using phenol-chloroform-isoamyl alcohol, followed by ice-cold absolute ethanol precipitation.
For Cy3 labeling of cDNA, the Nimblegen One-Color DNA Labeling kit was used according to the Gene Expression Analysis protocol (Nimblegen Systems). One microgram of ds-cDNA was incubated for 10 min at 98°C with 2 OD of Cy3-9mer primer. Then, 100 pmol of deoxyribonucleotide triphosphate and 100 units of the Klenow fragment (New England Biolabs) were added, and the mix was incubated at 37°C for 2.5 h. The reaction was stopped by adding 0.1 volume of 0.5 m EDTA, and the labeled ds-cDNA was purified by isopropanol/ice-cold 80% ethanol precipitation.
Microarray Processing and Analysis
A flax high-density oligomicroarray platform (Gene Expression Omnibus no. GPL10419), based on the Nimblegen 385K chip, was used for gene expression analysis (Fénart et al., 2010). Microarrays were hybridized with 6 μg of Cy3-labeled ds-cDNA in Nimblegen hybridization buffer/hybridization component A at 38°C during 16 to 18 h in a hybridization chamber (Hybridization System; Nimblegen Systems). Following hybridization, washing was performed using the Nimblegen Wash buffer kit (Nimblegen Systems), and slides were immediately scanned at 5 μm pixel−1 resolution using an Axon GenePix 4000B scanner (Molecular Devices) piloted by GenePix Pro 6.0 software (Axon). Scanned images were then imported into NimbleScan software (Nimblegen Systems) for grid alignment and expression data analyses. Expression data were normalized through quantile normalization and the Robust Multichip Average algorithm included in the NimbleScan software (Irizarry et al., 2003). Two technical replicates were analyzed for both biological replicates. The complete microarray data set is publicly available at the National Center for Biotechnology Information Gene Expression Omnibus database (http://ncbi.nlm.nih.gov/geo/) with accession number GSE29345.
Identification of genes displaying a change in expression over repetitions was accomplished with a script utilizing library functions in R with a false discovery rate of less than 5%. The SAM (Tusher et al., 2001) was used to identify differentially expressed genes over different conditions, and log2 (ratio) was used for filtering gene expression profiles.
Analyses were performed for whole inner versus whole outer stem tissues and for each stem region (upper, middle, and lower) versus whole inner or outer tissue.
Supplemental Data
The following materials are available in the online version of this article.
Supplemental Data S1. Identification of novel flax oligolignols based upon MS2 and MS3 fragmentation patterns.
Supplemental Data S2. List of genes differentially expressed between flax inner and outer stem tissues.
Supplemental Data S3. GO (biological processes) classification of genes differentially expressed in stem inner and outer tissues.
Supplemental Data S4. List of genes differentially expressed between upper, middle, and lower inner stem tissues.
Supplemental Data S5. List of genes differentially expressed between upper, middle, and lower outer stem tissues.
Supplemental Data S6. GO (biological processes) classification of genes differentially expressed within inner and outer stem tissues.
Supplemental Data S7. List of flax TFs and cell wall-related genes used for K-means analyses and transcript profiling.
Supplementary Material
References
- Attoumbré J, Bienaimé C, Dubois F, Fliniaux M-A, Chabbert B, Baltora-Rosset S. (2010) Development of antibodies against secoisolariciresinol: application to the immunolocalization of lignans in Linum usitatissimum seeds. Phytochemistry 71: 1979–1987 [DOI] [PubMed] [Google Scholar]
- Beejmohun V, Fliniaux O, Grand É, Lamblin F, Bensaddek L, Christen P, Kovensky J, Fliniaux M-A, Mesnard F. (2007) Microwave-assisted extraction of the main phenolic compounds in flaxseed. Phytochem Anal 18: 275–282 [DOI] [PubMed] [Google Scholar]
- Berthet S, Demont-Caulet N, Pollet B, Bidzinski P, Cézard L, Le Bris P, Borrega N, Hervé J, Blondet E, Balzergue S, et al. (2011) Disruption of LACCASE4 and 17 results in tissue-specific alterations to lignification of Arabidopsis thaliana stems. Plant Cell 23: 1124–1137 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boerjan W, Ralph J, Baucher M. (2003) Lignin biosynthesis. Annu Rev Plant Biol 54: 519–546 [DOI] [PubMed] [Google Scholar]
- Brown DM, Zeef LA, Ellis J, Goodacre R, Turner SR. (2005) Identification of novel genes in Arabidopsis involved in secondary cell wall formation using expression profiling and reverse genetics. Plant Cell 17: 2281–2295 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Charlet S, Bensaddek L, Raynaud S, Gillet F, Mesnard F, Fliniaux M-A. (2002) An HPLC procedure for the quantification of anhydrosecoisolariciresinol: application to the evaluation of flax lignan content. Plant Physiol Biochem 40: 225–229 [Google Scholar]
- Charlton A, Donarski J, Harrison M, Jones S, Godward J, Oehlschlager S, Arques J, Ambrose M, Chinoy C, Mullineaux P, et al. (2008) Responses of the pea (Pisum sativum L.) leaf metabolome to drought stress assessed by nuclear magnetic resonance spectroscopy. Metabolomics 4: 312–327 [Google Scholar]
- Chen SX, Schopfer P. (1999) Hydroxyl-radical production in physiological reactions: a novel function of peroxidase. Eur J Biochem 260: 726–735 [DOI] [PubMed] [Google Scholar]
- Cloutier S, Ragupathy R, Niu Z, Duguid S. (2010) SSR-based linkage map of flax (Linum usitatissimum L.) and mapping of QTLs underlying fatty acid composition traits. Mol Breed 28: 437–451 [Google Scholar]
- Dauwe R, Morreel K, Goeminne G, Gielen B, Rohde A, Van Beeumen J, Ralph J, Boudet A-M, Kopka J, Rochange SF, et al. (2007) Molecular phenotyping of lignin-modified tobacco reveals associated changes in cell-wall metabolism, primary metabolism, stress metabolism and photorespiration. Plant J 52: 263–285 [DOI] [PubMed] [Google Scholar]
- Davidson RM, Reeves PA, Manosalva PM, Leach JE. (2009) Germins: a diverse protein family important for crop improvement. Plant Sci 177: 499–510 [Google Scholar]
- Davin LB, Lewis NG. (2003) An historical perspective on lignan biosynthesis: monolignol, allyphenol and hydroxycinnamic acid coupling and downstream metabolism. Phytochem Rev 7: 257–288 [Google Scholar]
- Day A, Addi M, Kim W, David H, Bert F, Mesnage P, Rolando C, Chabbert B, Neutelings G, Hawkins S. (2005a) ESTs from the fibre-bearing stem tissues of flax (Linum usitatissimum L.): expression analyses of sequences related to cell wall development. Plant Biol 7: 23–32 [DOI] [PubMed] [Google Scholar]
- Day A, Ruel K, Neutelings G, Crônier D, David H, Hawkins S, Chabbert B. (2005b) Lignification in the flax stem: evidence for an unusual lignin in bast fibers. Planta 222: 234–245 [DOI] [PubMed] [Google Scholar]
- del Río JC, Rencoret J, Gutiérrez A, Nieto L, Jiménez-Barbero J, Martínez AT. (2011) Structural characterization of guaiacyl-rich lignins in flax (Linum usitatissimum) fibers and shives. J Agric Food Chem 59: 11088–11099 [DOI] [PubMed] [Google Scholar]
- Demura T, Fukuda H. (2007) Transcriptional regulation in wood formation. Trends Plant Sci 12: 64–70 [DOI] [PubMed] [Google Scholar]
- De Pauw MA, Vidmar JJ, Collins J, Bennett RA, Deyholos MK. (2007) Microarray analysis of bast fibre producing tissues of Cannabis sativa identifies transcripts associated with conserved and specialised processes of secondary cell wall development. Funct Plant Biol 34: 737–749 [DOI] [PubMed] [Google Scholar]
- Dharmawardhana P, Brunner AM, Strauss SH. (2010) Genome-wide transcriptome analysis of the transition from primary to secondary stem development in Populus trichocarpa. BMC Genomics 11: 150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ehlting J, Mattheus N, Aeschliman DS, Li E, Hamberger B, Cullis IF, Zhuang J, Kaneda M, Mansfield SD, Samuels L, et al. (2005) Global transcript profiling of primary stems from Arabidopsis thaliana identifies candidate genes for missing links in lignin biosynthesis and transcriptional regulators of fiber differentiation. Plant J 42: 618–640 [DOI] [PubMed] [Google Scholar]
- Faix O. (1991) Classification of lignins from different botanical origins by FT-IR spectroscopy. Holzforschung 45: 21–27 [Google Scholar]
- Fénart S, Ndong Y-P, Duarte J, Rivière N, Wilmer J, van Wuytswinkel O, Lucau A, Cariou E, Neutelings G, Gutierrez L, et al. (2010) Development and validation of a flax (Linum usitatissimum L.) gene expression oligo microarray. BMC Genomics 11: 592. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fornalé S, Shi X, Chai C, Encina A, Irar S, Capellades M, Fuguet E, Torres J-L, Rovira P, Puigdomènech P, et al. (2010) ZmMYB31 directly represses maize lignin genes and redirects the phenylpropanoid metabolic flux. Plant J 64: 633–644 [DOI] [PubMed] [Google Scholar]
- Fornalé S, Sonbol FM, Maes T, Capellades M, Puigdomènech P, Rigau J, Caparrós-Ruiz D. (2006) Down-regulation of the maize and Arabidopsis thaliana caffeic acid O-methyl-transferase genes by two new maize R2R3-MYB transcription factors. Plant Mol Biol 62: 809–823 [DOI] [PubMed] [Google Scholar]
- Gang DR, Kasahara H, Xia Z-Q, Vander Mijnsbrugge K, Bauw G, Boerjan W, Van Montagu M, Davin LB, Lewis NG. (1999) Evolution of plant defense mechanisms: relationships of phenylcoumaran benzylic ether reductases to pinoresinol-lariciresinol and isoflavone reductases. J Biol Chem 274: 7516–7527 [DOI] [PubMed] [Google Scholar]
- Gorshkova T, Morvan C. (2006) Secondary cell-wall assembly in flax phloem fibres: role of galactans. Planta 223: 149–158 [DOI] [PubMed] [Google Scholar]
- Hano C, Addi M, Bensaddek L, Crônier D, Baltora-Rosset S, Doussot J, Maury S, Mesnard F, Chabbert B, Hawkins S, et al. (2006a) Differential accumulation of monolignol-derived compounds in elicited flax (Linum usitatissimum) cell suspension cultures. Planta 223: 975–989 [DOI] [PubMed] [Google Scholar]
- Hano C, Martin I, Fliniaux O, Legrand B, Gutierrez L, Arroo RR, Mesnard F, Lamblin F, Lainé E. (2006b) Pinoresinol-lariciresinol reductase gene expression and secoisolariciresinol diglucoside accumulation in developing flax (Linum usitatissimum) seeds. Planta 224: 1291–1301 [DOI] [PubMed] [Google Scholar]
- Hemmati S, von Heimendahl CB, Klaes M, Alfermann AW, Schmidt TJ, Fuss E. (2010) Pinoresinol-lariciresinol reductases with opposite enantiospecificity determine the enantiomeric composition of lignans in the different organs of Linum usitatissimum L. Planta Med 76: 928–934 [DOI] [PubMed] [Google Scholar]
- Holmborn B, Eckerman C, Eklund P, Hemming J, Nisula L, Reunanen M, Sjöholm R, Sundberg A, Sundberg K, Willför S. (2003) Knots in trees: a new rich source of lignans. Phytochem Rev 2: 331–340 [Google Scholar]
- Irizarry RA, Hobbs B, Collin F, Beazer-Barclay YD, Antonellis KJ, Scherf U, Speed TP. (2003) Exploration, normalization, and summaries of high density oligonucleotide array probe level data. Biostatistics 4: 249–264 [DOI] [PubMed] [Google Scholar]
- Jin H, Cominelli E, Bailey P, Parr A, Mehrtens F, Jones J, Tonelli C, Weisshaar B, Martin C. (2000) Transcriptional repression by AtMYB4 controls production of UV-protecting sunscreens in Arabidopsis. EMBO J 19: 6150–6161 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kacurakova M, Capek P, Sasinkova V, Wellner N, Ebringerova A. (2000) FT-IR study of plant cell wall model compounds: pectic polysaccharides and hemicelluloses. Carbohydr Polym 43: 195–203 [Google Scholar]
- Kaneda M, Schuetz M, Lin BSP, Chanis C, Hamberger B, Western TL, Ehlting J, Samuels AL. (2011) ABC transporters coordinately expressed during lignification of Arabidopsis stems include a set of ABCBs associated with auxin transport. J Exp Bot 62: 2063–2077 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ko J-H, Prassinos C, Han K-H. (2006) Developmental and seasonal expression of PtaHB1, a Populus gene encoding a class III HD-Zip protein, is closely associated with secondary growth and inversely correlated with the level of microRNA (miR166). New Phytol 169: 469–478 [DOI] [PubMed] [Google Scholar]
- Kvavadze E, Bar-Yosef O, Belfer-Cohen A, Boaretto E, Jakeli N, Matskevich Z, Meshveliani T. (2009) 30,000-year-old wild flax fibers. Science 325: 1359–1359 [DOI] [PubMed] [Google Scholar]
- Kwon M, Davin LB, Lewis NG. (2001) In situ hybridization and immunolocalization of lignan reductases in woody tissues: implications for heartwood formation and other forms of vascular tissue preservation. Phytochemistry 57: 899–914 [DOI] [PubMed] [Google Scholar]
- Lapierre C, Monties B, Rolando C, Chirale L. (1985) Thioacidolysis of lignin: comparison with acidolysis. J Wood Chem Technol 5: 277–292 [Google Scholar]
- Legay S, Sivadon P, Blervacq A-S, Pavy N, Baghdady A, Tremblay L, Levasseur C, Ladouce N, Lapierre C, Séguin A, et al. (2010) EgMYB1, an R2R3 MYB transcription factor from eucalyptus negatively regulates secondary cell wall formation in Arabidopsis and poplar. New Phytol 188: 774–786 [DOI] [PubMed] [Google Scholar]
- Leplé JC, Dauwe R, Morreel K, Storme V, Lapierre C, Pollet B, Naumann A, Kang KY, Kim H, Ruel K, et al. (2007) Downregulation of cinnamoyl-coenzyme A reductase in poplar: multiple-level phenotyping reveals effects on cell wall polymer metabolism and structure. Plant Cell 19: 3669–3691 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lu F, Ralph J. (2003) Non-degradative dissolution and acetylation of ball-milled plant cell walls: high-resolution solution-state NMR. Plant J 35: 535–544 [DOI] [PubMed] [Google Scholar]
- Marechal Y, Chanzy H. (2000) The hydrogen bond network in Ibeta cellulose as observed by infrared spectrometry. J Mol Struct 523: 183–196 [Google Scholar]
- McDougall GJ. (1992) Changes in cell wall-associated peroxidases during the lignification of flax fibres. Phytochemistry 31: 3385–3389 [Google Scholar]
- Miao YC, Liu CJ. (2010) ATP-binding cassette-like transporters are involved in the transport of lignin precursors across plasma and vacuolar membranes. Proc Natl Acad Sci USA 107: 22728–22733 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morreel K, Dima O, Kim H, Lu F, Niculaes C, Vanholme R, Dauwe R, Goeminne G, Inzé D, Messens E, et al. (2010a) Mass spectrometry-based sequencing of lignin oligomers. Plant Physiol 153: 1464–1478 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morreel K, Kim H, Lu F, Dima O, Akiyama T, Vanholme R, Niculaes C, Goeminne G, Inzé D, Messens E, et al. (2010b) Mass spectrometry-based fragmentation as an identification tool in lignomics. Anal Chem 82: 8095–8105 [DOI] [PubMed] [Google Scholar]
- Morreel K, Ralph J, Kim H, Lu F, Goeminne G, Ralph S, Messens E, Boerjan W. (2004) Profiling of oligolignols reveals monolignol coupling conditions in lignifying poplar xylem. Plant Physiol 136: 3537–3549 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moss G. (2000) Nomenclature of lignans and neolignans. Pure Appl Chem 72: 1493–1523 [Google Scholar]
- Niedźwiedź-Siegień I. (1998) Cyanogenic glucosides in Linum usitatissimum. Phytochemistry 49: 59–63 [Google Scholar]
- Passardi F, Penel C, Dunand C. (2004) Performing the paradoxical: how plant peroxidases modify the cell wall. Trends Plant Sci 9: 534–540 [DOI] [PubMed] [Google Scholar]
- Paux E, Tamasloukht M, Ladouce N, Sivadon P, Grima-Pettenati J. (2004) Identification of genes preferentially expressed during wood formation in Eucalyptus. Plant Mol Biol 55: 263–280 [DOI] [PubMed] [Google Scholar]
- Prassinos C, Ko JH, Yang J, Han KH. (2005) Transcriptome profiling of vertical stem segments provides insights into the genetic regulation of secondary growth in hybrid aspen trees. Plant Cell Physiol 46: 1213–1225 [DOI] [PubMed] [Google Scholar]
- Prigge MJ, Otsuga D, Alonso JM, Ecker JR, Drews GN, Clark SE. (2005) Class III homeodomain-leucine zipper gene family members have overlapping, antagonistic, and distinct roles in Arabidopsis development. Plant Cell 17: 61–76 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qin YM, Hu CY, Zhu YX. (2008) The ascorbate peroxidase regulated by H(2)O(2) and ethylene is involved in cotton fiber cell elongation by modulating ROS homeostasis. Plant Signal Behav 3: 194–196 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ragupathy R, Rathinavelu R, Cloutier S. (2011) Physical mapping and BAC-end sequence analysis provide initial insights into the flax (Linum usitatissimum L.) genome. BMC Genomics 12: 217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ralph J, Akiyama T, Kim H, Lu F, Schatz PF, Marita JM, Ralph SA, Reddy MS, Chen F, Dixon RA. (2006) Effects of coumarate 3-hydroxylase down-regulation on lignin structure. J Biol Chem 281: 8843–8853 [DOI] [PubMed] [Google Scholar]
- Ramires EC, Megiatto JD, Jr, Gardrat C, Castellan A, Frollini E. (2010) Valorization of an industrial organosolv-sugarcane bagasse lignin: characterization and use as a matrix in biobased composites reinforced with sisal fibers. Biotechnol Bioeng 107: 612–621 [DOI] [PubMed] [Google Scholar]
- Ros Barcelo A. (1998) The generation of H2O2 in the xylem of Zinnia elegans is mediated by an NADPH-oxidase-like enzyme. Planta 207: 207–216 [Google Scholar]
- Shadle G, Chen F, Srinivasa Reddy MS, Jackson L, Nakashima J, Dixon RA. (2007) Down-regulation of hydroxycinnamoyl CoA:shikimate hydroxycinnamoyl transferase in transgenic alfalfa affects lignification, development and forage quality. Phytochemistry 68: 1521–1529 [DOI] [PubMed] [Google Scholar]
- Singh KK, Mridula D, Rehal J, Barnwal P. (2011) Flaxseed: a potential source of food, feed and fiber. Crit Rev Food Sci Nutr 51: 210–222 [DOI] [PubMed] [Google Scholar]
- Sterjiades R, Dean JFD, Gamble G, Himmelsbach DS, Eriksson K-EL. (1993) Extracellular laccases and peroxidases from sycamore maple (Acer pseudoplatanus) cell-suspension cultures. Planta 190: 75–87 [Google Scholar]
- Sterky F, Bhalerao RR, Unneberg P, Segerman B, Nilsson P, Brunner AM, Charbonnel-Campaa L, Lindvall JJ, Tandre K, Strauss SH, et al. (2004) A Populus EST resource for plant functional genomics. Proc Natl Acad Sci USA 101: 13951–13956 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Struijs K, Vincken JP, Doeswijk TG, Voragen AG, Gruppen H. (2009) The chain length of lignan macromolecule from flaxseed hulls is determined by the incorporation of coumaric acid glucosides and ferulic acid glucosides. Phytochemistry 70: 262–269 [DOI] [PubMed] [Google Scholar]
- Summerscales J, Dissanayake N, Virk AS, Hall W. (2010) A review of bast fibres and their composites: I. Fibres as reinforcements. Compos Part A Appl Sci Manuf 41: 1329–1335 [Google Scholar]
- Syrjanen K, Brunow G. (1998) Oxidative cross coupling of p-hydroxycinnamic alcohols with dimeric arylglycerol β-aryl ether lignin model compounds: the effect of oxidation potentials. J Chem Soc Perkin Trans 1: 3425–3430 [Google Scholar]
- Toure A, Xu XM. (2010) Flaxseed lignans: source, biosynthesis, metabolism, antioxidant activity, bio-active components, and health benefits. Compr Rev Food Sci Food Saf 9: 261–269 [DOI] [PubMed] [Google Scholar]
- Touzel JP, Chabbert B, Monties B, Debeire P, Cathala B. (2003) Synthesis and characterization of dehydrogenation polymers in Gluconacetobacter xylinus cellulose and cellulose/pectin composite. J Agric Food Chem 51: 981–986 [DOI] [PubMed] [Google Scholar]
- Tusher VG, Tibshirani R, Chu G. (2001) Significance analysis of microarrays applied to the ionizing radiation response. Proc Natl Acad Sci USA 98: 5116–5121 [DOI] [PMC free article] [PubMed] [Google Scholar]
- van den Broeck HC, Maliepaard C, Ebskamp MJM, Toonen MAJ, Koops AJ. (2008) Differential expression of genes involved in C1 metabolism and lignin biosynthesis in wooden core and bast tissues of fibre hemp (Cannabis sativa L.). Plant Sci 174: 205–220 [Google Scholar]
- Vander Mijnsbrugge K, Beeckman H, De Rycke R, Van Montagu M, Engler G, Boerjan W. (2000) Phenylcoumaran benzylic ether reductase, a prominent poplar xylem protein, is strongly associated with phenylpropanoid biosynthesis in lignifying cells. Planta 211: 502–509 [DOI] [PubMed] [Google Scholar]
- Vanholme R, Morreel K, Ralph J, Boerjan W. (2008) Lignin engineering. Curr Opin Plant Biol 11: 278–285 [DOI] [PubMed] [Google Scholar]
- Vanholme R, Ralph J, Akiyama T, Lu FC, Pazo JR, Kim H, Christensen JH, Van Reusel B, Storme V, De Rycke R, et al. (2010) Engineering traditional monolignols out of lignin by concomitant up-regulation of F5H1 and down-regulation of COMT in Arabidopsis. Plant J 64: 885–897 [DOI] [PubMed] [Google Scholar]
- Venglat P, Xiang D, Qiu S, Stone SL, Tibiche C, Cram D, Alting-Mees M, Nowak J, Cloutier S, Deyholos M, et al. (2011) Gene expression analysis of flax seed development. BMC Plant Biol 11: 74. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wagner A, Ralph J, Akiyama T, Flint H, Phillips L, Torr K, Nanayakkara B, Te Kiri L. (2007) Exploring lignification in conifers by silencing hydroxycinnamoyl-CoA:shikimate hydroxycinnamoyltransferase in Pinus radiata. Proc Natl Acad Sci USA 104: 11856–11861 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weng J-K, Chapple C. (2010) The origin and evolution of lignin biosynthesis. New Phytol 187: 273–285 [DOI] [PubMed] [Google Scholar]
- Westcott N, Muir A. (2003) Flax seed lignan in disease prevention and health promotion. Phytochem Rev 2: 401–417 [Google Scholar]
- Whetten R, Sederoff R. (1995) Lignin biosynthesis. Plant Cell 7: 1001–1013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamaguchi M, Ohtani M, Mitsuda N, Kubo M, Ohme-Takagi M, Fukuda H, Demura T. (2010) VND-INTERACTING2, a NAC domain transcription factor, negatively regulates xylem vessel formation in Arabidopsis. Plant Cell 22: 1249–1263 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yazaki K. (2006) ABC transporters involved in the transport of plant secondary metabolites. FEBS Lett 580: 1183–1191 [DOI] [PubMed] [Google Scholar]
- Zhang J, Elo A, Helariutta Y. (2011) Arabidopsis as a model for wood formation. Curr Opin Biotechnol 22: 293–299 [DOI] [PubMed] [Google Scholar]
- Zhao C, Avci U, Grant EH, Haigler CH, Beers EP. (2008) XND1, a member of the NAC domain family in Arabidopsis thaliana, negatively regulates lignocellulose synthesis and programmed cell death in xylem. Plant J 53: 425–436 [DOI] [PubMed] [Google Scholar]
- Zhao Q, Dixon RA. (2011) Transcriptional networks for lignin biosynthesis: more complex than we thought? Trends Plant Sci 16: 227–233 [DOI] [PubMed] [Google Scholar]
- Zhong R, Lee C, Ye Z-H. (2010) Evolutionary conservation of the transcriptional network regulating secondary cell wall biosynthesis. Trends Plant Sci 15: 625–632 [DOI] [PubMed] [Google Scholar]
- Zhou J, Lee C, Zhong R, Ye ZH. (2009) MYB58 and MYB63 are transcriptional activators of the lignin biosynthetic pathway during secondary cell wall formation in Arabidopsis. Plant Cell 21: 248–266 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.