Abstract
The antiterminator Q gene of bacteriophage 933W (Q933) was identified upstream of the stx2 gene in 90% of human disease–origin Escherichia coli O157:H7 isolates and in 44.5% of bovine isolates. Shiga toxin production was higher in Q933-positive isolates than Q933-negative isolates. This genetic marker may provide a useful molecular tool for epidemiologic studies.
Keywords: E. coli O157, Shiga-toxin production, stx2-encoding phages, dispatch
Escherichia coli O157 is recognized worldwide as an important cause of diarrheal disease, which in some patients is followed by hemolytic uremic syndrome and death (1). A primary virulence factor of this pathogen is the prophage-encoded Shiga toxin (2). Greater Shiga toxin production per bacterium is associated with increasing severity of human disease (3,4). Because of its location in the phage genome, the stx-gene variant dubbed stx2 is under similar regulatory control as other phage late-genes, as it is governed by the interaction of the transcription antiterminator Q with the late promoter PR´ (5).
Although cattle and other ruminants appear to be the natural reservoir for E. coli O157 and other Shiga toxin–producing E. coli (STEC), only a small fraction of STEC serotypes routinely present in cattle are frequently isolated from human patients. Mounting evidence suggests that considerable genetic, phenotypic, and pathogenic diversity exists among these pathogens (6–8). Furthermore, genetic subtypes or lineages of E. coli O157 do not appear to be equally distributed among isolates of bovine and human origin (7). The purpose of this study was to examine the distribution of specific sequences upstream of the stx2 gene among E. coli O157:H7 of human and bovine origin, along with corresponding magnitudes of Shiga toxin production.
The Study
A total of 158 stx2-encoding E. coli O157:H7 isolates were assayed, 91 isolates of bovine origin and 67 originally isolated from ill persons (Tables A1 and A2). All isolates demonstrated unique banding patterns on pulsed-field gel electrophoresis (PFGE). For polymerase chain reaction (PCR) analysis, 5 µL of DNA obtained from boiled stationary-phase bacteria was added to a 50-µL PCR master mix containing a final concentration of 1.5 (Q933) or 2.5 (Q21) mmol MgCl2, 200 µmol/L each deoxynucleoside triphosphate, 1 U Taq polymerase, 0.6 pg/µL of primer 595 (5´-CCGAAGAAAAACCCAGTAACAG-3´) (9), and 0.6 pg/µL of either primer Q933 (5´-CGGAGGGGATTGTTGAAGGC-3´;QStxf) (9) or primer Q21 (5´-GAAATCCTCAATGCCTCGTTG-3´; this study). PCR consisted of an initial denaturation at 94°C for 5 min; 30 cycles of 94°C for 30 s, 52°C (Q933) or 55°C (Q21) for 1 min, and 72°C for 1 min; and a final 10-min extension step at 72°C. E. coli strain 933 or FAHRP88 was used as a positive control and master mix alone as a negative control. All PCR products were separated by gel electrophoresis (100 V) in 1% agarose gels, stained with ethidium bromide, and visualized by using UV illumination.
Shiga toxin production was determined by using a commercially available enzyme-linked immunosorbent assay (ELISA) kit (Premiere EHEC, Meridian Diagnostics, Cincinnati, OH). Briefly, log-phase cells from Luria-Bertani broth enrichments were diluted to 0.6 optical density (OD) at 600 nm, subsequently pelleted, resuspended in phosphate-buffered saline, and induced by exposure to UV light (240 nm) for 3 s (10). A 1:9 volume of a 10x concentrate of brain heart infusion broth was added to each culture and shaken at 37°C for 2.5 h. Replicate cultures that were not exposed to UV light (noninduced controls) were maintained at 4°C. Two hundred microliters of each induced and noninduced enrichment was subsequently used as the specimen in the EHEC ELISA, as described (11). OD results were recorded for each isolate both with and without UV induction. The relative change in Shiga toxin production after induction was calculated for each isolate; (ODinduced)/ODnoninduced). E. coli O157 (EDL933) and a toxin-negative control isolate were assayed as positive and negative controls each time the assay was repeated.
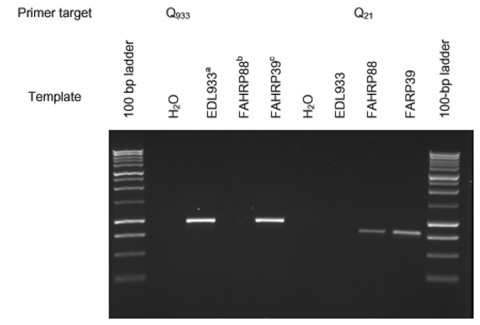
E. coli O157 isolates were classified on the basis of the presence or absence of bands of the predicted size on the Q933-595 and Q21-595 PCR reactions (Figure). A chi-square test was used to determine whether different PCR genotypes were equally distributed among isolates of bovine and human origin. Likewise, a chi-square test was used to assess the equality of distribution of PCR genotypes among bovine isolates from different countries. One-way analysis of variance for nonparametric data (Kruskal-Wallis test) was used to identify differences in ranked-transformed toxin production among noninduced and induced E. coli O157 isolates as well as to determine significant differences in the percent increase in toxin following induction.
Figure.
Ethidium bromide–stained gel of the amplification products obtained from Q933-595 and Q21-595 polymerase chain reactions. aEDL933, human isolate (ATCC43895). Obtained from the STEC Center, Michigan State University. bFAHRP88, isolated from Ohio dairy cow. cFAHRP39, human isolate (E29962) (12).
Previously, Kim et al. described a nonrandom distribution of E. coli O157 subtypes among cattle and humans by using an octamer-based genome-scanning method (7). We tested several of the isolates that had been previously characterized. Nine had been previously identified as belonging to the lineage I genotype and seven isolates as belonging to the lineage II genotype. We found that all nine lineage I isolates consistently amplified the Q933 target, regardless of species of origin. All four bovine isolates classified as lineage II by Kim et al. amplified the Q21 target. One lineage II human isolate (NE015) amplified the Q933 target, and another lineage II isolate (NE037) produced no amplicons in either PCR reaction. One human isolate classified as lineage II (ATCC 43889) amplified both target sequences, presumably because of polylysogeny.
The distribution of the specific Q-gene alleles found upstream of the prophage stx region among bovine isolates may have a geographic component. The distribution of E. coli O157 phage genotypes collected from healthy cattle from diverse geographic areas is consistent with the variable incidences of human disease in different countries (Table 1). For example, six (75%) of eight Scottish bovine isolates examined amplified the Q933 target, the same target that is frequently present in human isolates of human disease origin. Scotland reports some of the highest incidence rates of human E. coli O157–related diseases and hemolytic uremic syndrome (13). In contrast, none of the seven Australian E. coli O157 bovine isolates amplified the 1750-bp fragment. Contrary to the situation in Scotland and the United States, E. coli O157 infection of humans is rarely reported in Australia (14).
Table 1. Distribution of polymerase chain reaction results from bovine Escherichia coli O157 isolates based on geographic origina.
| Country of origin | No. tested | Q allele | ||
|---|---|---|---|---|
| 933 | 21 | Both | ||
| N (%) | N (%) | N (%) | ||
| USA | 46 | 20 (44) | 25 (54) | 1 (2) |
| Scotland | 8 | – (0) | 2 (25) | 6 (75) |
| Australia | 7 | – (0) | 7 (100) | – (0) |
| Japan | 17 | 3 (18) | 14 (82) | – (0) |
| Total | 78 | 23 (29) | 48 (62) | 7 (9) |
a–, not detected. Percentages are read across rows, not down columns. Significant difference in proportion of Q alleles isolated from different countries (p < 0.05, chi-square test for homogeneity).
Conclusions
The Q933 gene target was more commonly identified in human disease–associated strains of E. coli O157 than from strains of bovine origin. Amplification of the Q933 target, either alone or in combination with amplification of the Q21 target from the same isolate, was identified in 60 (9%) of 66 (55/66 alone and 5/66 in combination with Q21; 1 isolate amplified neither target) compared to 40 (44%) of 91 (32/91 alone, and 8/91 in combination with Q21) of bovine isolates (p < 0.001). Furthermore, these genetic subtypes were nonrandomly distributed among the E. coli O157 isolates of bovine origin obtained from different countries (p < 0.05) (Table 1).
These limited data suggest that the distribution of E. coli O157 strains in cattle may differ between countries or regions, thereby providing an explanation for geographic differences in the incidence of human E. coli O157 infection. More isolates from cattle need to be analyzed with these methods to better characterize the E. coli O157 in the bovine reservoir of each country.
A positive reaction with the Q933 target was significantly associated with higher OD results on the Shiga toxin ELISA (both noninduced and induced) and higher-fold increases in toxin production following induction than isolates amplifying the Q21 target alone (p < 0.0001) (Table 2). Despite these differences, we did not identify any clinical associations between the magnitude of Shiga toxin production and severity of human disease could be identified in this study. Other, non–Shiga toxin–related virulence factors and host susceptibility are also believed to play essential roles in the outcome of clinical STEC infections. The Q933-negative isolates obtained from human disease might have lost this Q933-containing prophage by the time of isolation, or these isolates might have been recovered from patients also infected with STEC containing Q933-type prophage (15). Whether specific Q-gene alleles directly correlate with the magnitude of Shiga-toxin production or whether other (unstudied) factors within the phage lytic cascade genetically linked to specific Q alleles instead are responsible for the magnitude of toxin production is not known.
Table 2. Shiga toxin production by Escherichia coli O157:H7 by Q allele.
| Assay | Q allele | Response | ||
| Median | Minimum | Maximum | ||
| OD600nm noninduced | Q 933 | 0.442 | 0.153 | 2.814 |
| Q 21 | 0.170 | 0.120 | 0.413 | |
| OD600nm induced | Q 933 | 1.228 | 0.172 | 2.896 |
| Q 21 | 0.165 | 0.084 | 1.210 | |
| Fold increase in OD600nm after inductiona | Q 933 | 2.2 | 0.3 | 7.7 |
| Q 21 | 0.9 | 0.4 | 5.1 | |
a(ODinduced)/(ODnoninduced). The maximum and minimum optical density readings at 600 nm listed in each row are not necessarily from the same isolate; therefore, the maximum- and minimum-fold increase cannot be calculated directly from the table.
The antiterminator Q, the protein product of the Q gene, and PR´, the late promoter, are reputed to be involved in regulating phage late-genes and, because of the location of PR´ in prophage genome, of Shiga toxin production as well (5). In E. coli O157 phage 933W (GenBank no. 9632466) and E. coli O157 stx2vhd (GenBank no. 15718404), the 359-bp sequence immediately upstream of the stx2 gene is nearly identical (>95% nucleotide identity). However, further upstream of this area of identity, DNA sequences differ significantly. In E. coli O157 933W, this gene is identified as the antiterminator Q gene. In contrast, in E. coli O157 stx2vhd this area is occupied by a gene with >95% sequence identity with the antiterminator Q gene of bacteriophage 21 (gi 4539472). The Q gene of bacteriophage 21 does not share DNA sequence homology with the Q gene of bacteriophage 933W, and only 36% predicted amino acid homology. Since the Q gene is reputed to play an important role in regulating toxin production, our results provide a plausible explanation (differential regulation of Shiga toxin production) of why certain E. coli O157 genotypes are more commonly isolated from human patients (7).
Acknowledgments
We thank the state departments of health of Ohio, Washington, and Idaho for many of the human isolates used in this study and the SETC Center, Michigan State University, for providing two of the strains we tested.
This project was funded by beef and veal producers and importers through their $1-per-head checkoff and was produced for the Cattlemen's Beef Board and state beef councils by the National Cattlemen's Beef Association. Research in S.S. and J.T.L. laboratories is also supported by state and federal funds appropriated to the Ohio Agricultural Research and Development Center.
Biography
Dr. LeJeune is an assistant professor in the Food Animal Health Research Program, in the Department of Veterinary Preventive Medicine, Ohio State University. His research interests include the epidemiology and ecology of foodborne pathogens in the preharvest stages of food production.
Table A1. Source of human isolates used in this studya.
| FAHRP ID | Source ID | Country | Year | Clinical signs and symptoms | References or source |
|---|---|---|---|---|---|
| 6 | FRIK 528 | USA | 1998 | Diarrhea | 16 |
| 7 | FRIK 579 | USA | 1998 | Diarrhea | 16 |
| 8 | 93-001 | USA | 1999 | Hemorrhagic colitis | 17 |
| 9 | ATCC 35150 | USA | 1999 | Hemorrhagic colitis | 17 |
| 16 | 91671 | USA | 1999 | Hemorrhagic colitis | 17 |
| 17 | ATCC 43889 | USA | 1999 | Hemorrhagic colitis | 17 |
| 18 | NE 037 | USA | 1999 | Hemorrhagic colitis | 17 |
| 19 | NE 15 | USA | 1999 | Hemorrhagic colitis | 17 |
| 39 | E29962 | UK | 1991 | NR | 18 |
| 54 | CL56 | Canada | 1991 | NR | 18 |
| 60 | E32511 | USA | 2002 | HUS | 19 |
| 58 | EDL933 | USA | 1982 | Hemorrhagic colitis | 20 |
| 126 | 02 5225 | USA | 2002 | NR | Washingtonb |
| 127 | 02 4857 | USA | 2002 | NR | Washington |
| 128 | 02 6776 | USA | 2002 | NR | Washington |
| 129 | 02 6579 | USA | 2002 | NR | Washington |
| 130 | 02 6546 | USA | 2002 | NR | Washington |
| 131 | 02 6722 | USA | 2002 | NR | Washington |
| 132 | 02 6598 | USA | 2002 | NR | Washington |
| 133 | 02 6696 | USA | 2002 | NR | Washington |
| 134 | 02 6791 | USA | 2002 | NR | Washington |
| 135 | 02 6829 | USA | 2002 | NR | Washington |
| 136 | 02 6755 | USA | 2002 | NR | Washington |
| 137 | 02 6644 | USA | 2002 | NR | Washington |
| 138 | 06 781 | USA | 2002 | Diarrhea | Idahoc |
| 139 | 06 852 | USA | 2002 | NR | Idaho |
| 140 | 06 854 | USA | 2002 | Watery diarrhea, vomiting | Idaho |
| 141 | 06 856 | USA | 2002 | Diarrhea | Idaho |
| 142 | 06 855 | USA | 2002 | NR | |
| 143 | 06 886 | USA | 2002 | Diarrhea, abdominal pain | Idaho |
| 144 | 06 889 | USA | 2002 | Abdominal pain | Idaho |
| 145 | 06 988 | USA | 2002 | Gastrointestinal bleeding | Idaho |
| 146 | 07 004 | USA | 2002 | Bloody stool | Idaho |
| 147 | 07 007 | USA | 2002 | Bloody stool | Idaho |
| 148 | 07 023 | USA | 2002 | Bloody stool | Idaho |
| 149 | 07 085 | USA | 2002 | NR | Idaho |
| 150 | 07 147 | USA | 2002 | NR | Idaho |
| 151 | 07 154 | USA | 2002 | NR | Idaho |
| 152 | O2191230 | USA | 2002 | Diarrhea | Ohiod |
| 153 | O2191229 | USA | 2002 | Diarrhea | Ohio |
| 154 | O2191231 | USA | 2002 | Diarrhea | Ohio |
| 155 | O2191294 | USA | 2002 | Diarrhea | Ohio |
| 156 | O2190819 | USA | 2002 | Diarrhea | Ohio |
| 157 | O2190864 | USA | 2002 | Diarrhea | Ohio |
| 158 | O2191309 | USA | 2002 | Diarrhea | Ohio |
| 159 | O2191311 | USA | 2002 | Diarrhea | Ohio |
| 160 | O2191313 | USA | 2002 | Diarrhea | Ohio |
| 161 | O2191361 | USA | 2002 | Diarrhea | Ohio |
| 162 | O2191602 | USA | 2002 | Diarrhea | Ohio |
| 163 | O2191624 | USA | 2002 | Diarrhea | Ohio |
| 164 | O2191541 | USA | 2002 | Diarrhea | Ohio |
| 165 | O2191546 | USA | 2002 | Diarrhea | Ohio |
| 166 | O2191423 | USA | 2002 | Diarrhea | Ohio |
| 167 | O2191509 | USA | 2002 | Diarrhea | Ohio |
| 168 | O2191363 | USA | 2002 | Diarrhea | Ohio |
| 169 | O2191364 | USA | 2002 | Diarrhea | Ohio |
| 170 | O2191365 | USA | 2002 | Diarrhea | Ohio |
| 171 | O2191366 | USA | 2002 | Diarrhea | Ohio |
| 172 | O2190889 | USA | 2002 | Diarrhea | Ohio |
| 173 | O2190893 | USA | 2002 | Diarrhea | Ohio |
| 174 | O2191176 | USA | 2002 | Diarrhea | Ohio |
| 175 | O2191177 | USA | 2002 | Diarrhea | Ohio |
| 176 | O2191623 | USA | 2002 | Diarrhea | Ohio |
| 177 | O2191625 | USA | 2002 | Diarrhea | Ohio |
| 178 | O2191645 | USA | 2002 | Diarrhea | Ohio |
| 179 | O2191675 | USA | 2002 | Diarrhea | Ohio |
| 180 | O2191765 | USA | 2002 | Diarrhea | Ohio |
| 181 | O2191831 | USA | 2002 | Diarrhea | Ohio |
aFAHRP, Food Animal Health Research Program, Ohio State University; NR, not reported; HUS, hemolytic uremic syndrome. bWashington State Department of Health isolates. cIdaho Department of Health and Welfare isolates. dOhio Department of Health isolates.
Table A2. Source of bovine isolates used in this study.
| FAHRPa ID | Source ID | Country | Year | References or source |
|---|---|---|---|---|
| 1 | FRIK 1986 | USA | 1991 | 21 |
| 2 | FRIK 1997 | USA | 1991 | 21 |
| 3 | FRIK 1994 | USA | 1991 | 21 |
| 4 | FRIK 2002 | USA | 1991 | 21 |
| 5 | FRIK 1987 | USA | 1991 | 21 |
| 10 | FRIK 920 | USA | 1998 | 22 |
| 11 | FRIK 1054 | USA | 1998 | 22 |
| 12 | FRIK 1540 | USA | 1998 | 22 |
| 13 | FRIK 1988 | USA | 1998 | 21 |
| 22 | LCDC 87-2930 | Canada | 1991 | 23 |
| 27 | OARDC1 | USA | 2002 | FAHRP |
| 29 | OARDC2 | USA | 2002 | FAHRP |
| 31 | OARDC3 | USA | 2002 | FAHRP |
| 35 | P673 | UK | 1987 | 24 |
| 37 | P277 | UK | 1987 | 24 |
| 47 | c1526-77 | Argentina | 1991 | 23 |
| 50 | CDC B9253-DMS1 | USA | 1991 | 23 |
| 51 | A39 | Canada | 1991 | 23 |
| 52 | A43 | Canada | 1991 | 23 |
| 56 | LCDC 87-2924 | Canada | 1991 | 23 |
| 57 | LCDC 87-1799 | Canada | 1991 | 23 |
| 62 | CDC B6830-MS1/0 | USA | 1991 | 23 |
| 63 | CDCB7205-MS1/0 | USA | 1991 | 23 |
| 64 | CDC B8038-MS1/0 | USA | 1991 | 23 |
| 65 | 8832 | USA | 2002 | 25 |
| 66 | EC66 | USA | 2002 | FAHRP |
| 67 | EC 67 | USA | 2002 | FAHRP |
| 82 | 8833 | USA | 2002 | 25 |
| 83 | EC 83 | USA | 2002 | FAHRP |
| 84 | EC 84 | USA | 2002 | FAHRP |
| 85 | 8834 | USA | 2002 | 25 |
| 87 | EC87 | USA | 2002 | FAHRP |
| 88 | EC88 | USA | 2002 | FAHRP |
| 93 | EC 93 | USA | 2002 | FAHRP |
| 94 | EC94 | USA | 2002 | FAHRP |
| 95 | EC95 | USA | 2002 | FAHRP |
| 96 | EC96 | USA | 2002 | FAHRP |
| 97 | EC97 | USA | 2002 | FAHRP |
| 98 | EC98 | USA | 2002 | FAHRP |
| 99 | EC99 | USA | 2002 | FAHRP |
| 100 | EC100 | USA | 2002 | FAHRP |
| 102 | EC102 | USA | 2002 | FAHRP |
| 103 | EC103 | USA | 2002 | FAHRP |
| 104 | EC104 | USA | 2002 | FAHRP |
| 113 | 8837 | USA | 2002 | 25 |
| 115 | EC115 | USA | 2002 | FAHRP |
| 116 | EC116 | USA | 2002 | FAHRP |
| 117 | EC117 | USA | 2002 | FAHRP |
| 120 | EC120 | USA | 2002 | FAHRP |
| 122 | EC122 | USA | 2002 | FAHRP |
| 182 | 757 | USA | 1994 | 25 |
| 183 | 817 | USA | 1994 | 25 |
| 185 | 1104 | USA | 1994 | 25 |
| 186 | 1119 | USA | 1994 | 25 |
| 187 | 1124 | USA | 1994 | 25 |
| 188 | 1136 | USA | 1994 | 25 |
| 189 | 1273 | USA | 1994 | 25 |
| 190 | 3735 | USA | 1996 | 25 |
| 191 | 4048 | USA | 1996 | 25 |
| 192 | 7407 | Japan | 1996 | 25 |
| 193 | 7409 | Japan | 1996 | 25 |
| 194 | 7416 | Japan | 1996 | 25 |
| 195 | 7420 | Japan | 1996 | 25 |
| 196 | 7421 | Japan | 1996 | 25 |
| 197 | 7423 | Japan | 1996 | 25 |
| 198 | 7433 | Japan | 1996 | 25 |
| 199 | 7436 | Japan | 1996 | 25 |
| 200 | 7439 | Japan | 1996 | 25 |
| 201 | 7460 | Japan | 1996 | 25 |
| 202 | 7469 | Japan | 1996 | 25 |
| 203 | 7478 | Japan | 1996 | 25 |
| 204 | 7484 | Japan | 1996 | 25 |
| 205 | 7488 | Japan | 1996 | 25 |
| 206 | 7495 | Japan | 1996 | 25 |
| 207 | 7500 | Japan | 1996 | 25 |
| 208 | 7505 | Japan | 1996 | 25 |
| 209 | 7622 | Scotland | 1996 | 25 |
| 210 | 7630 | Scotland | 1999 | 25 |
| 211 | 7632 | Scotland | 1999 | 25 |
| 213 | 7637 | Scotland | 1999 | 25 |
| 214 | 7638 | Scotland | 1999 | 25 |
| 217 | 7648 | Scotland | 1999 | 25 |
| 218 | 7649 | Scotland | 1999 | 25 |
| 219 | 7653 | Scotland | 1999 | 25 |
| 220 | 8176 | Australia | 1999 | 25 |
| 221 | 8177 | Australia | 1996 | 25 |
| 222 | 8179 | Australia | 1997 | 25 |
| 223 | 8182 | Australia | 1997 | 25 |
| 224 | 8183 | Australia | 1997 | 25 |
| 225 | 8184 | Australia | 1998 | 25 |
| 226 | 8185 | Australia | 1999 | 25 |
aFAHRP, Food Animal Health Research Program, Ohio State University.
Footnotes
Suggested citation for this article: LeJeune JT, Abedon ST, Takemura K, Christie NP, Sreevatsan S. Human Escherichia coli O157:H7 genetic marker in isolates of bovine origin. Emerg Infect Dis [serial on the Internet]. 2004 Aug [date cited]. http://dx.doi.org/10.3201/eid1008.030784
References
- 1.Karmali MA, Arbus GS, Petric M, Patrick ML, Roscoe M, Shaw J, et al. Hospital-acquired Escherichia coli O157:H7 associated haemolytic uraemic syndrome in a nurse [letter]. Lancet. 1988;1:526. 10.1016/S0140-6736(88)91310-4 [DOI] [PubMed] [Google Scholar]
- 2.O'Loughlin EV, Robins-Browne RM. Effect of Shiga toxin and Shiga-like toxins on eukaryotic cells. Microbes Infect. 2001;3:493–507. 10.1016/S1286-4579(01)01405-8 [DOI] [PubMed] [Google Scholar]
- 3.Kimura N, Watanabe M, Komatsubara A. [Verotoxin producing ability of verotoxin-producing Escherichia coli strains isolated from fecal specimens of healthy persons is lower than that of patients]. Kansenshogaku Zasshi. 2000;74:849–51. [DOI] [PubMed] [Google Scholar]
- 4.Pradel N, Boukhors K, Bertin Y, Forestier C, Martin C, Livrelli V. Heterogeneity of Shiga toxin-producing Escherichia coli strains isolated from hemolytic-uremic syndrome patients, cattle, and food samples in central France. Appl Environ Microbiol. 2001;67:2460–8. 10.1128/AEM.67.6.2460-2468.2001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Wagner PL, Neely MN, Zhang X, Acheson DW, Waldor MK, Friedman DI. Role for a phage promoter in Shiga toxin 2 expression from a pathogenic Escherichia coli strain. J Bacteriol. 2001;183:2081–5. 10.1128/JB.183.6.2081-2085.2001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Baker DR, Moxley RA, Francis DH. Variation in virulence in the gnotobiotic pig model of O157:H7 Escherichia coli strains of bovine and human origin. Adv Exp Med Biol. 1997;412:53–8. [DOI] [PubMed] [Google Scholar]
- 7.Kim J, Nietfeldt J, Benson AK. Octamer-based genome scanning distinguishes a unique subpopulation of Escherichia coli O157:H7 strains in cattle. Proc Natl Acad Sci U S A. 1999;96:13288–93. 10.1073/pnas.96.23.13288 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.McNally A, Roe AJ, Simpson S, Thomson-Carter FM, Hoey DE, Currie C, et al. Differences in levels of secreted locus of enterocyte effacement proteins between human disease-associated and bovine Escherichia coli O157. Infect Immun. 2001;69:5107–14. 10.1128/IAI.69.8.5107-5114.2001 [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 9.Unkmeir A, Schmidt H. Structural analysis of phage-borne stx genes and their flanking sequences in shiga toxin-producing Escherichia coli and Shigella dysenteriae type 1 strains. Infect Immun. 2000;68:4856–64. 10.1128/IAI.68.9.4856-4864.2000 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Arber W, Enquist L, Hohn B, Murray K, Murray N. Experimental methods. In: Hendrix R, Roberts J, Stahl F, Weisberg R, editors. Lambda II. Cold Springs Harbor (NY): Cold Spring Harbor Laboratory; 1983. [Google Scholar]
- 11.Schmidt H, Scheef J, Morabito S, Caprioli A, Wieler LH, Karch H. A new Shiga toxin 2 variant (Stx2f) from Escherichia coli isolated from pigeons. Appl Environ Microbiol. 2000;66:1205–8. 10.1128/AEM.66.3.1205-1208.2000 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Dorn CR, Angrick E. Serotype O157:H7 Escherichia coli from bovine and meat sources. J Clin Microbiol. 1991;29:1225–31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Locking M. HUS rates in Scotland. Glasgow: Scottish Centre for Infection & Environmental Health; 2002. [Google Scholar]
- 14.Elliott EJ, Robins-Browne RM, O'Loughlin EV, Bennett-Wood V, Bourke J, Henning P, et al. Nationwide study of haemolytic uraemic syndrome: clinical, microbiological, and epidemiological features. Arch Dis Child. 2001;85:125–31. 10.1136/adc.85.2.125 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Karch H, Russmann H, Schmidt H, Schwarzkopf A, Heesemann J. Long-term shedding and clonal turnover of enterohemorrhagic Escherichia coli O157 in diarrheal diseases. J Clin Microbiol. 1995;33:1602–5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Gouveia S, Proctor ME, Lee M-S, Luchansky JB, Kaspar CW. Genomic comparisons and Shiga toxin production among Escherichia coli O157:H7 Isolates from a day care center outbreak and sporadic cases in southeastern Wisconsin. J Clin Microbiol. 1998;36:727–33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Kim J, Nietfeldt J, Benson A. Octamer-based genome scanning distinguishes a unique subpopulation of Escherichia coli O157:H7 strains in cattle. Proc Natl Acad Sci U S A. 1999;96:13288–93. 10.1073/pnas.96.23.13288 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Dorn CR, Angrick E. Serotype O157:H7 Escherichia coli from bovine and meat sources. J Clin Microbiol. 1991;29:1225–31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Schmitt CK, McKee ML, O'Brien AD. Two copies of Shiga-like toxin II-related genes common in enterohemorrhagic Escherichia coli strains are responsible for the antigenic heterogeneity of the O157:H- strain E32511. [Obtained from the STEC Center, Michigan State University.]. Infect Immun. 1991;59:1065–73. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Perna NT, Mayhew GF, Posfai G, Elliott S, Donnenberg MS, Kaper JB, et al. Genome sequence of enterohemorrhagic Escherichia coli O157:H7. Nature. 2001;409:529–33. 10.1038/35054089 [DOI] [PubMed] [Google Scholar]
- 21.Kim J, Nietfeldt J, Benson A. Octamer-based genome scanning distinguishes a unique subpopulation of Escherichia coli O157:H7 strains in cattle. Proc Natl Acad Sci U S A. 1999;96:13288–93. 10.1073/pnas.96.23.13288 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Shere JA, Bartlett KJ, Kaspar CW. Longitudinal study of Escherichia coli O157:H7 dissemination on four dairy farms in Wisconsin. Appl Environ Microbiol. 1998;64:1390–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Dorn CR, Angrick E. Serotype O157:H7 Escherichia coli from bovine and meat sources. J Clin Microbiol. 1991;29:1225–31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Huck LG, Dorn CR, Angrick EJ. DNA probe for detection of serogroup O157 enterohemorrhagic Escherichia coli. Int J Food Microbiol. 1995;25:277–87. 10.1016/0168-1605(94)00084-J [DOI] [PubMed] [Google Scholar]
- 25.Davis MA, Hancock DD, Besser TE, Rice DH, Hovde CJ, Digiacomo R, et al. Correlation between geographic distance and genetic similarity in an international collection of bovine faecal Escherichia coli O157:H7 isolates. Epidemiol Infect. 2003;131:923–30. 10.1017/S0950268803008884 [DOI] [PMC free article] [PubMed] [Google Scholar]