Abstract
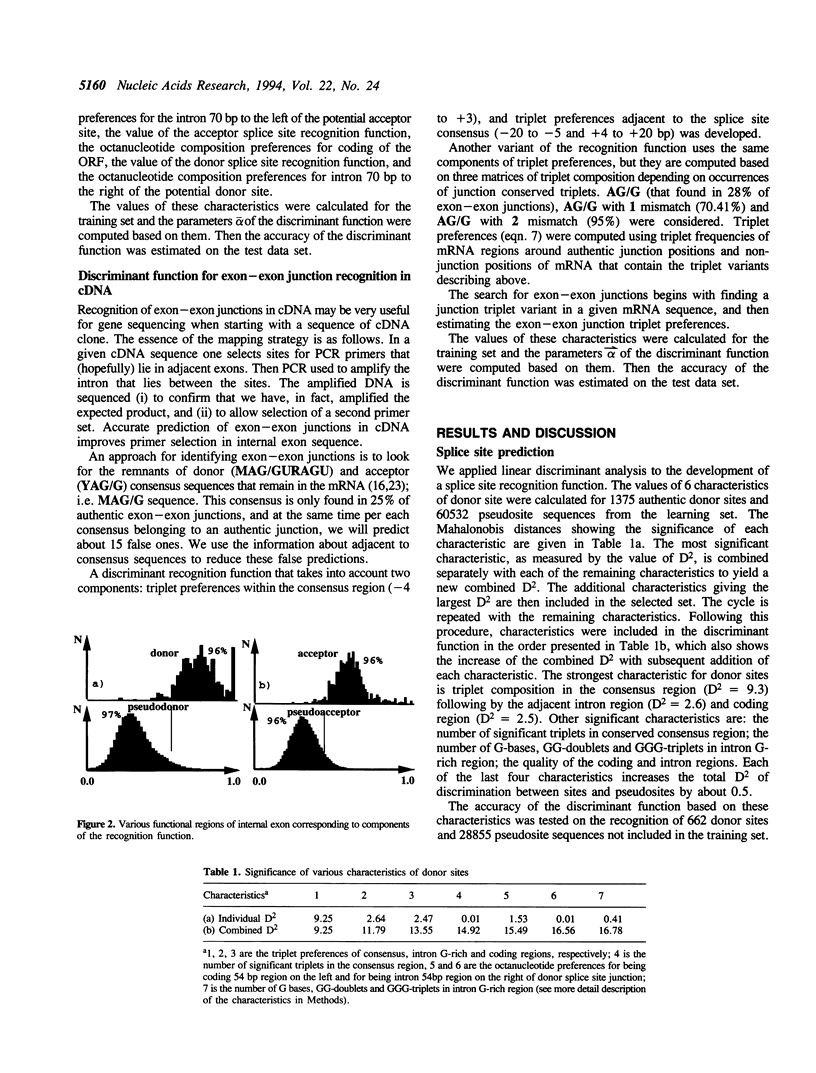
A new method which predicts internal exon sequences in human DNA has been developed. The method is based on a splice site prediction algorithm that uses the linear discriminant function to combine information about significant triplet frequencies of various functional parts of splice site regions and preferences of oligonucleotides in protein coding and intron regions. The accuracy of our splice site recognition function is 97% for donor splice sites and 96% for acceptor splice sites. For exon prediction, we combine in a discriminant function the characteristics describing the 5'-intron region, donor splice site, coding region, acceptor splice site and 3'-intron region for each open reading frame flanked by GT and AG base pairs. The accuracy of precise internal exon recognition on a test set of 451 exon and 246693 pseudoexon sequences is 77% with a specificity of 79%. The recognition quality computed at the level of individual nucleotides is 89% for exon sequences and 98% for intron sequences. This corresponds to a correlation coefficient for exon prediction of 0.87. The precision of this approach is better than other methods and has been tested on a larger data set. We have also developed a means for predicting exon-exon junctions in cDNA sequences, which can be useful for selecting optimal PCR primers.
Full text
PDF






Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Brunak S., Engelbrecht J., Knudsen S. Prediction of human mRNA donor and acceptor sites from the DNA sequence. J Mol Biol. 1991 Jul 5;220(1):49–65. doi: 10.1016/0022-2836(91)90380-o. [DOI] [PubMed] [Google Scholar]
- Cinkosky M. J., Fickett J. W., Gilna P., Burks C. Electronic data publishing and GenBank. Science. 1991 May 31;252(5010):1273–1277. doi: 10.1126/science.1925538. [DOI] [PubMed] [Google Scholar]
- Farber R., Lapedes A., Sirotkin K. Determination of eukaryotic protein coding regions using neural networks and information theory. J Mol Biol. 1992 Jul 20;226(2):471–479. doi: 10.1016/0022-2836(92)90961-i. [DOI] [PubMed] [Google Scholar]
- Fickett J. W., Tung C. S. Assessment of protein coding measures. Nucleic Acids Res. 1992 Dec 25;20(24):6441–6450. doi: 10.1093/nar/20.24.6441. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fields C. A., Soderlund C. A. gm: a practical tool for automating DNA sequence analysis. Comput Appl Biosci. 1990 Jul;6(3):263–270. doi: 10.1093/bioinformatics/6.3.263. [DOI] [PubMed] [Google Scholar]
- Guigó R., Knudsen S., Drake N., Smith T. Prediction of gene structure. J Mol Biol. 1992 Jul 5;226(1):141–157. doi: 10.1016/0022-2836(92)90130-c. [DOI] [PubMed] [Google Scholar]
- Hutchinson G. B., Hayden M. R. The prediction of exons through an analysis of spliceable open reading frames. Nucleic Acids Res. 1992 Jul 11;20(13):3453–3462. doi: 10.1093/nar/20.13.3453. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lawrence C. B., Solovyev V. V. Assignment of position-specific error probability to primary DNA sequence data. Nucleic Acids Res. 1994 Apr 11;22(7):1272–1280. doi: 10.1093/nar/22.7.1272. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matthews B. W. Comparison of the predicted and observed secondary structure of T4 phage lysozyme. Biochim Biophys Acta. 1975 Oct 20;405(2):442–451. doi: 10.1016/0005-2795(75)90109-9. [DOI] [PubMed] [Google Scholar]
- Nakata K., Kanehisa M., DeLisi C. Prediction of splice junctions in mRNA sequences. Nucleic Acids Res. 1985 Jul 25;13(14):5327–5340. doi: 10.1093/nar/13.14.5327. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Penotti F. E. Human pre-mRNA splicing signals. J Theor Biol. 1991 Jun 7;150(3):385–420. doi: 10.1016/s0022-5193(05)80436-9. [DOI] [PubMed] [Google Scholar]
- Senapathy P., Shapiro M. B., Harris N. L. Splice junctions, branch point sites, and exons: sequence statistics, identification, and applications to genome project. Methods Enzymol. 1990;183:252–278. doi: 10.1016/0076-6879(90)83018-5. [DOI] [PubMed] [Google Scholar]
- Snyder E. E., Stormo G. D. Identification of coding regions in genomic DNA sequences: an application of dynamic programming and neural networks. Nucleic Acids Res. 1993 Feb 11;21(3):607–613. doi: 10.1093/nar/21.3.607. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Staden R. Finding protein coding regions in genomic sequences. Methods Enzymol. 1990;183:163–180. doi: 10.1016/0076-6879(90)83012-x. [DOI] [PubMed] [Google Scholar]
- Stephens R. M., Schneider T. D. Features of spliceosome evolution and function inferred from an analysis of the information at human splice sites. J Mol Biol. 1992 Dec 20;228(4):1124–1136. doi: 10.1016/0022-2836(92)90320-j. [DOI] [PubMed] [Google Scholar]
- Uberbacher E. C., Mural R. J. Locating protein-coding regions in human DNA sequences by a multiple sensor-neural network approach. Proc Natl Acad Sci U S A. 1991 Dec 15;88(24):11261–11265. doi: 10.1073/pnas.88.24.11261. [DOI] [PMC free article] [PubMed] [Google Scholar]



