Summary
A growing body of evidence indicates that serine/threonine kinases (STK) and phosphatases (STP) regulate gene expression in prokaryotic organisms. As prokaryotic STKs and STPs are not DNA binding proteins, regulation of gene expression is accomplished through post-translational modification of their targets. These include two-component response regulators, DNA binding proteins and proteins that mediate transcription and translation. This review summarizes our current understanding of how STKs and STPs mediate gene expression in prokaryotes. Further studies to identify environmental signals that trigger the signaling cascade and elucidation of mechanisms that regulate cross-talk between eukaryotic-like signaling enzymes, two-component systems, and components of the transcriptional and translational machinery will facilitate a greater understanding of prokaryotic gene regulation.
Introduction
Coordinated gene expression is critical for all living organisms to adapt to their environment. In prokaryotes, timely expression of factors important for environmental adaptation was thought to be primarily achieved by the action of two-component systems (TCSs) consisting of sensor histidine kinases (HKs) and cognate DNA binding response regulators (RRs) (for reviews see [1–3]). However, recent studies have shown that prokaryotes encode signaling enzymes commonly found in eukaryotes and include serine/threonine kinases (STKs) and phosphatases (STPs) (for recent reviews see [4,5]). Analysis of mutants that lack these signaling enzymes has facilitated our understanding of how serine/threonine kinases and phosphatases contribute to regulation of gene expression in prokaryotes. STK and STP mediated gene expression is important for cellular processes such as growth, virulence, antibiotic resistance and secondary metabolite production. Although they are not DNA binding proteins, STK and STPs mediate prokaryotic gene expression through post-translational modification of a variety of targets, including two-component response regulators or critical components of prokaryotic transcriptional and translational machinery. This review summarizes how reversible phosphorylation by STKs and STPs facilitate prokaryotic gene expression.
Interaction with response regulators and transcription factors
Cross-talk between eukaryotic-like serine/threonine kinases (STKs) and phosphatases (STPs) with two-component systems (TCSs) is known to occur in a number of prokaryotes [6–13]. Post-translational modifications of TCSs by STKs affect virulence, iron transport, antibiotic production and antibiotic resistance [6–13]. In some prokaryotes, it is thought that the membrane-associated STK senses external signals and regulates the DNA-binding activity of response regulators (RR) (see Figure 1A). This mechanism provides an additional level of control of two-component mediated gene expression and increases the versatility of the organism for environmental adaptation. Below, we have included examples of how STK mediated post-translational modification of response regulators affects gene expression.
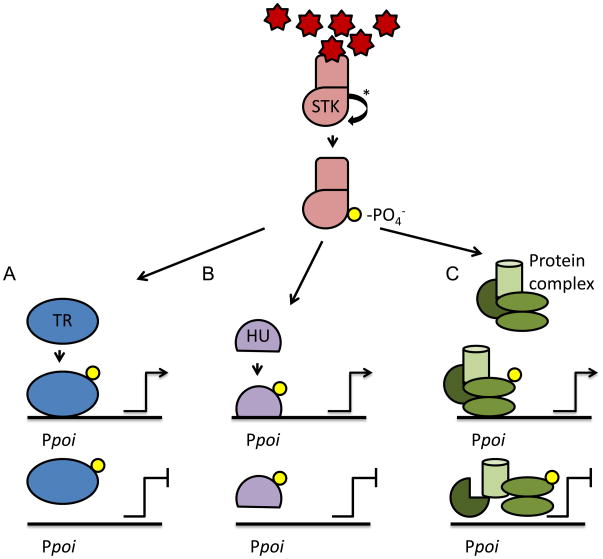
Figure 1.
Schematic representation of known mechanisms of STK-mediated regulation of gene expression. Prokaryotic serine/threonine kinases (STK) are membrane bound protein kinases that autophosphorylate in response to an activating environmental signal. This phosphate group can then be transferred to different targets within the bacterial cell to modulate gene expression. Known targets include A) transcriptional regulators (TR) which include DNA-binding response regulators (RR) of two-component systems (TCS), B) DNA-binding histone-like proteins (HU), and C) proteins of transcriptional or translational machinery. Phosphorylation of these targets has been shown to induce or repress gene expression allowing the bacteria to adapt appropriately to their changing external environment.
Streptococcus agalactiae (also known as Group B Streptococcus or GBS) is an important human pathogen. The TCS known as CovR/CovS (Cov = control of virulence) regulates the expression of over 100 GBS genes [14–16]. Thirty genes comprise the core regulon [16,17], including CovR/S repression of an important GBS toxin known as β-hemolysin/cytolysin (β-H/C) [14,15]. Aspartate phosphorylation of CovR by its sensor kinase (CovS) enhances CovR binding to the β-H/C promoter PcylE [6,14,15]. Conversely, Stk1 phosphorylation of CovR at a threonine residue (T65) decreases promoter DNA binding and alleviates repression of β-H/C [6]. Thus, Stk1 phosphorylation of CovR allows GBS to fine tune the expression of an important virulence factor and possibly facilitates the transition of GBS from commensal to invasive niches.
The TCS CovR/S was originally identified in the human pathogen Streptococcus pyogenes (also known as Group A Streptococcus or GAS) [18,19]. In GAS, CovR/S regulates the expression of a number of virulence genes [20,21] and Bugrysheva et al described that the S. pyogenes STK (SP-STK) regulates the expression of genes that are linked to and are independent of CovR/S [22]. Agarwal et al recently showed that SP-STK phosphorylates GAS CovR at threonine residues [23], similar to GBS. SP-STK and SP-STP mutants exhibit changes in capsule and hemolysin expression [23,24], presumably due to a link with the CovR/S system. Additionally, SP-STK was also shown to phosphorylate WalR of the WalRK TCS [23] which may explain the decrease in expression of a cell division protein cdhA in GAS mutants lacking SP-STK [24,25]. Although the role of SP-STK phosphorylation on WalR and CovR promoter binding and gene regulation remains to be elucidated [23], these observations provide evidence for the interaction between TCS and eukaryotic-like signaling enzymes in bacterial pathogens.
In the human pathogen Streptococcus pneumoniae, RitR (Rit = repressor of iron transport) is a stand alone transcriptional regulator that negatively regulates expression of an iron uptake ATP-binding cassette (ABC) transporter known as Piu. RitR is homologous to CovR of GBS (45% homology, see [26]), is important for virulence of S. pneumoniae and is reversibly phosphorylated by the kinase StkP and the phosphatase PhpP [7]. STK phosphorylation was localized to the DNA-binding domain of RitR suggesting that phosphorylation might regulate binding to the piu promoter (Ppiu, [26]), though this was not examined. Surprisingly, both PhpP and StkP positively regulate Piu as mutations in either decreased piu expression [7]. The exact mechanism of how StkP and PhpP regulate RitR function and Piu expression is unclear [7]. As the complex consisting of RitR and PhpP bound to Ppiu was dissolved upon addition of StkP, Ulijaz et al hypothesize that phosphorylation of RitR by StkP blocks the formation of a ternary complex between PhpP, RitR and Ppiu [7]. How S/T phosphorylation regulates RitR mediated gene expression and iron acquisition of S. pneumoniae remains to be clarified.
Similar to CovR and RitR, EmbR is a transcriptional regulator belonging to the OmpR-like family and is important for cell wall biosynthesis of the human pathogen Mycobacterium tuberculosis (MTB, [8,10]). EmbR regulates expression of three arabinosyltransferases (embC, A, and B) that are important for modulation of the mycobacterial cell wall, MTB virulence and ethambutol resistance [8–10]). EmbR contains a DNA binding domain, transcriptional activator domain and a forkhead-associated (FHA) phosphoprotein recognition domain [8,10]. Mutations in three residues of the FHA domain (Arg312, Ser326 and Asn348) abrogated phosphorylation of EmbR by the STK PknH [8]. While threonine phosphorylation inhibited CovR DNA binding [6], phosphorylation of EmbR by the kinase PknH enhanced its ability to bind to promoter DNA [10]. PknH expression increased during macrophage infection and the kinase promotes virulence of MTB through its ability to control expression of the embCAB genes [10]. Interestingly, EmbR was also described to be phosphorylated by other mycobacterial serine/threonine kinases known as PknA and PknB and is also dephosphorylated by the phosphatase Mstp [9]. Collectively, these observations indicate that regulation of virulence through post-translational modification of EmbR is important for regulation of gene expression in MTB infections.
AfsR of Streptomyces coelicolor is a homolog of EmbR of MTB [8]. AfsR is a global regulatory protein that controls the expression of two pigmented antibiotics namely, actinorhodin and undecylprodigiosin [27]. AfsR was found to be phosphorylated at serine and threonine residues by the kinase AfsK [11]. Loss of AfsK resulted in a significant decrease in actinorhodin production suggesting that phosphorylation of AfsR is necessary for its function [11]. Paradoxically, other STKs are predicted to regulate AfsR function as AfsR was still found to be phosphorylated in an AfsK mutant strain [11]. Whether phosphorylation of AfsR occurs at different amino acid residues in the absence of AfsK and the consequent effect on gene expression requires additional study. Collectively, these studies indicate that multiple STKs control the expression of secondary metabolite production in S. coelicolor through phosphorylation of the global regulator AfsR.
A global transcriptional regulator important for virulence of Staphylococcus aureus is the Staphylococcus accessory regulator, SarA (for a recent review see [28]). SarA is known to be involved in the regulation of over 100 S. aureus genes including positive regulation of the agr locus and negative regulation of itself [28–30]. SarA was recently suggested to be phosphorylated at threonine residues by the STK (Stk1) that is conserved in all S. aureus strains [12]. Phosphorylation of SarA by Stk1 led to a four fold increase in binding to P2agr and a four fold decrease in binding to P1sar [12]. The in vivo relevance of Stk1 regulation of SarA is unclear because the observations of Didier et al imply that the absence of Stk1 should lead to decreased sarA transcription and decreased expression of the Agr locus. However, microarray analysis of a pknB (stk1) mutant in S. aureus 8325 indicated an increase in expression of both agrA and agrC [31]. Also, expression of Agr genes in a stk1 mutant of S. aureus Newman was similar to the WT [32]. In addition, there was no change in sarA expression in the stk1 mutants from either 8325 or Newman strains [31,32]. Didier et al also suggest that SarA is phosphorylated by a second serine threonine kinase Stk2 (found only in certain S. aureus strains such as N315) at serine residues (SA0077, [12]). However, further studies are essential to understand the downstream implications of SarA phosphorylation on regulation of gene expression and S. aureus virulence.
In addition to SarA, S. aureus Stk1 has also been described to phosphorylate another global transcriptional regulator, MgrA [13,33,34]. MgrA regulates expression of the multi-drug efflux pump, NorA [35,36]. Phosphorylation by PknB prevents MgrA binding to the norA promoter, thus increasing norA transcription and resistance to certain fluoroquinolone antibiotics [13]. The link between PknB and MgrA regulation was discovered because of conflicting results on whether MgrA is a positive or negative regulator of norA expression [13]. Truong-Bolduc et al discovered that MgrA is a positive regulator of norA in rsbU- strains and a negative regulator of norA in rsbU+ strains [13]. rsbU is required for activation of the sigB regulon which controls the expression of multiple genes in response to stress [37]. Transcription of pknB is similar in both rsbU+ and rsbU− strains [13]. Whether RsbU and/or SigB expression affects PknB regulation of MgrA is not known. Regardless, MgrA mediated regulation of S. aureus resistance to certain fluoroquinolone antibiotics is controlled through PknB phosphorylation.
One of the first STKs characterized was from M. xanthus, a gram-negative soil bacterium that exhibits vegetative growth and forms fruiting bodies in response to environmental stress [38,39]. Pkn8 and Pkn14 are two serine/threonine kinases of M. xanthus that regulate MrpC, an essential transcription factor necessary for activating fruA expression during fruiting body development [40]. The membrane kinase Pkn8 phosphorylates the cytoplasmic kinase Pkn14 which in turn phosphorylates MrpC [40]. MrpC binds and activates its own promoter and phosphorylation by Pkn14 prevents promoter DNA binding [41]. Pkn8/Pkn14-mediated phosphorylation of MrpC represses mrpC expression during vegetative growth and allows for timely expression of fruA and fruiting body development [40]. Thus, the membrane bound STK Pkn8 enables survival of M. xanthus by sensing an external signal and initiating a phosphorylation cascade that regulates fruiting body development in response to environmental stress.
Interaction with transcription/translation components
In this section, we provide examples of the role of STKs and STPs in regulation of prokaryotic transcriptional and translational machinery. Similar to the role of histones in eukaryotes, histone-like proteins have been described to regulate DNA transcription and replication in bacteria [42–46]. In E. coli, the DNA-binding histone-like protein HU regulates transcription of approximately 8% of the genome [47]. It is thought that binding of histone-like proteins can introduce structural changes to the DNA which facilitate or prevent binding of other regulatory proteins to DNA. Recent studies have indicated that histone-like proteins are phosphorylated by STKs such as Stk1 of S. aureus [32] and SP-STK of S. pyogenes [24] (see Figure 1B). While the role of phosphorylation on the DNA binding ability of histone-like proteins in S. aureus and S. pneumoniae is not known, the serine/threonine kinase Prk2 of M. xanthus was described to phosphorylate HUα and HUβ of E. coli and phosphorylation of HUα at threonine 59 (T59) prevented DNA binding [48]. HUα and HUβ are highly conserved in bacteria and form a heterodimer for regulation of gene expression [49,50]. These observations indicate that post-translational modification of histone-like proteins by STKs may affect DNA binding or introduce structural changes to DNA that either directly regulates transcription or affects the binding of regulators that mediate transcription.
In addition to phosphorylating histone-like proteins to control transcription, prokaryotic STK and STP enzymes have also been found to reversibly phosphorylate RNA and DNA polymerases directly (see Figure 1C). Some transcriptional activators interact with the C-terminal domain of the alpha subunit of RNA polymerase (RpoA) in order to function properly [51,52]. RpoA was identified as a substrate of S. pneumoniae StkP [52], suggesting that StkP may regulate gene expression by controlling the interaction of certain transcription factors, like RitR [7], with RpoA. In L. monocytogenes, the catalytic domain of the STK PrkA was shown to interact with the alpha subunit of DNA polymerase III PolC, the DNA-directed RNA polymerase subunits alpha and beta (RpoA and RpoB) and the recombination protein RecA [53]. The interaction of these proteins implies a role for PrkA in transcriptional regulation of L. monocytogenes. Understanding the in vivo relevance (e.g. changes in gene expression) due to of S/T phosphorylation of transcription machinery will further establish the relevance of these in vitro observations.
A number of prokaryotic STKs and STPs have been found to phosphorylate elongation factors. In prokaryotes, elongation factors play a critical role in protein biosynthesis. Given that transcription and translation is coupled in prokaryotes, these observations provide an additional mechanism by which STKs and STPs regulate prokaryotic gene expression. Translation elongation is initiated by three elongation factors: EF-Tu, EF-Ts and EF-G [54,55]. EF-Tu delivers aminoacyl-tRNAs to the ribosome and associates with GTP [54,55]. EF-Ts acts as a guanine nucleotide exchange factor (GEF) on EF-Tu and EF-G is an additional essential GTPase involved in mRNA and tRNA translocation [54,55]. Phosphorylation of EF-Tu has been found to prevent binding to aminoacyl-tRNAs and thus inhibits translation elongation [54]. In eukaryotes, the functional homologs of EF-Tu, EF-Ts and EF-G (eEF1A, eEF1B and eEF2 respectively) are known to be regulated via phosphorylation in order to maintain the appropriate rate of protein synthesis under a variety of conditions including starvation and growth stimulation [54,56–59]. Therefore, mechanisms that regulate prokaryotic translational elongation may also serve to control the rate of protein synthesis in response to the environment.
Pereira et al hypothesized that post-translational modification of elongation factors allows bacteria to direct protein translation to specific mRNAs required for growth phase transition or in response to different growth environments sensed by the kinase [4]. In the spore-forming soil bacterium B. subtilis, EF-G and EF-Tu are reversibly phosphorylated by PrpC (STP) and PrkC (STK), which regulates bacterial germination from dormant spores [60–63]. Thus, exit from dormancy may be controlled through specific expression of germination genes via phosphorylation of elongation factors. Similarly, EF-Tu was found to be a substrate of PknB in MTB [64]. Sajid et al found that phosphorylation of EF-Tu by PknB reduced its interaction with GTP, increased resistance to EF-Tu specific antibiotics, and caused an overall decrease in protein synthesis which can promote dormancy in MTB [64]. EF-Tu is also a substrate for Stp in L. monocytogenes [54], and PrkA (STK) was found to interact with both EF-Tu and EF-G [53]. Archambaud et al suggested that phosphorylation of EF-Tu in L. monocytogenes allows the bacterium to adapt to the stressful environments encountered in the host by regulating the expression of specific genes [54]. Recently, EF-Tu and ATP-dependent RNA and DNA helicases were found to be phosphorylated by Stk1 in GBS Stp1 mutants [65]. As increased phosphorylation of GBS proteins were observed in the absence of Stp1, the authors predict that Stp1 regulates Stk1 phosphorylation of its targets for appropriate regulation of gene expression and GBS autolysis [65]. Taken together, STK-directed phosphorylation of prokaryotic factors could either lead to a specific change in protein expression and/or function. These changes enable the organism to respond to environment signals for regulation of gene expression and survival.
Conclusion
Regulation of gene expression by eukaryotic-like serine/threonine kinases and phosphatases appears to be a conserved function in prokaryotes. Although some STK and STP mediated gene regulation can be explained through the post-translational modifications of TCSs, DNA binding proteins, transcription and translation machinery (summarized in Table 1 and Figure 1), further studies will be essential for a complete understanding of the signaling mechanisms. In S. mutans approximately 4% of the genome was shown to be regulated by the STK PknB, but no targets have been identified to date [66]. In addition, microarray analysis of kinase mutants in Staphylococcus aureus [31,32], Streptococcus pyogenes [22], Streptococcus pneumoniae [67] and GBS (L. Rajagopal et al, unpublished) revealed that a large number of genes are regulated by STKs in these organisms, which may not be limited to the mechanisms described to date. Therefore, our understanding of prokaryotic STP and STK signaling is still in its infancy.
Table 1.
Targets of eukaryotic-like enzymes that mediate gene expression in prokaryotes
| Strain | Targets |
|---|---|
| Streptococcus agalactiae/ Group B Streptococcus | DNA-binding Response regulator, CovR |
| Elongation factor Tu, EF-Tu | |
| ATP-dependent RNA helicase | |
| ATP-dependent DNA helicase | |
|
| |
| Streptococcus pyogenes/ Group A Streptococcus | DNA-binding histone-like protein, SP-HLP |
| DNA-binding Response regulator, CovR | |
| DNA-binding Response regulator, WalR | |
|
| |
| Streptococcus pneumoniae | DNA-binding Response regulator, RitR |
| DNA-directed RNA polymerase subunitα, RpoA | |
|
| |
| Mycobacterium tuberculosis | DNA-binding Response regulator, EmbR |
| Elongation factor Tu, EF-Tu | |
|
| |
| Streptomyces coelicolor | DNA-binding Response regulator, AfsR |
|
| |
| Staphylococcus aureus | DNA-binding histone-like protein, HU |
| Global transcriptional regulator, MgrA | |
| Global transcriptional regulator, SarA | |
|
| |
| Myxococcus xanthus | Transcription factor, MrpC |
| DNA-binding histone-like protein, Huα | |
| DNA-binding histone-like protein, Huβ | |
|
| |
| Listeria monocytogenes | DNA Polymerase III PolC, α subunit |
| DNA-directed RNA polymerase subunit α, RpoA | |
| DNA-directed RNA polymerase subunit β, RpoB | |
| Recombinant protein, RecA | |
| Elongation factor Tu, EF-Tu | |
| Elongation factor G, EF-G | |
|
| |
| Bacillus subtilis | Elongation factor G, EF-G |
| Elongation factor Tu, EF-Tu | |
It is noteworthy that signals that activate prokaryotic STK and STP enzymes have not been extensively described. In B. subtilis, unlinked peptidoglycan was shown to be an activating ligand for PrkC and mediates exit from dormancy [61]. More recently, a link between peptidoglycan biosynthesis and/or remodeling has been described for StkP of S. pneumoniae (see Sham et al review this issue). Therefore, understanding the activating signals for STK in prokaryotes will provide further insight into signaling mechanisms that mediate prokaryotic gene expression.
Highlights.
Serine/threonine kinases and phosphatases regulate prokaryotic gene expression
Signaling mediates growth, virulence and antibiotic resistance
Targets include DNA-binding regulators, transcriptional and translational machinery
Acknowledgments
We would like to extend our apologies to researchers whose work on gene regulation by serine/threonine kinases and phosphatases has not been reviewed here. We would like to thank Laura K. DeMaster for critical review of the manuscript. Research in Lakshmi Rajagopal’s laboratory is supported by funding from the NIH/NIAID (R01A1070749) and Seattle Children’s Research Institute. Kellie Burnside is a postdoctoral fellow in Lakshmi Rajagopal’s laboratory and is supported by an NIH training grant (5 T32 HD07233-26, PI: Lisa Frenkel). The authors have no other relevant affiliations or financial involvement with any organization or entity with a financial interest in or financial conflict with the subject matter or materials discussed in the manuscript apart from those disclosed. No writing assistance was utilized in the production of this manuscript.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Foussard M, Cabantous S, Pedelacq J, Guillet V, Tranier S, Mourey L, Birck C, Samama J. The molecular puzzle of two-component signaling cascades. Microbes Infect. 2001;3(5):417–424. doi: 10.1016/s1286-4579(01)01390-9. [DOI] [PubMed] [Google Scholar]
- 2.Stock AM, Robinson VL, Goudreau PN. Two-component signal transduction. Annu Rev Biochem. 2000;69:183–215. doi: 10.1146/annurev.biochem.69.1.183. [DOI] [PubMed] [Google Scholar]
- 3.Hoch J, Silhavy T, editors. Two-component Signal Transduction. American Society for Microbiology; Washington DC: 1995. [Google Scholar]
- 4▪.Pereira SF, Goss L, Dworkin J. Eukaryote-like serine/threonine kinases and phosphatases in bacteria. Microbiol Mol Biol Rev. 2011;75(1):192–212. doi: 10.1128/MMBR.00042-10. A comprehensive review article detailing the current state of research into eukaryotic-like serine/threonine kinases and phosphatases in bacteria. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5▪.Burnside K, Rajagopal L. Aspects of eukaryotic-like signaling in Gram-positive cocci: a focus on virulence. Future Microbiol. 2011;6:747–761. doi: 10.2217/fmb.11.62. An interesting review article focused on the current understanding of the role eukaryotic-like serine/threonine kinases and phosphatases play in the virulence of gram-positive cocci. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Lin WJ, Walthers D, Connelly JE, Burnside K, Jewell KA, Kenney LJ, Rajagopal L. Threonine phosphorylation prevents promoter DNA binding of the Group B Streptococcus response regulator CovR. Mol Microbiol. 2009;71(6):1477–1495. doi: 10.1111/j.1365-2958.2009.06616.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Ulijasz AT, Falk SP, Weisblum B. Phosphorylation of the RitR DNA-binding domain by a Ser-Thr phosphokinase: implications for global gene regulation in the streptococci. Mol Microbiol. 2009;71(2):382–390. doi: 10.1111/j.1365-2958.2008.06532.x. [DOI] [PubMed] [Google Scholar]
- 8.Molle V, Kremer L, Girard-Blanc C, Besra GS, Cozzone AJ, Prost JF. An FHA phosphoprotein recognition domain mediates protein EmbR phosphorylation by PknH, a Ser/Thr protein kinase from Mycobacterium tuberculosis. Biochemistry (Mosc) 2003;42(51):15300–15309. doi: 10.1021/bi035150b. [DOI] [PubMed] [Google Scholar]
- 9.Sharma K, Gupta M, Krupa A, Srinivasan N, Singh Y. EmbR, a regulatory protein with ATPase activity, is a substrate of multiple serine/threonine kinases and phosphatase in Mycobacterium tuberculosis. FEBS J. 2006;273(12):2711–2721. doi: 10.1111/j.1742-4658.2006.05289.x. [DOI] [PubMed] [Google Scholar]
- 10.Sharma K, Gupta M, Pathak M, Gupta N, Koul A, Sarangi S, Baweja R, Singh Y. Transcriptional control of the mycobacterial embCAB operon by PknH through a regulatory protein, EmbR, in vivo. J Bacteriol. 2006;188(8):2936–2944. doi: 10.1128/JB.188.8.2936-2944.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Matsumoto A, Hong SK, Ishizuka H, Horinouchi STB. Phosphorylation of the AfsR protein involved in secondary metabolism in Streptomyces species by a eukaryotic-type protein kinase. Gene. 1994;146(1):47–56. doi: 10.1016/0378-1119(94)90832-x. [DOI] [PubMed] [Google Scholar]
- 12.Didier JP, Cozzone AJ, Duclos B. Phosphorylation of the virulence regulator SarA modulates its ability to bind DNA in Staphylococcus aureus. FEMS Microbiol Lett. 2010;306(1):30–36. doi: 10.1111/j.1574-6968.2010.01930.x. [DOI] [PubMed] [Google Scholar]
- 13.Truong-Bolduc QC, Ding Y, Hooper DC. Posttranslational modification influences the effects of MgrA on norA expression in Staphylococcus aureus. J Bacteriol. 2008;190(22):7375–7381. doi: 10.1128/JB.01068-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Jiang SM, Cieslewicz MJ, Kasper DL, Wessels MR. Regulation of virulence by a two-component system in group B streptococcus. J Bacteriol. 2005;187(3):1105–1113. doi: 10.1128/JB.187.3.1105-1113.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Lamy MC, Zouine M, Fert J, Vergassola M, Couve E, Pellegrini E, Glaser P, Kunst F, Msadek T, Trieu-Cuot P, Poyart C. CovS/CovR of group B streptococcus: a two-component global regulatory system involved in virulence. Mol Microbiol. 2004;54(5):1250–1268. doi: 10.1111/j.1365-2958.2004.04365.x. [DOI] [PubMed] [Google Scholar]
- 16▪▪.Lembo A, Gurney MA, Burnside K, Banerjee A, de los Reyes M, Connelly JE, Lin WJ, Jewell KA, Vo A, Renken CW, Doran KS, et al. Regulation of CovR expression in Group B Streptococcus impacts blood-brain barrier penetration. Mol Microbiol. 2010;77(2):431–443. doi: 10.1111/j.1365-2958.2010.07215.x. One of a series of publications from this group detailing the cross-talk between two-component systems and eukaryotic-like signaling enzymes and the implications of this cross-talk on virulence of GBS. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Jiang SM, Ishmael N, Hotopp JD, Puliti M, Tissi L, Kumar N, Cieslewicz MJ, Tettelin H, Wessels MR. Variation in the group B Streptococcus CsrRS regulon and effects on pathogenicity. J Bacteriol. 2008;190(6):1956–1965. doi: 10.1128/JB.01677-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Federle MJ, McIver KS, Scott JR. A response regulator that represses transcription of several virulence operons in the group A streptococcus. J Bacteriol. 1999;181(12):3649–3657. doi: 10.1128/jb.181.12.3649-3657.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Gryllos I, Levin JC, Wessels MR. The CsrR/CsrS two-component system of group A Streptococcus responds to environmental Mg2+ Proc Natl Acad Sci U S A. 2003;100(7):4227–4232. doi: 10.1073/pnas.0636231100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Dalton TL, Scott JR. CovS inactivates CovR and is required for growth under conditions of general stress in Streptococcus pyogenes. J Bacteriol. 2004;186(12):3928–3937. doi: 10.1128/JB.186.12.3928-3937.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Gryllos I, Grifantini R, Colaprico A, Jiang S, Deforce E, Hakansson A, Telford JL, Grandi G, Wessels MR. Mg(2+) signalling defines the group A streptococcal CsrRS (CovRS) regulon. Mol Microbiol. 2007;65(3):671–683. doi: 10.1111/j.1365-2958.2007.05818.x. [DOI] [PubMed] [Google Scholar]
- 22.Bugrysheva J, Froehlich BJ, Freiberg JA, Scott JR. Serine/Threonine protein kinase Stk is required for virulence, stress response and penicillin tolerance in Streptococcus pyogenes. Infect Immun. 2011 doi: 10.1128/IAI.05360-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Agarwal S, Pancholi P, Pancholi V. Role of Serine/Threonine Phosphatase (SP-STP) in Streptococcus pyogenes Physiology and Virulence. J Biol Chem. 2011;286(48):41368–41380. doi: 10.1074/jbc.M111.286690. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Jin H, Pancholi V. Identification and biochemical characterization of a eukaryotic-type serine/threonine kinase and its cognate phosphatase in Streptococcus pyogenes: their biological functions and substrate identification. J Mol Biol. 2006;357(5):1351–1372. doi: 10.1016/j.jmb.2006.01.020. [DOI] [PubMed] [Google Scholar]
- 25.Pancholi V, Boel G, Jin H. Streptococcus pyogenes Ser/Thr kinase-regulated cell wall hydrolase is a cell division plane-recognizing and chain-forming virulence factor. J Biol Chem. 2010;285(40):30861–30874. doi: 10.1074/jbc.M110.153825. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Ulijasz AT, Andes DR, Glasner JD, Weisblum B. Regulation of iron transport in Streptococcus pneumoniae by RitR, an orphan response regulator. J Bacteriol. 2004;186(23):8123–8136. doi: 10.1128/JB.186.23.8123-8136.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Horinouchi S, Hara O, Beppu T. Cloning of a pleiotropic gene that positively controls biosynthesis of A-factor, actinorhodin, and prodigiosin in Streptomyces coelicolor A3(2) and Streptomyces lividans. J Bacteriol. 1983;155(3):1238–1248. doi: 10.1128/jb.155.3.1238-1248.1983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Cheung AL, Nishina KA, Trotonda MP, Tamber S. The SarA protein family of Staphylococcus aureus. Int J Biochem Cell Biol. 2008;40(3):355–361. doi: 10.1016/j.biocel.2007.10.032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Chien Y, Manna AC, Projan SJ, Cheung AL. SarA, a global regulator of virulence determinants in Staphylococcus aureus, binds to a conserved motif essential for sar-dependent gene regulation. J Biol Chem. 1999;274(52):37169–37176. doi: 10.1074/jbc.274.52.37169. [DOI] [PubMed] [Google Scholar]
- 30.Cheung AL, Nishina K, Manna AC. SarA of Staphylococcus aureus binds to the sarA promoter to regulate gene expression. J Bacteriol. 2008;190(6):2239–2243. doi: 10.1128/JB.01826-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Donat S, Streker K, Schirmeister T, Rakette S, Stehle T, Liebeke M, Lalk M, Ohlsen K. Transcriptome and functional analysis of the eukaryotic-type serine/threonine kinase PknB in Staphylococcus aureus. J Bacteriol. 2009;191(13):4056–4069. doi: 10.1128/JB.00117-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Burnside K, Lembo A, de Los Reyes M, Iliuk A, Binhtran NT, Connelly JE, Lin WJ, Schmidt BZ, Richardson AR, Fang FC, Tao WA, et al. Regulation of hemolysin expression and virulence of Staphylococcus aureus by a serine/threonine kinase and phosphatase. PLoS ONE. 2010;5(6):e11071. doi: 10.1371/journal.pone.0011071. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Fournier B, Aras R, Hooper DC. Expression of the multidrug resistance transporter NorA from Staphylococcus aureus is modified by a two-component regulatory system. J Bacteriol. 2000;182(3):664–671. doi: 10.1128/jb.182.3.664-671.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Fournier B, Truong-Bolduc QC, Zhang X, Hooper DC. A mutation in the 5' untranslated region increases stability of norA mRNA, encoding a multidrug resistance transporter of Staphylococcus aureus. J Bacteriol. 2001;183(7):2367–2371. doi: 10.1128/JB.183.7.2367-2371.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Kaatz GW, Thyagarajan RV, Seo SM. Effect of promoter region mutations and mgrA overexpression on transcription of norA, which encodes a Staphylococcus aureus multidrug efflux transporter. Antimicrob Agents Chemother. 2005;49(1):161–169. doi: 10.1128/AAC.49.1.161-169.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Luong TT, Dunman PM, Murphy E, Projan SJ, Lee CY. Transcription Profiling of the mgrA Regulon in Staphylococcus aureus. J Bacteriol. 2006;188(5):1899–1910. doi: 10.1128/JB.188.5.1899-1910.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Shaw LN, Aish J, Davenport JE, Brown MC, Lithgow JK, Simmonite K, Crossley H, Travis J, Potempa J, Foster SJ. Investigations into sigmaB-modulated regulatory pathways governing extracellular virulence determinant production in Staphylococcus aureus. J Bacteriol. 2006;188(17):6070–6080. doi: 10.1128/JB.00551-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Munoz-Dorado J, Inouye S, Inouye M. A gene encoding a protein serine/threonine kinase is required for normal development of M. xanthus, a gram-negative bacterium. Cell. 1991;67(5):995–1006. doi: 10.1016/0092-8674(91)90372-6. [DOI] [PubMed] [Google Scholar]
- 39.Dworkin M. Recent advances in the social and developmental biology of the myxobacteria. Microbiol Rev. 1996;60(1):70–102. doi: 10.1128/mr.60.1.70-102.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Nariya H, Inouye S. Identification of a protein Ser/Thr kinase cascade that regulates essential transcriptional activators in Myxococcus xanthus development. Molec Microbiol. 2005;58(2):367–379. doi: 10.1111/j.1365-2958.2005.04826.x. [DOI] [PubMed] [Google Scholar]
- 41.Nariya H, Inouye S. A protein Ser/Thr kinase cascade negatively regulates the DNA-binding activity of MrpC, a smaller form of which may be necessary for the Myxococcus xanthus development. Mol Microbiol. 2006;60 (5):1205–1217. doi: 10.1111/j.1365-2958.2006.05178.x. [DOI] [PubMed] [Google Scholar]
- 42.Dorman CJ, Deighan P. Regulation of gene expression by histone-like proteins in bacteria. Curr Opin Genet Dev. 2003;13(2):179–184. doi: 10.1016/s0959-437x(03)00025-x. [DOI] [PubMed] [Google Scholar]
- 43.Rimsky S. Structure of the histone-like protein H-NS and its role in regulation and genome superstructure. Curr Opin Microbiol. 2004;7(2):109–114. doi: 10.1016/j.mib.2004.02.001. [DOI] [PubMed] [Google Scholar]
- 44.Peterson CL, Laniel MA. Histones and histone modifications. Curr Biol. 2004;14 (14):R546–551. doi: 10.1016/j.cub.2004.07.007. [DOI] [PubMed] [Google Scholar]
- 45.Aki T, Choy HE, Adhya S. Histone-like protein HU as a specific transcriptional regulator: co-factor role in repression of gal transcription by GAL repressor. Genes Cells. 1996;1(2):179–188. doi: 10.1046/j.1365-2443.1996.d01-236.x. [DOI] [PubMed] [Google Scholar]
- 46.Cerutti H, Casas-Mollano JA. Histone H3 phosphorylation: universal code or lineage specific dialects? Epigenetics. 2009;4(2):71–75. doi: 10.4161/epi.4.2.7781. [DOI] [PubMed] [Google Scholar]
- 47.Oberto J, Nabti S, Jooste V, Mignot H, Rouviere-Yaniv J. The HU regulon is composed of genes responding to anaerobiosis, acid stress, high osmolarity and SOS induction. PLoS ONE. 2009;4(2):e4367. doi: 10.1371/journal.pone.0004367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Udo H, Lam CK, Mori S, Inouye M, Inouye S. Identification of a substrate for Pkn2, a protein Ser/Thr kinase from Myxococcus xanthus by a novel method for substrate identification. J Mol Microbiol Biotechnol. 2000;2 (4):557–563. [PubMed] [Google Scholar]
- 49.Kano Y, Wada M, Nagase T, Imamoto F. Genetic characterization of the gene hupB encoding the HU-1 protein of Escherichia coli. Gene. 1986;45(1):37–44. doi: 10.1016/0378-1119(86)90129-0. [DOI] [PubMed] [Google Scholar]
- 50.Lewis DE, Geanacopoulos M, Adhya S. Role of HU and DNA supercoiling in transcription repression: specialized nucleoprotein repression complex at gal promoters in Escherichia coli. Mol Microbiol. 1999;31(2):451–461. doi: 10.1046/j.1365-2958.1999.01186.x. [DOI] [PubMed] [Google Scholar]
- 51.Busby S, Ebright RH. Transcription activation by catabolite activator protein (CAP) J Mol Biol. 1999;293(2):199–213. doi: 10.1006/jmbi.1999.3161. [DOI] [PubMed] [Google Scholar]
- 52.Novakova L, Saskova L, Pallova P, Janecek J, Novotna J, Ulrych A, Echenique J, Trombe MC, Branny P. Characterization of a eukaryotic type serine/threonine protein kinase and protein phosphatase of Streptococcus pneumoniae and identification of kinase substrates. Febs J. 2005;272(5):1243–1254. doi: 10.1111/j.1742-4658.2005.04560.x. [DOI] [PubMed] [Google Scholar]
- 53▪.Lima A, Duran R, Schujman GE, Marchissio MJ, Portela MM, Obal G, Pritsch O, de Mendoza D, Cervenansky C. Serine/threonine protein kinase PrkA of the human pathogen Listeria monocytogenes: Biochemical characterization and identification of interacting partners through proteomic approaches. J Proteomics. 2011 doi: 10.1016/j.jprot.2011.03.005. Using affinity chromatography and proteomics, this study identified the “interactome” of PrkA revealing a global role for this STK in numerous fundamental biological processes from L. monocytogenes. [DOI] [PubMed] [Google Scholar]
- 54.Archambaud C, Gouin E, Pizarro-Cerda J, Cossart P, Dussurget O. Translation elongation factor EF-Tu is a target for Stp, a serine-threonine phosphatase involved in virulence of Listeria monocytogenes. Mol Microbiol. 2005;56 (2):383–396. doi: 10.1111/j.1365-2958.2005.04551.x. [DOI] [PubMed] [Google Scholar]
- 55.Rodnina MV, Wintermeyer W. Recent mechanistic insights into eukaryotic ribosomes. Curr Opin Cell Biol. 2009;21(3):435–443. doi: 10.1016/j.ceb.2009.01.023. [DOI] [PubMed] [Google Scholar]
- 56.Hershey JW. Protein phosphorylation controls translation rates. J Biol Chem. 1989;264(35):20823–20826. [PubMed] [Google Scholar]
- 57.Browne GJ, Proud CG. Regulation of peptide-chain elongation in mammalian cells. Eur J Biochem. 2002;269(22):5360–5368. doi: 10.1046/j.1432-1033.2002.03290.x. [DOI] [PubMed] [Google Scholar]
- 58.Rhoads RE. Signal transduction pathways that regulate eukaryotic protein synthesis. J Biol Chem. 1999;274(43):30337–30340. doi: 10.1074/jbc.274.43.30337. [DOI] [PubMed] [Google Scholar]
- 59.Sans MD, Xie Q, Williams JA. Regulation of translation elongation and phosphorylation of eEF2 in rat pancreatic acini. Biochem Biophys Res Commun. 2004;319(1):144–151. doi: 10.1016/j.bbrc.2004.04.164. [DOI] [PubMed] [Google Scholar]
- 60.Gaidenko TA, Kim TJ, Price CW. The PrpC serine-threonine phosphatase and PrkC kinase have opposing physiological roles in stationary-phase Bacillus subtilis cells. J Bacteriol. 2002;184(22):6109–6114. doi: 10.1128/JB.184.22.6109-6114.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61▪▪.Shah IM, Laaberki MH, Popham DL, Dworkin J. A eukaryotic-like Ser/Thr kinase signals bacteria to exit dormancy in response to peptidoglycan fragments. Cell. 2008;135(3):486–496. doi: 10.1016/j.cell.2008.08.039. The first study to reveal the activating signal for a prokaryotic STK as well as describe the downstream implications. This study describes how PrkC of B. subtilis senses unlinked peptidoglycan and induces germination. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62▪.Absalon C, Obuchowski M, Madec E, Delattre D, Holland IB, Seror SJ. CpgA EF-Tu and the stressosome protein YezB are substrates of the Ser/Thr kinase/phosphatase couple, PrkC/PrpC, in Bacillus subtilis. Microbiology. 2009;155(Pt 3):932–943. doi: 10.1099/mic.0.022475-0. This elegant study describes a mechanism for how PrkC and PrpC of B. subtilis are able to control growth and germination by sensing extracellular unlinked peptidoglycan. [DOI] [PubMed] [Google Scholar]
- 63.Shah IM, Dworkin J. Induction and regulation of a secreted peptidoglycan hydrolase by a membrane Ser/Thr kinase that detects muropeptides. Mol Microbiol. 2010;75(5):1232–1243. doi: 10.1111/j.1365-2958.2010.07046.x. [DOI] [PubMed] [Google Scholar]
- 64▪▪.Sajid A, Arora G, Gupta M, Singhal A, Chakraborty K, Nandicoori VK, Singh Y. Interaction of Mycobacterium tuberculosis Elongation Factor Tu with GTP is regulated by phosphorylation. J Bacteriol. 2011 doi: 10.1128/JB.05469-11. The first study to describe how STK phosphorylation of an elongation factor is able to regulate protein synthesis. This study shows that phosphorylation of EF-Tu by PknB of M. tuberulosis prevents its interaction with GTP which is essential for function and protein synthesis. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Burnside K, Lembo A, Harrell MI, Gurney M, Xue L, Binh Tran NT, Connelly JE, Jewell KA, Schmidt BZ, de Los Reyes M, Tao WA, et al. The serine/threonine phosphatase Stp1 mediates post transcriptional regulation of hemolysin, autolysis and virulence of Group B Streptococcus. J Biol Chem. 2011 doi: 10.1074/jbc.M111.313486. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Banu LD, Conrads G, Rehrauer H, Hussain H, Allan E, van der Ploeg JR. The Streptococcus mutans serine/threonine kinase, PknB, regulates competence development, bacteriocin production, and cell wall metabolism. Infect Immun. 2010;78(5):2209–2220. doi: 10.1128/IAI.01167-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Saskova L, Novakova L, Basler M, Branny P. Eukaryotic-type serine/threonine protein kinase StkP is a global regulator of gene expression in Streptococcus pneumoniae. J Bacteriol. 2007;189(11):4168–4179. doi: 10.1128/JB.01616-06. [DOI] [PMC free article] [PubMed] [Google Scholar]