Abstract
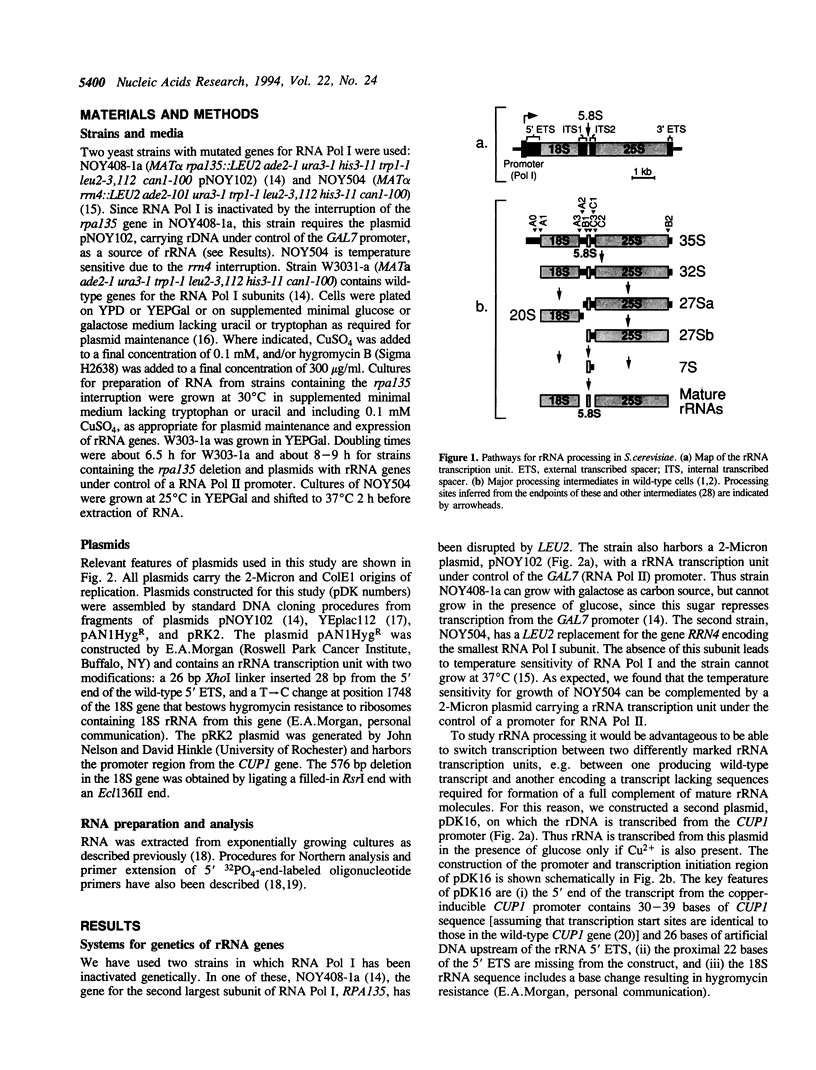
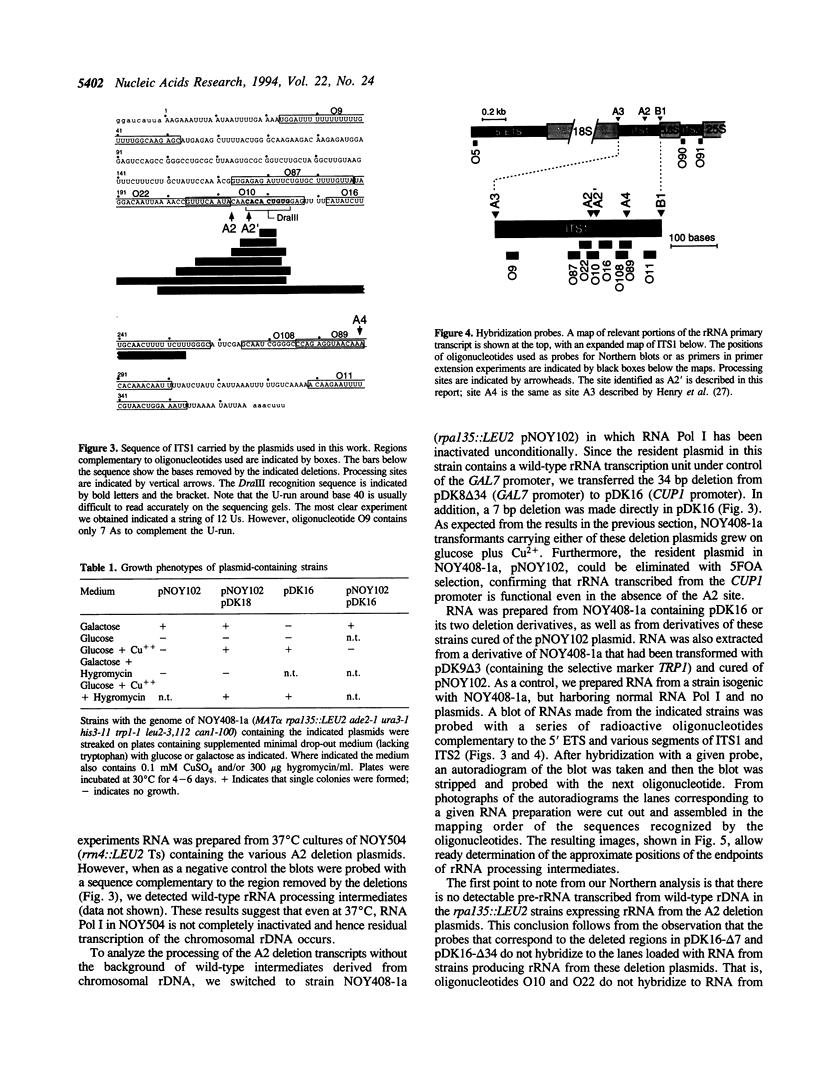
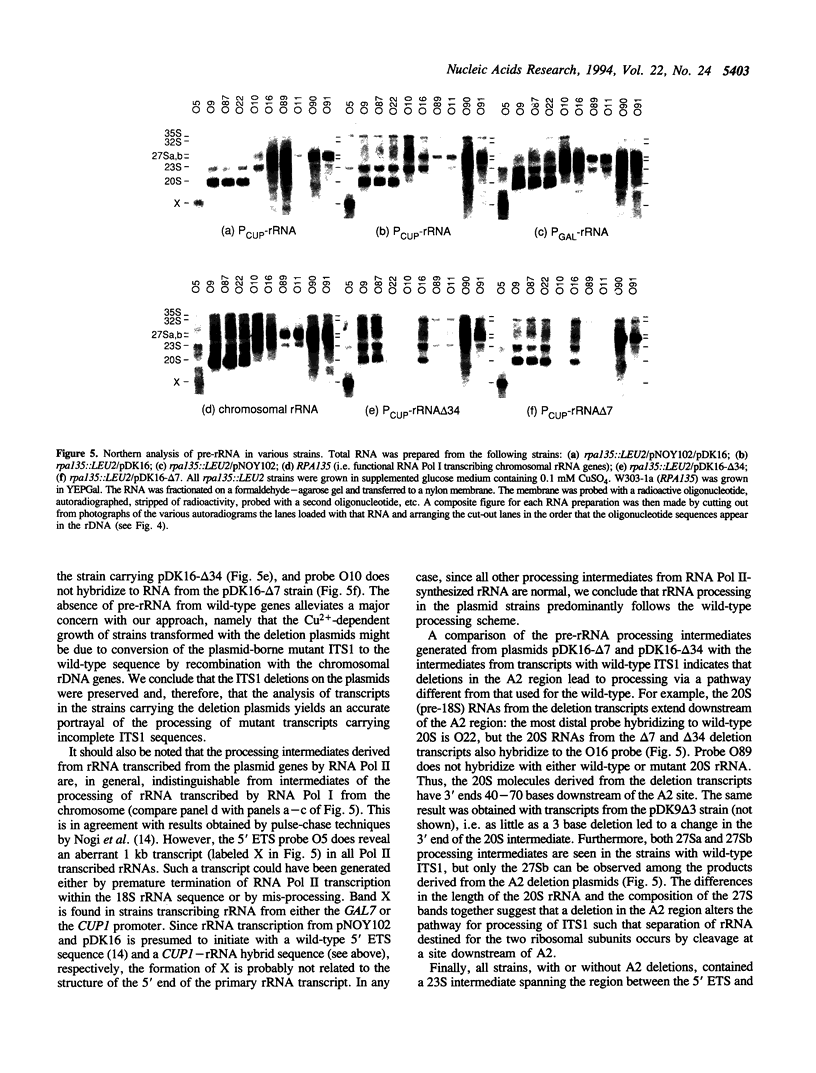
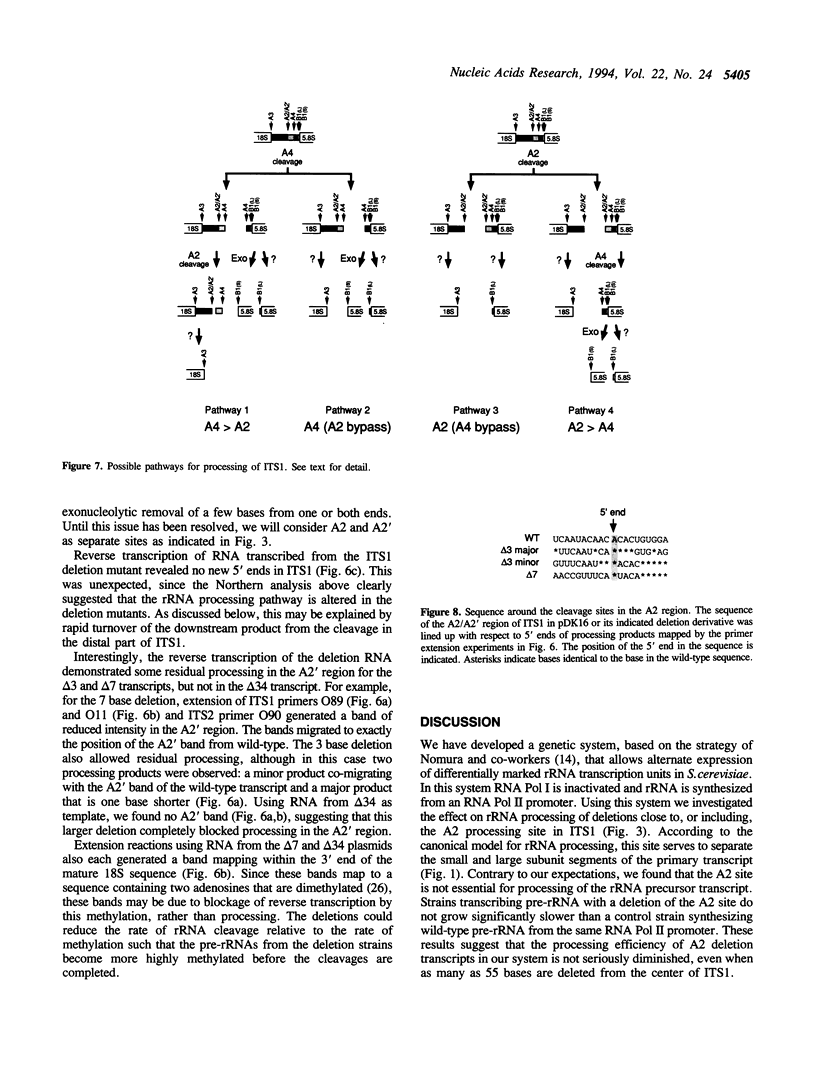
We have extended the system of Nogi et al. (Proc. Natl. Acad. Sci. USA 88, 1991, 3962-3966) for transcription of rRNA from an RNA polymerase II promoter in strains lacking functional RNA polymerase I. In our strains two differentially marked rRNA transcription units can be expressed alternately. Using this system we have shown that the A2 processing site in the internal transcribed spacer 1 (ITS1) of the pre-rRNA is dispensable. According to the accepted processing scheme, the A2 site serves to separate the parts of the primary rRNA transcript that are destined for incorporation into the two ribosomal subunits. However, we have found that, when A2 is impaired, separation of the small and large subunit rRNAs occurs at a processing site further downstream in ITS1, indicating that alternate pathways for ITS1 processing exist. Short deletions in the A2 region still allow residual processing at the A2 site. Mapping of the cleavage sites in such deletion transcripts suggests that sequences downstream of the A2 site are used for determining the position of the cleavage.
Full text
PDF




Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Beltrame M., Tollervey D. Identification and functional analysis of two U3 binding sites on yeast pre-ribosomal RNA. EMBO J. 1992 Apr;11(4):1531–1542. doi: 10.1002/j.1460-2075.1992.tb05198.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boeke J. D., Trueheart J., Natsoulis G., Fink G. R. 5-Fluoroorotic acid as a selective agent in yeast molecular genetics. Methods Enzymol. 1987;154:164–175. doi: 10.1016/0076-6879(87)54076-9. [DOI] [PubMed] [Google Scholar]
- Chu S., Archer R. H., Zengel J. M., Lindahl L. The RNA of RNase MRP is required for normal processing of ribosomal RNA. Proc Natl Acad Sci U S A. 1994 Jan 18;91(2):659–663. doi: 10.1073/pnas.91.2.659. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Craig N., Kass S., Sollner-Webb B. Sequence organization and RNA structural motifs directing the mouse primary rRNA-processing event. Mol Cell Biol. 1991 Jan;11(1):458–467. doi: 10.1128/mcb.11.1.458. [DOI] [PMC free article] [PubMed] [Google Scholar]
- De Jonge P., Klootwijk J., Planta R. J. Terminal nucleotide sequences of 17-S ribosomal RNA and its immediate precursor 18-S RNA in yeast. Eur J Biochem. 1977 Jan;72(2):361–369. doi: 10.1111/j.1432-1033.1977.tb11260.x. [DOI] [PubMed] [Google Scholar]
- Fournier M. J., Maxwell E. S. The nucleolar snRNAs: catching up with the spliceosomal snRNAs. Trends Biochem Sci. 1993 Apr;18(4):131–135. doi: 10.1016/0968-0004(93)90020-n. [DOI] [PubMed] [Google Scholar]
- Gietz R. D., Sugino A. New yeast-Escherichia coli shuttle vectors constructed with in vitro mutagenized yeast genes lacking six-base pair restriction sites. Gene. 1988 Dec 30;74(2):527–534. doi: 10.1016/0378-1119(88)90185-0. [DOI] [PubMed] [Google Scholar]
- Henry Y., Wood H., Morrissey J. P., Petfalski E., Kearsey S., Tollervey D. The 5' end of yeast 5.8S rRNA is generated by exonucleases from an upstream cleavage site. EMBO J. 1994 May 15;13(10):2452–2463. doi: 10.1002/j.1460-2075.1994.tb06530.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hughes J. M., Ares M., Jr Depletion of U3 small nucleolar RNA inhibits cleavage in the 5' external transcribed spacer of yeast pre-ribosomal RNA and impairs formation of 18S ribosomal RNA. EMBO J. 1991 Dec;10(13):4231–4239. doi: 10.1002/j.1460-2075.1991.tb05001.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Karin M., Najarian R., Haslinger A., Valenzuela P., Welch J., Fogel S. Primary structure and transcription of an amplified genetic locus: the CUP1 locus of yeast. Proc Natl Acad Sci U S A. 1984 Jan;81(2):337–341. doi: 10.1073/pnas.81.2.337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lindahl L., Archer R. H., Zengel J. M. A new rRNA processing mutant of Saccharomyces cerevisiae. Nucleic Acids Res. 1992 Jan 25;20(2):295–301. doi: 10.1093/nar/20.2.295. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maden B. E. The numerous modified nucleotides in eukaryotic ribosomal RNA. Prog Nucleic Acid Res Mol Biol. 1990;39:241–303. doi: 10.1016/s0079-6603(08)60629-7. [DOI] [PubMed] [Google Scholar]
- Mattaj I. W., Tollervey D., Séraphin B. Small nuclear RNAs in messenger RNA and ribosomal RNA processing. FASEB J. 1993 Jan;7(1):47–53. doi: 10.1096/fasebj.7.1.8422974. [DOI] [PubMed] [Google Scholar]
- McCormick J. R., Zengel J. M., Lindahl L. Intermediates in the degradation of mRNA from the lactose operon of Escherichia coli. Nucleic Acids Res. 1991 May 25;19(10):2767–2776. doi: 10.1093/nar/19.10.2767. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morrissey J. P., Tollervey D. Yeast snR30 is a small nucleolar RNA required for 18S rRNA synthesis. Mol Cell Biol. 1993 Apr;13(4):2469–2477. doi: 10.1128/mcb.13.4.2469. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nogi Y., Yano R., Dodd J., Carles C., Nomura M. Gene RRN4 in Saccharomyces cerevisiae encodes the A12.2 subunit of RNA polymerase I and is essential only at high temperatures. Mol Cell Biol. 1993 Jan;13(1):114–122. doi: 10.1128/mcb.13.1.114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nogi Y., Yano R., Nomura M. Synthesis of large rRNAs by RNA polymerase II in mutants of Saccharomyces cerevisiae defective in RNA polymerase I. Proc Natl Acad Sci U S A. 1991 May 1;88(9):3962–3966. doi: 10.1073/pnas.88.9.3962. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Raué H. A., Planta R. J. Ribosome biogenesis in yeast. Prog Nucleic Acid Res Mol Biol. 1991;41:89–129. doi: 10.1016/s0079-6603(08)60007-0. [DOI] [PubMed] [Google Scholar]
- Ripmaster T. L., Vaughn G. P., Woolford J. L., Jr A putative ATP-dependent RNA helicase involved in Saccharomyces cerevisiae ribosome assembly. Proc Natl Acad Sci U S A. 1992 Dec 1;89(23):11131–11135. doi: 10.1073/pnas.89.23.11131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rubin G. M. Three forms of the 5.8-S ribosomal RNA species in Saccharomyces cerevisiae. Eur J Biochem. 1974 Jan 3;41(1):197–202. doi: 10.1111/j.1432-1033.1974.tb03260.x. [DOI] [PubMed] [Google Scholar]
- Schmitt M. E., Clayton D. A. Nuclear RNase MRP is required for correct processing of pre-5.8S rRNA in Saccharomyces cerevisiae. Mol Cell Biol. 1993 Dec;13(12):7935–7941. doi: 10.1128/mcb.13.12.7935. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shuai K., Warner J. R. A temperature sensitive mutant of Saccharomyces cerevisiae defective in pre-rRNA processing. Nucleic Acids Res. 1991 Sep 25;19(18):5059–5064. doi: 10.1093/nar/19.18.5059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van Nues R. W., Rientjes J. M., van der Sande C. A., Zerp S. F., Sluiter C., Venema J., Planta R. J., Raué H. A. Separate structural elements within internal transcribed spacer 1 of Saccharomyces cerevisiae precursor ribosomal RNA direct the formation of 17S and 26S rRNA. Nucleic Acids Res. 1994 Mar 25;22(6):912–919. doi: 10.1093/nar/22.6.912. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van der Sande C. A., Kwa M., van Nues R. W., van Heerikhuizen H., Raué H. A., Planta R. J. Functional analysis of internal transcribed spacer 2 of Saccharomyces cerevisiae ribosomal DNA. J Mol Biol. 1992 Feb 20;223(4):899–910. doi: 10.1016/0022-2836(92)90251-e. [DOI] [PubMed] [Google Scholar]