Background: The structure of intraflagellar transport complex A is poorly understood.
Results: Interactions between IFT A proteins are identified.
Conclusion: Three of the IFT A proteins can form a stable subcomplex.
Significance: Determining the structure of IFT A will be crucial to understanding the molecular basis of its ciliary function.
Keywords: Chlamydomonas, Cilia, Microtubules, Protein Purification, Protein-Protein Interactions, Ciliopathies, Flagella, Intraflagellar Transport
Abstract
Chlamydomonas reinhardtii intraflagellar transport (IFT) particles can be biochemically resolved into two smaller assemblies, complexes A and B, that contain up to six and 15 protein subunits, respectively. We provide here the proteomic and immunological analyses that verify the identity of all six Chlamydomonas A proteins. Using sucrose density gradient centrifugation and antibody pulldowns, we show that all six A subunits are associated in a 16 S complex in both the cell bodies and flagella. A significant fraction of the cell body IFT43, however, exhibits a much slower sedimentation of ∼2 S and is not associated with the IFT A complex. To identify interactions between the six A proteins, we combined exhaustive yeast-based two-hybrid analysis, heterologous recombinant protein expression in Escherichia coli, and analysis of the newly identified complex A mutants, ift121 and ift122. We show that IFT121 and IFT43 interact directly and provide evidence for additional interactions between IFT121 and IFT139, IFT121 and IFT122, IFT140 and IFT122, and IFT140 and IFT144. The mutant analysis further allows us to propose that a subset of complex A proteins, IFT144/140/122, can form a stable 12 S subcomplex that we refer to as the IFT A core. Based on these results, we propose a model for the spatial arrangement of the six IFT A components.
Introduction
Cilia and flagella are cellular appendages that serve as sensory organelles for a wide variety of eukaryotic organisms. A subset of cilia are dynamic motile structures that act to propel cells through their environment or to move liquid and other materials across cell surfaces such as the ciliated trachea and cerebral ventricles found in higher animals (1, 2). Most, if not all, cilia act to transmit environmental cues that can range from light perception in visual cells to volatile chemical recognition in olfactory tissues, whereas other cilia mediate developmental signaling pathways such as sonic hedgehog and Wnt signaling (3–5). Defects in the assembly and function of cilia and flagella can perturb one or more of these critical motile or sensory functions that can give rise to over a dozen human diseases known as ciliopathies (6–12). Central to the formation and function of these structures is a conserved transport of materials in and out of the organelle known as intraflagellar transport or IFT (13–15).
First identified in Chlamydomonas reinhardtii, intraflagellar transport (IFT)3 is characterized by the bidirectional transport of large protein particles along the axonemal microtubules of eukaryotic cilia and flagella (16, 17). The anterograde movement of the particles out to the distal tip is the responsibility of the plus end-directed microtubule motor Kinesin-2 (17–21), whereas the retrograde transport back to the cell body is powered by the minus end-directed cytoplasmic dynein 1b/2 (22, 23). First isolated from the flagella of Chlamydomonas, the IFT particles were found to contain multiple copies of ∼20 proteins, each of which was associated with one of two complexes, IFT A and IFT B (20, 24, 25, reviewed in Ref. 26). With a predicted mass of ∼750 kDa, IFT A contains six protein subunits, IFT144, IFT140, IFT139, IFT122, IFT121, and IFT43 (20, 25, 27–29). When intact, IFT B has a predicted mass close to 1 MDa and contains IFT172, IFT88, IFT81×2, IFT80, IFT74, IFT72, IFT70, IFT57, IFT52, IFT46, IFT27, IFT25, IFT22, and IFT20 (20, 25, 27–39).
Functional analysis of the IFT motors and particle subunits in diverse model organisms has progressed rapidly in the past decade to reveal a conserved role in the assembly and function of cilia (reviewed in Refs. 13–15 and 40–42). Mutations that cause disruptions in many, but not all, IFT B subunits result in severe ciliogenic phenotypes similar to those observed in Kinesin-2 mutants, contributing to the model that IFT B is closely associated with anterograde transport. The loss of IFT52 or IFT88, for example, effectively blocks anterograde IFT resulting in little or no assembly of the organelle (28, 30, 31, 43–45). Mutations affecting IFT A genes, however, often result in less severe ciliary phenotypes. Mutations affecting IFT122, for example, typically result in partially or completely assembled organelles that contain accumulations of IFT B proteins (46–49).
Structural analysis of IFT has not progressed rapidly but elegant high resolution EM tomographic reconstructions of intact IFT particles reveal convoluted structures that bridge from the ciliary membrane to the axonemal microtubules (50). To complement this top-down approach, other studies have focused on specific interactions within the IFT motors and complexes. Direct interactions, for example, have been demonstrated for the following pairs of IFT B components IFT20-IFT57, IFT25-IFT27, IFT46-IFT52, IFT46-IFT70, IFT46-IFT88, IFT52-IFT70, IFT52-IFT88, IFT70-IFT74/72; IFT74/72-IFT81, and IFT81-IFT81 (29, 34, 39, 51–54). Biochemical analysis of complex B isolated from Chlamydomonas flagella has provided additional architectural information that includes the stepwise removal of peripheral subunits revealing a stable 11 S subcomplex known as the IFT B core (34).
By comparison to IFT B, much less is known about the architecture of the A complex. Unlike complex B, IFT A is a stable complex that has resisted partial dissociation to reveal stable subcomplexes. In addition, the large size and relative insolubility of five of the six IFT A components has slowed the analysis of individual components expressed in heterologous systems such as E. coli or yeast. Indeed, as reported here, heterologous expression revealed only one convincing interaction between the IFT A subunits, IFT121 and IFT43. To better understand the A complex, we isolated two Chlamydomonas IFT A mutants, ift121 and ift122, that carried disruptions in the IFT121 and IFT122 genes, respectively. As observed with many IFT mutants, these two strains were nonmotile with cells being either aflagellate or possessing very short flagella. To determine how the loss of IFT121 or IFT122 affects complex A, the remaining IFT A proteins were analyzed by antibody pull-downs and sucrose density gradient centrifugation. The loss of IFT122 had a severe effect on IFT A with three of the polypeptides, IFT122, IFT121, and IFT139 being undetectable, whereas the levels of IFT140 and IFT144 were significantly lower than in the parental strain; IFT43 was present but appeared to be unassociated with IFT140 and IFT144. The loss of IFT121, however, resulted in the formation of a stable IFT A subcomplex or core of IFT122, IFT140, and IFT144 that sedimented at ∼12 S. IFT43 was present but unassociated with the IFT A core, which was consistent with IFT121-IFT43 interactions observed using heterologous expression. This dissociation coupled with the absence of IFT139 indicated that IFT121 was required for both IFT139 and IFT43 to be stably associated with complex A. Thus, both IFT121 and IFT122 were essential in keeping complex A intact, but the loss of IFT122 resulted in nearly complete disruption of the complex.
EXPERIMENTAL PROCEDURES
Algal Strains and Media
The Chlamydomonas cell wall-deficient strain CC-503 (cw92) and temperature-sensitive flagellar assembly strain CC-1390 (fla2) were obtained from the Chlamydomonas Center. Chlamydomonas strains were grown and maintained on solid or in liquid TAP medium (55).
Antibody Generation
With the exception of the pre-existing monoclonal 139.1 (20), polyclonal antisera were generated against IFT A proteins using a variety of host animals (supplemental Table S1). Antigens were expressed as recombinant chimeric proteins in E. coli and purified by affinity chromatography according to the protocols provided by the suppliers. Recombinant His6-tagged proteins were derived from the pTrcHis expression vector (Invitrogen) and purified on nickel affinity resin (His-Bind; Novagen). Recombinant maltose-binding protein (MBP) chimeric proteins were derived from the pMalC2X expression vector (New England Biolabs) and purified on amylose resin (New England Biolabs). Aliquots of purified proteins were supplied to the company for antiserum production. Rabbits were immunized with 200 μg of antigen initially and 100 μg at 2-week intervals for a total of six injections. Chickens were immunized following a similar schedule. Rats were initially immunized and subsequently boosted every 3 weeks with 100 μg of antigen. Aliquots of immune sera (rabbits, rats) or IgY fractions (chickens) were tested against transfer blots of the respective recombinant IFT A protein and sucrose gradient-purified native IFT proteins to verify the avidity and specificity of each serum.
Insertional Mutagenesis
Chlamydomonas motility mutants were generated as described previously (52) employing insertional mutagenesis of the cell wall-deficient strain, CC-503, using the pHyg3 plasmid carrying an aminoglycoside phosphotransferase (aph7″) gene that confers resistance to hygromycin B (56–58). Transformed strains that displayed motility and flagellar assembly defects were screened for disruptions in IFT genes using gene-specific PCR amplification with genomic DNA as the template; PCR primer sequences and amplification conditions are available upon request. After strains were identified that failed to yield IFT121 or IFT122 PCR products, the genomic region containing each gene was screened using a variety of PCR primer sets to better define the disrupted region.
Flagellar Isolation and Sucrose Density Gradient Centrifugation
Intact flagella were isolated from C. reinhardtii cells by low pH treatment of actively swimming cells as described previously (20, 59). Isolated flagella were stored as frozen pellets at −80 °C in HMDEK buffer (10 mm HEPES, 5 mm MgSO4, 1 mm DTT, 0.5 mm EDTA, 25 mm KCl, pH 7.2) containing a protease inhibitor mixture (HMDEK + PI: HMDEK plus protease inhibitors, 2 mm PMSF, 50 μg/ml of soybean trypsin inhibitor, 1 μg/ml of pepstatin A, 2 μg/ml of aprotinin, 1 μg/ml of leupeptin). To extract the soluble membrane plus matrix containing soluble IFT complexes, frozen flagella were pipetted repeatedly in 0.2–0.5 ml of HMDEK + PI prior to centrifugation in a microcentrifuge at 4 °C for 15 min at 16,000 × g. In some, but not all cases, the supernatant was further clarified for 10 min in an Airfuge A-100/18 rotor (Beckman Instruments) at 25 p.s.i. Linear sucrose density gradients (10–25% sucrose in HMDEK buffer) were prepared in 14 × 89-mm tubes appropriate for a SW41Ti rotor (Beckman Instruments); 0.5 ml of clarified membrane plus matrix protein solution was applied to the top of the gradient, and the sample was centrifuged at 37,000 rpm (169,000 × g) for 16 h at 4 °C. Following centrifugation, the gradient was partitioned into 24 fractions that were resolved on 7.5–15% SDS-polyacrylamide gels (Idea Scientific) and stained with Coomassie Brilliant Blue; any particles with sedimentation values much greater than 20 S would have passed completely through the sucrose density gradient and would not have been further analyzed. Gel images were obtained by scanning the gel on a LICOR Odyssey Infrared Scanner at 680 nm. Additional samples of each fraction were resolved on 10% SDS-polyacrylamide gels, electrophoretically transferred onto nitrocellulose membrane, and analyzed as detailed below under “Immunoblot Analysis.”
Proteomic Identification of IFT A Subunits
IFT proteins were isolated from Chlamydomonas flagella, fractionated by 10–25% sucrose density gradient centrifugation in HMDEK buffer (SW55 or SW41 rotor, Beckman), and separated by two-dimensional or SDS-PAGE as described previously (20, 29). To identify IFT144, IFT140, IFT139, and IFT122, the corresponding protein spots were excised from Coomassie Blue-stained two-dimensional gels prior to tryptic digestion and LC-mass spectrometry; pure peptide fractions were subjected to Edman degradation to determine peptide sequence. To identify IFT121, the 16 S sucrose density gradient fraction containing complex A was fractionated by SDS-PAGE as described previously (29) to separate IFT121 and IFT122, which were excised from the gel and digested with trypsin; isolated peptides were analyzed by Edman degradation. To identify IFT43, complex A was immunoprecipitated from the 16 S fraction using an anti-IFT139 resin where 139.1 was covalently bound to CNBr-activated Sepharose. To minimize contamination by IgG subunits, complex A was eluted from the anti-IFT139 resin for 2 min using 100 mm glycine, pH 2.0, followed by neutralization with equimolar Tris, pH 8.5. Fractionation and Coomassie Blue staining of the resulting IFT A eluate on a 10% acrylamide SDS-PAGE gel revealed a visible IFT43 protein band. IFT43 was excised and digested with trypsin followed by peptide identification using MALDI mass spectrometry as described previously (34).
Cloning of IFT A Subunits
The coding regions of the six Chlamydomonas IFT A subunits were isolated and cloned into the pBS KS− vector. DNA was amplified by PCR using as the template either specific cDNA plasmid clones obtained from the Kazusa DNA Research Institute (Kisarazu, Chiba, Japan; (60, 61) or from a pool of cDNA harvested from a λ cDNA library generously provided by George Witman (62). Mammalian IFT43 and IFT121 cDNA clones were obtained from Open Biosystems (Thermo Scientific): human IFT121, Hs121, clone ID 5266940, accession BI460483; human IFT43, Hs43, clone ID 3608220, accession BE515088; murine IFT43, Mm43, clone ID 5148723, accession BI248497. Plasmid DNA was isolated from bacterial hosts and purified using a standard alkaline lysis protocol (63) followed by treatment with Miniprep Express Matrix (MP Biochemicals); λ template DNA was isolated by two sequential phenol:chloroform:isoamyl alcohol (25:24:1) extractions, followed by chloroform extraction and ethanol precipitation. Phusion HF DNA polymerase (Finnzymes; New England Biolabs) was utilized to amplify specific cDNA fragments by PCR, following the directions supplied by the manufacturer; primers for PCR incorporated the proper restriction endonuclease sites to facilitate subsequent cloning into a variety of vectors, and were of sufficiently high Tm to allow for two-cycle PCR. In some cases, amplification of the 5′ end of the full-length cDNA was accomplished by utilizing the RT-PCR reagents in the BD Matchmaker Kit (Clontech) and total RNA template isolated from re-flagellating C. reinhardtii as described previously (34). DNA encoding selected portions of specific IFT A proteins were amplified and cloned as described above. Putative conserved protein domains were identified by multispecies sequence analysis utilizing the multiple alignment program MACAW (NCBI) (64). Primers were designed to amplify these regions using Gene Runner (Hastings Software), and Phusion HF DNA polymerase was employed as described for the above PCR. The resulting DNA fragments were cloned, sequenced, and analyzed as previously detailed. Following standard ligation into prepared pBS KS− vectors and transformation into TOP-10 E. coli cells (Invitrogen Corp. (63)), plasmid DNA was purified as above, and the DNA was exhaustively sequenced (Washington State University Center for Reproductive Biology Molecular Biology Core). PCR primer sequences and amplification conditions are available upon request. Analysis of sequence data were facilitated by use of the software programs Chromas Lite (Technelysium Pty Ltd.) and Gene Runner.
Complementation of C. reinhardtii Motility Mutants
To confirm the identity of the ift121 and ift122 mutant lesions, these strains were transformed with Chlamydomonas genomic BAC clones 14N19 and 3B16, respectively (Clemson University Genomics Institute, Clemson, SC), as described previously (52). To verify that the observed phenotypes were due to the disruption of a single gene, DNA fragments encoding only IFT121 or IFT122 genes were PCR amplified and used for transformation. In brief, PCR primers were designed to specifically amplify the coding region of each gene, along with 1–1.5 kb of genomic DNA beyond the 5′ and 3′ ends of the coding region (to allow for promoter and terminator sequences). The amplified regions did not contain parts of any other genes, as determined by examination of the C. reinhardtii genomic sequence data base at the DOE Joint Genome Institute. The PCR utilized Phusion HF DNA polymerase and BAC genomic DNA fragments (65) containing the appropriate genes as template. PCR product was used directly from the reaction tube as DNA for transformation of the ift121 and ift122 strains, using the protocols detailed above. Complemented cells were chosen based on their recovery of flagellar assembly and ability to swim; subsequent genomic DNA analysis verified the incorporation of the exogenous IFT121 or IFT122 genes.
Yeast-based Two-hybrid Analysis
Potential interactions between IFT A subunits were surveyed with yeast-based two-hybrid analysis. DNA fragments encoding full-length and partial proteins were cloned into vectors pAD-MCS (a derivative of pAD-GAL4; Stratagene) and pGBKT7 (Clontech); these vectors express fusion proteins incorporating the GAL4 activation domain (AD) and the DNA binding domain (BD) as N-terminal fusions to the cloned exogenous proteins. The integrity of each plasmid construct was verified by restriction analysis and DNA sequencing. Yeast strain AH109 (Clontech) was co-transformed with both AD and BD plasmids, utilizing a standard lithium acetate/PEG-8000/sheared salmon sperm DNA protocol as described previously (34). Following co-transformation, yeast cells containing plasmids expressing both AD and BD fusion proteins were plated on selective media to screen for potential interacting proteins; the presence of both plasmids was verified by growth on minimal medium lacking leucine and tryptophan (SD, -Leu, -Trp), whereas interacting AD and BD fusion proteins produced a yeast clone capable of growing on minimal medium lacking leucine, tryptophan, histidine, and adenine (SD, -Leu, -Trp, -His, -Ade).
A pooled library approach was designed to quickly screen for interacting IFT A subunits. Full-length copies of each of the six A subunits were cloned into the AD and BD vectors. Aliquots of competent AH109 yeast cells were transformed with each individual AD plasmid construct; transformed yeast were selected for on solid medium (SD, -Leu). In an analogous fashion, competent Y187 yeast cells were transformed with each individual BD plasmid construct, and transformed yeast were selected on solid medium (SD, -Trp). Yeast cultures expressing each subunit in the AD vector were grown and mixed. An analogous library was prepared with BD vector yeast cultures, and the two mixtures were combined and the yeast were allowed to mate, following instructions in the BD Matchmaker Kit (BD Biosciences, Clontech Laboratories). The resulting library mating mixture was plated on a selective medium (SD, -Leu, -Trp, -His, -Ade) and screened for interacting proteins. Yeast colonies that grew on this selective medium were analyzed by PCR to determine the identity of the interacting proteins. Individual colonies were suspended in 20 μl of water and heated at 95 °C for 5 min. Insoluble material was removed by centrifugation at 13,000 × g for 5 min, and 1.0 μl of the clarified yeast extract was utilized as template in an 11-μl PCR. To expedite the screening process, two PCR master mixtures were prepared: each contained a specific vector primer, for either the AD or BD vector; in addition, each mixture contained six unique subunit-specific primers, and all the other reagents necessary for amplification. Each vector/subunit primer pair generated a differently sized DNA fragment in the presence of its corresponding template DNA. Electrophoretic analysis of the resulting DNA identified two recombinant proteins in each yeast colony.
Bacterial Protein Expression and Purification
DNA fragments encoding proteins of interest were cloned into two bacterial protein expression systems. Co-transformation of appropriate E. coli strains with two protein-expressing plasmids allowed for the simultaneous expression of two target proteins in a single host. Two different expression plasmids were utilized in this study. The pMalC2X plasmid (New England Biolabs) was designed to produce an N-terminal fusion of the E. coli MBP onto an exogenous protein; the resulting chimeric protein bound specifically and reversibly to amylose resin. The Duet family of vectors (Novagen) comprises a group of similar plasmids, all of which were designed to express one or two recombinant proteins. We created a novel dual chimeric expression vector, pRXS, based on the pRSF Duet plasmid. In this construct, the existing N-terminal epitope tag and the multicloning site (MCS) of MCS2 were replaced by the Strep-II tag sequence (WSHPQFEK) and the MCS (MfeI-BglII-XhoI-STOP); in addition, MCS1, which is preceded by a His6 epitope tag, was replaced by the following MCS (EcoRI-BamHI-SalI-STOP). The restriction sites of EcoRI and MfeI, and SalI and XhoI, generate compatible cohesive ends upon digestion, thereby facilitating the cloning of the various subunits. The pMalC2X plasmid contains an ampicillin resistance gene and the pBR322 (ColE1) origin of replication; pRXS contains a kanamycin resistance gene and the RSF1030 origin of replication. PCR primer sequences and amplification conditions are available upon request.
DNA fragments encoding selected proteins were cloned into expression plasmids using standard protocols. Initial transformations utilized chemically competent BL21(DE3) E. coli (Novagen) followed by electroporation of the second plasmid using standard protocols (63). One hundred-ml aliquots of LB media, supplemented with the appropriate antibiotics, were inoculated with 1 ml of overnight culture. The cells were grown to an A600 of 0.3–0.6 (typically 2–3 h at 37 °C), and recombinant protein production was induced with 1 mm isopropyl β-d-thiogalactopyranoside. Cultures were harvested after an additional 2–2.5 h of growth by centrifugation at 10,000 × g and 4 °C in an SL-1500 rotor (Sorvall). Cell pellets were resuspended in 7–10 ml of ice-cold MBP column buffer (MCB: 20 mm Tris-Cl, 0.2 m NaCl, pH 7.4) supplemented with protease inhibitors (1 μg/ml of leupeptin, 1 μg/ml of pepstatin A, 2 μg/ml of aprotinin, 2 mm PMSF). The resulting suspension was stored at −27 °C until used.
Thawed E. coli cell suspensions containing recombinant proteins were lysed by sonication, using a Branson microtip connected to a Branson 450 power unit. Cells were subjected to a total of 2.5 min of sonication, with a power level of 3 and a duty cycle of 50%. The cell suspensions were kept in an ice-water bath, and the temperature during sonication was monitored carefully. Following sonication, the lysed cell suspension was clarified by centrifugation for 10 min at 10,000 × g and 4 °C in an SS-34 rotor (Sorvall). The supernatant was carefully decanted and stored on ice; an additional aliquot of 2 mm PMSF was added at this point. The insoluble pellet was resuspended in MCB, and an aliquot taken for subsequent SDS-PAGE analysis.
Typical protein chromatography utilized a 0.4-ml (packed) column, either amylose resin (New England BioLabs) or StrepTactin resin (GE Healthcare) prepared according to the supplier's instructions. The clarified cell lysate, containing recombinant proteins, was loaded onto the column three times. For amylose chromatography, nonbinding proteins were removed by extensive washing (20 column volumes) with MCB. Specifically bound protein(s) were eluted from the amylose resin with 4 sequential bed volumes of MCB containing 10 mm maltose. Nonbinding proteins were washed from the StrepTactin resin with 5 bed volumes of MCB. The Strep-II-tagged proteins were eluted with 4 sequential bed volumes of 2.5 mm desthiobiotin and 0.15 m NaCl in 0.1 m Tris-HCl, pH 8.0. In some instances, tandem protein purification was employed. The soluble protein fraction was first loaded onto StrepTactin resin, washed, and the specifically bound protein(s) were eluted with desthiobiotin as described above. A sample of the eluted protein(s) was then applied to an amylose column, washed, and the bound protein(s) eluted with maltose and analyzed by SDS-PAGE on 8-cm 10% acrylamide gels (Idea Scientific).
Total Protein Extraction from Whole Chlamydomonas Cells
Wild-type and mutant strains of C. reinhardtii were grown on solid TAP media (100 mm plates) for 2–3 days under constant light (2300 lux average). Each plate was flooded with 15 ml of liquid TAP medium, incubated under light (1200 lux average) for 60–90 min prior to gentle cell trituration (10 ml pipette), and transferred to a 15-ml conical tube. Cells were counted and the suspension was centrifuged at 2300 × g for 3 min in a Beckman Allegra 6R centrifuge with low brake. A final volume (Vf) was calculated to achieve a final cell density of 2 × 106 cells/5 μl. The cell pellet was resuspended in 0.7 Vf of TAP, 0.1 Vf of 20% SDS, and 0.2 Vf of 5× SDS sample buffer. The cell suspension was vortexed and incubated at 95 °C for 4 min. The cell suspension was mixed gently and centrifuged for 8 min at 13,000 rpm (16,000 × g) in a microcentrifuge. The top ¾ of the supernatant was transferred to another tube and stored at −27 °C. Protein samples were analyzed using SDS-PAGE and immunoblots as detailed below.
Soluble Protein Extraction from Chlamydomonas Cells
Wild-type and mutant Chlamydomonas strains were grown on solid TAP media (5–10 100-mm plates) for 2–3 days under constant light (2300 lux average). Following flooding with 15 ml of TAP medium (1–2 h), cells were gently triturated and the cells were shaken in a 125-ml flask at 100–150 rpm under constant light (1200 lux average) for 90 min at room temperature. Tween 20 (1:2000 volume of 10% Tween 20) was added to the cell suspension to enhance cell centrifugation. After brief cooling on ice, the cells were concentrated by centrifugation at 2300–2600 rpm (1300–1500 × g) in a Beckman Allegra 6R at 4 °C with no braking. Cell pellets were resuspended in 5–10 ml of HMDEK + PI.
Two methods were used to lyse cells and release the soluble proteins. Under one method, the cell suspension was sonicated with a Branson Sonifier 450 set to 50% cycle, power level 3, for 2 min on ice with a microtip. The second method utilized an ESGE Biohomogenizer for 2-min bursts at high speed. In either case, the disrupted cell suspension was clarified in a Sorvall SS-34 rotor for 15 min at 15,000 × g and 4 °C. The resulting supernatant was further clarified by ultracentrifugation in either a Beckman 50.2Ti rotor (45 min; 40,000 rpm; 146,000 × g) or a Beckman SW55Ti rotor (24 min; 50,000 rpm; 237,000 × g) in a Beckman Optima XL ultracentrifuge at 4 °C. The resulting supernatant served as the source of whole cell IFT complex A for subsequent subunit composition, sizing, and immunoprecipitation experiments.
Hydrodynamic Analysis of IFT A Complexes
Soluble protein was extracted from Chlamydomonas whole cells as detailed above using the Biohomogenizer, except the final ultracentrifugation step was omitted. Aliquots (0.35 ml) of extract were loaded onto 10–25% sucrose density gradients prepared in HMDEK; for high-salt gradients, NaCl was added to a final concentration of 0.30 m. All gradients were prepared in tubes that fit the Beckman SW55-Ti rotor. Samples were centrifuged at 4 °C for 17 h at 32,000 rpm (97,000 × g). Following centrifugation, the gradients were fractionated into 16 fractions. Protein samples were analyzed on 8% SDS-PAGE gels; total proteins were visualized with Coomassie Blue and imaged on a LICOR Odyssey infrared scanner, and individual IFT subunits were characterized by corresponding immunoblots as detailed below. Approximate S values were derived using the sedimentation behavior of the following standards: thyroglobulin (19.2 S), apoferritin (17.1 S), catalase (11.3 S), β-amylase (9.6 S), alcohol dehydrogenase (7.2 S), BSA (4.6 S), ovalbumin (3.5 S), and cytochrome c (1.9 S).
Immunoprecipitation of Chlamydomonas IFT A Proteins
Antibody resins were prepared according to the manufacturer's protocols. Briefly, polyclonal sera were applied to a DEAE-Affi-Gel Blue resin (Bio-Rad) to remove serum albumin; the remaining serum proteins were cross-linked to cyanogen bromide-activated Sepharose 4B (Sigma). Aliquots of clarified C. reinhardtii cell extracts (0.2–0.5 ml) were mixed with subunit-specific antibody resin (0.05–0.1 ml of packed resin); 5 m NaCl was added to a final concentration of 0.15 m. This suspension was incubated overnight at 4 °C on a slow rotatory shaker (<10 rpm). Following incubation, the resin was consolidated by a brief centrifugation (16,000 × g, 15 s), and the nonbound proteins were removed for analysis. The resin was washed with 0.5 ml of HMDEK containing 0.15 m NaCl four times at room temperature. Antibody-bound proteins were released with the incubation of 2 resin volumes of 1% SDS at room temperature for 10 min, followed by consolidation of the resin as above. The supernatant was removed and saved for analysis. Any proteins still bound to the resin were released by the addition of 0.2 resin volumes of 5× SDS sample buffer and 1 resin volume of 1× SDS sample buffer, followed by heating at 95 °C for 5 min. Following centrifugation (2 min at 16,000 × g), the supernatant was saved for SDS-PAGE analysis.
Immunoblot Analysis
Protein samples were resolved by SDS-PAGE using 8 or 10% polyacrylamide gels. The proteins in the gels were transferred electrophoretically to nitrocellulose membranes with a Mini-Genie blotter (Idea Scientific), using 0.5× Towbin buffer (12.5 mm Tris base, 81 mm glycine) (66). The membranes were incubated with blocking buffer (1.3% coldwater fish gelatin in PBS) for 30–60 min at room temperature. Primary antibodies were diluted into blocking buffer containing 0.1% Tween 20, and incubated with the membranes overnight at 4 °C on a rocking platform. The membranes were washed four times for 5 min each with PBS plus 0.1% Tween 20. Fluorescently labeled secondary antibodies (LICOR, Molecular Probes, and Rockland Immunochemicals) were diluted into blocking buffer containing 0.1% Tween 20 and 0.01% SDS, and incubated with the membranes for 60 min at room temperature on a shaker. The membranes were washed as above, and rinsed in PBS before being visualized on a LICOR Odyssey infrared scanner.
RESULTS
Proteomic Identification of the Six Chlamydomonas IFT A Subunits
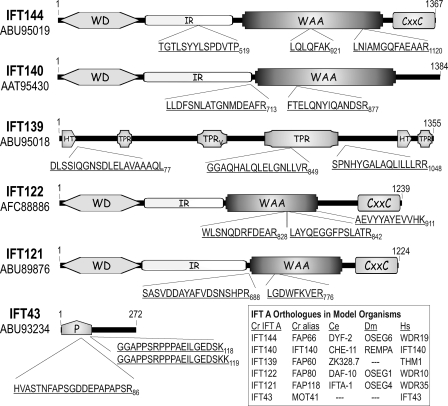
As isolated from flagella, the 16 S IFT complex A contains six protein subunits that have been identified as IFT144, IFT140, IFT139, IFT122, IFT121, and IFT43 (20, 24, 25, 29). To identify the genes that encode the IFT A proteins, each of the subunits was separated from the others using either one- or two-dimensional gel electrophoresis of sucrose density gradient-purified IFT protein. In the case of IFT43, an anti-IFT139 resin was used to pull down and, thus, concentrate complex A from a 16 S fraction of a sucrose density gradient; subsequent low pH elution of IFT A from the immune resin resulted in pure IFT protein with minimal IgG protein present (supplemental Fig. S1). Each of the excised complex A proteins was trypsinized with the resulting peptides analyzed by mass spectrometry and, with the exception of IFT43, subjected to N-terminal Edman sequencing; precise masses for select IFT43 peptides were generated using MALDI-TOF mass spectrometry. The resulting IFT A peptides corresponded to the predicted Chlamydomonas proteins as summarized in Fig. 1. After extensive sequencing of corresponding Chlamydomonas cDNA clones that were amplified from total cDNA or obtained from the Kazusa DNA Research Institute, the cDNA, and protein sequences were submitted to GenBankTM; accession numbers for the encoded proteins are identified in Fig. 1.
FIGURE 1.
Chlamydomonas IFT A protein analysis. Sucrose density gradient-purified IFT A proteins were separated by electrophoresis with isolated tryptic peptides analyzed by Edman degradation as described under “Experimental Procedures.” IFT43 was excised from an SDS-PAGE gel of immunopurified IFT A (see supplemental Fig. S1) prior to trypsinization and MALDI-TOF mass spectroscopy. Three peptide masses resulting from the IFT43 analysis, 2122.9794, 1874.9612, and 2003.0562 Da, matched predicted masses of three peptide sequences, HVASTNFAPSGDDEPAPAPSR86, GGAPPSRPPPAEILGEDSK118, and GGAPPSRPPPAEILGEDSKK119, respectively, that came from a single ORF in the Chlamydomonas genome. The third IFT43 peptide resulted from an incomplete cleavage at Lys119. WD, WD repeat domain; IR, intervening region; WAA, degenerate TPR-like repeats; CXXC, Cys-Xaa-Xaa-Cys repeat domain; TPR, tetratricopeptide repeats; TPRV, TPR domain present in vertebrate THM1/IFT139; HT, half of a tetratricopeptide repeat; P, proline-rich domain.
Five of the six IFT A proteins were present in the Chlamydomonas flagellar proteome and, with the exception of IFT140, were given FAP designations; IFT43 was not identified in the extensive proteomic analysis (67). With the exception of IFT43 and IFT139, the IFT A proteins are well conserved in ciliated model organisms (Fig. 1, inset). And with the exception of IFT43, each of the complex A proteins has previously been experimentally linked to IFT (28, 46, 68–73). In the present study, biochemical evidence demonstrates that all six of these proteins belong to a common complex.
Predicted Domain Organization of IFT A Proteins
Sequence analysis of the IFT A proteins reveals that four share a very similar domain organization (Fig. 1). Like IFT172 of IFT B (35), four of the Chlamydomonas IFT A proteins contain N-terminal WD domains followed by an undefined intervening region and a domain containing irregularly spaced TPR-like degenerate repeats that were originally termed WAA repeats due to a conserved WXXAXXXA motif found in most of the IFT172 repeats. Although many of the degenerate repeats in the IFT A subunits show variation in the WXXAXXXA motif, we still refer to the motifs as WAA repeats as noted previously for the nematode IFT122 orthologue, DAF-10 (74). Within the C-terminal 200 amino acids of three of the WD-WAA subunits can be found multiple CXXC motifs. Like the β-subunit of heterotrimeric G-proteins, the IFT WD domains are expected to form β-propeller structures that are likely to mediate protein-protein interactions. The structures and functions of the intervening region, WAA, and CXXC domains are unknown. IFT139 contains tetratricopeptide repeats but only one domain (amino acids 767–1035) appears to contain enough sequential repeats (at least 5) to form a minimum of one TPR binding pocket similar to that characterized for other TPR proteins including protein phosphatase 5 (75). A putative central TPR domain (TPRV) containing three repeats is predicted for the human and fish (Danio rerio) IFT139 but the degenerate motif is not apparent in the Chlamydomonas sequence at this position (amino acids 531–635). Although IFT43 does not appear to contain any putative structural domains, the N-terminal half is rich in proline and is predicted by the PONDR-FIT algorithm (76) to likely be disordered if not bound to another protein (supplemental Fig. S2).
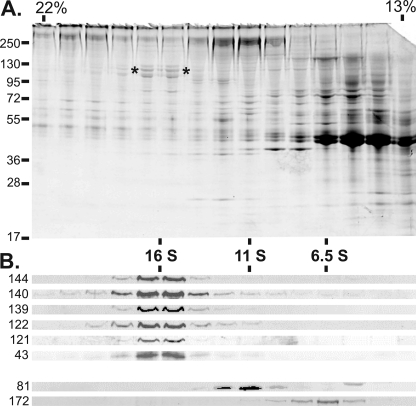
To generate IFT A-specific probes, polyclonal antisera were generated against each of the IFT A subunits with the exception of IFT139 for which a monoclonal antibody (139.1) had been previously generated (20). Using these anti-IFT A antibodies, we analyzed flagellar membrane plus matrix that was fractionated over a sucrose density gradient. As described previously (34) and shown in Fig. 2, centrifugation of membrane plus matrix in the presence of moderately strong ionic strength (300 mm NaCl in HMDEK buffer) results in a 16 S peak of proteins known as the IFT particle-derived complex A. Corresponding immunoblots probed with the anti-IFT A antibodies showed that all six of the complex A subunits sediment together, which verified their strong association with the A complex. As described previously, the algal A and B complexes do not associate with one another under these conditions and the B complex partially dissociates (34) as evidenced by the differential sedimentation of the IFT B core (IFT81) and the monomeric IFT172 at ∼11 and 6.5 S, respectively (Fig. 2).
FIGURE 2.
The IFT A complex is stable to increased ionic strength. The membrane plus matrix proteins were extracted from the flagella isolated from 16 liters of fla2 cells and fractionated through a 13-ml 10–25% sucrose density gradient (SW41Ti) in HMDEK buffer containing 300 mm NaCl. A, fractions 7–21 (21.5–13% sucrose) were resolved on a 7.5–15% acrylamide SDS-PAGE gel and stained with Coomassie Blue; the protein streaking visible in the upper half of the gel was due to the high salt present in the gradient fractions. The asterisks (*) denote the higher molecular mass IFT A subunits found at ∼16 S. Sucrose concentrations are shown at the top, whereas the positions of protein mass standards are indicated on the left. B, immunoblot analysis of fractions 7–21. Antibodies to specific IFT subunits are labeled on the left. All six of the IFT A subunits co-sediment at ∼16 S under these conditions. The anti-IFT81 (81.1) indicates the ∼11 S location of the IFT complex B core; the 6.5 S peak of IFT172 (172.1) serves as an example of a peripheral B component that dissociates from the B core in the presence of 300 mm NaCl.
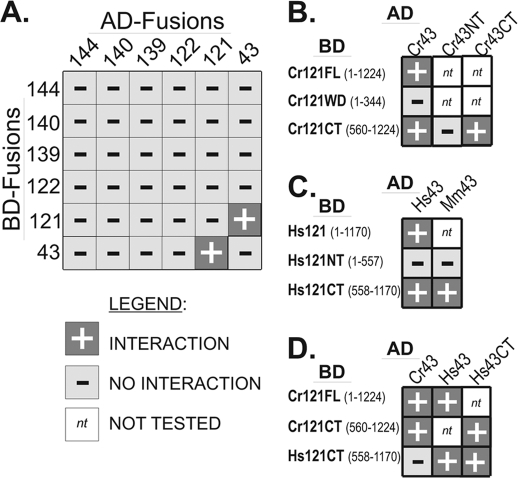
Exhaustive Yeast-based Two-hybrid Analysis Reveals an IFT121-IFT43 Interaction
To rapidly screen for interactions between IFT A subunits, we employed a pooled approach that involved mating an AD-vector based “library” of all six IFT A proteins with a BD-vector based “bait” pool that also represented all six IFT A proteins. This batch approach allowed for the simultaneous screening of all possible pairwise interactions, including homodimeric associations, within complex A. Only one strong interaction, that between IFT121 and IFT43, allowed the host yeast to survive multiple replatings on selective medium (Fig. 3A). To confirm the interaction, yeast were co-transformed with all combinations of IFT121 and IFT43 within the AD and BD vectors with and without appropriate control vectors. The combination of BD-IFT43 with the control vector that expresses only the AD protein (pADMCS) did allow some growth on selective media (data not shown); thus, all subsequent yeast-based analysis of IFT43 utilized only the AD-IFT43 vector.
FIGURE 3.
Directed yeast-based two-hybrid analysis reveals an IFT121-IFT43 interaction. IFT A proteins were fused with either the AD or BD of the GAL4 transcriptional activator and assayed for interaction in yeast. A, exhaustive two-hybrid analysis was carried out using all six of the Chlamydomonas IFT A proteins. All possible combinations were tested by mating a library of AH109 yeast cells with a library of Y187 cells. The AH109 library represented a mixture of cells that contained all six AD-IFT A fusions, whereas the Y187 library represented a mixture of cells that contained all six of the BD-IFT A fusions. Using this approach, only IFT43 and IFT121 showed an interaction. B, testing for interactions between the Chlamydomonas IFT43 and portions of IFT121 revealed that the N-terminal WD40 domain of IFT121 was neither sufficient nor required for the interaction with IFT43. C, the human and murine IFT43 interacted with the C-terminal half but not the N-terminal half of human IFT121. D, the Chlamydomonas IFT43 failed to interact with the C-terminal half of human IFT121 but human IFT43 did interact with both the human and algal IFT121. Interactions are denoted by +; no interaction is denoted by −; not tested is denoted by nt.
In silico analysis of the deduced amino acid sequences of IFT121 and IFT122 revealed similar domain organization for both proteins. Like IFT172, each protein possesses an ∼345 amino acid N-terminal WD40 domain followed by a poorly defined spacer region of several hundred amino acids and a “WAA” domain containing TPR-like degenerate repeats and a C-terminal domain with CXXC motifs (35). The deduced amino acid sequences of IFT121 and IFT122 from a range of organisms display a high level of sequence identity; comparisons of the algal and the orthologous mammalian proteins, WDR35 and WDR19/IFT122, revealed ∼50% amino acid identity for the full-length proteins. In contrast, IFT43 is poorly conserved and may not be present in all ciliated organisms. Because WD domains often serve to mediate protein-protein binding, we tested for a potential interaction between IFT43 and the WD domain of IFT121 (IFT121WD) but saw no interaction in the yeast-based assay (Fig. 3B). An interaction between IFT43 and the C-terminal half of IFT121 (Cr121CT), however, was observed. Thus, the WD domain of IFT121 was neither sufficient nor necessary to mediate the interaction with IFT43.
To test for a similar interaction between the orthologous mammalian IFT A proteins, the human and murine IFT43 were assayed against the human IFT121. As with the Chlamydomonas proteins, both the human IFT43 (Hs43) and the murine IFT43 (Mm43) showed interaction with the C-terminal half of the human IFT121 (Hs121CT) but neither interacted with the WD-containing N-terminal half of IFT121 (Fig. 3C). The poorly conserved nature of IFT43 raised speculation against the possible interaction of the algal and mammalian IFT A subunits. Although the algal AD-CrIFT43 did not interact with the human BD-HsIFT121CT, the human AD-HsIFT43 did interact with the algal BD-CrIFT121, suggesting at least a partial conservation of the interaction surfaces of these two IFT A proteins (Fig. 3D).
Heterologous Expression in Bacteria Confirms an IFT121-IFT43 Interaction
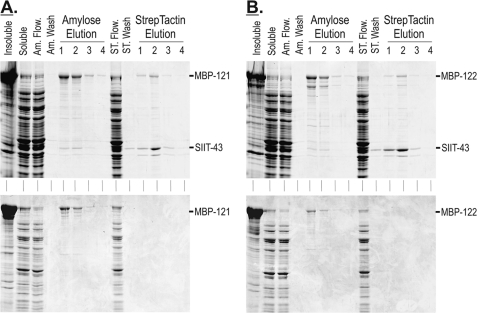
To verify the interactions observed with the yeast two-hybrid analyses, we utilized multiple expression plasmids in E. coli to co-express IFT A proteins. The larger IFT subunits were cloned into the expression vector, pMalC2X; proteins expressed from this plasmid incorporated an N-terminal fusion of the MBP, which served to increase solubility and subsequent yield of the large IFT A proteins. Constructs encoding the full-length IFT43 protein were cloned into MCS2 of pRXS, which incorporated an N-terminal Strep·TagII peptide. The MBP and Strep·TagII fusions allowed for parallel or tandem affinity chromatography using amylose and StrepTactin resins, respectively. As shown in Fig. 4A, MBP-tagged IFT121 (MBP-121) was co-expressed with Strep·TagII-tagged IFT43 (SIIT-43) with the resulting soluble proteins subjected to parallel affinity chromatography. The soluble MBP-tagged IFT121 bound to and was specifically eluted from the amylose resin in the presence (co-expression) or absence (control expression; no SIIT fusion) of IFT43. When co-expressed, a fraction of the total soluble IFT43 co-eluted with the MBP-121 from the amylose resin. In parallel, StrepTactin chromatography was used to selectively purify the soluble SIIT-43; a fraction of the soluble MBP-121 co-eluted with the IFT43. Because the MBP-121 did not bind the StrepTactin resin in the absence of SIIT-43, this verified the interaction between IFT121 and IFT43.
FIGURE 4.
Recombinant IFT43 co-purifies with recombinant IFT121 and IFT122. A, upper panel, parallel purification of co-expressed MBP-121 and SIIT-43 using amylose and StrepTactin chromatographies. A, lower panel, control expression of MBP-121 shows that MBP-121 did not bind to and elute from the StrepTactin resin when expressed in the absence of SIIT-IFT43. B, upper panel, parallel purification of co-expressed MBP-122 and SIIT-43 using amylose and StrepTactin chromatographies. B, lower panel, control expression of MBP-122 shows that MBP-122 did not bind to and elute from the StrepTactin resin when expressed in the absence of SIIT-43. Insoluble, insoluble bacterial proteins; soluble, soluble extract (column load); Am. Flow., amylose resin flow-through; Am. Wash, amylose resin final wash; ST Flow., StrepTactin resin flow-through; ST Wash, StrepTactin resin final wash.
To test for possible interactions between IFT43 and the remaining IFT A subunits, the SIIT-43 was co-expressed with MBP fusions of IFT144, IFT140, IFT139, and IFT122. The only additional interaction observed was between IFT122 and IFT43 as shown in Fig. 4B. As with IFT121, a fraction of the SIIT-43 co-purified with MBP-122 using amylose chromatography and a fraction of the MBP-122 copurified with the SIIT-43 but not the control expression when using StrepTactin chromatography. Because these results suggested that IFT43 was capable of interacting with IFT122, we turned to analysis of IFT A mutants to address how the loss of IFT121 or IFT122 would affect in vivo interactions of complex A.
Loss of IFT121 or IFT122 Disrupts Ciliogenesis
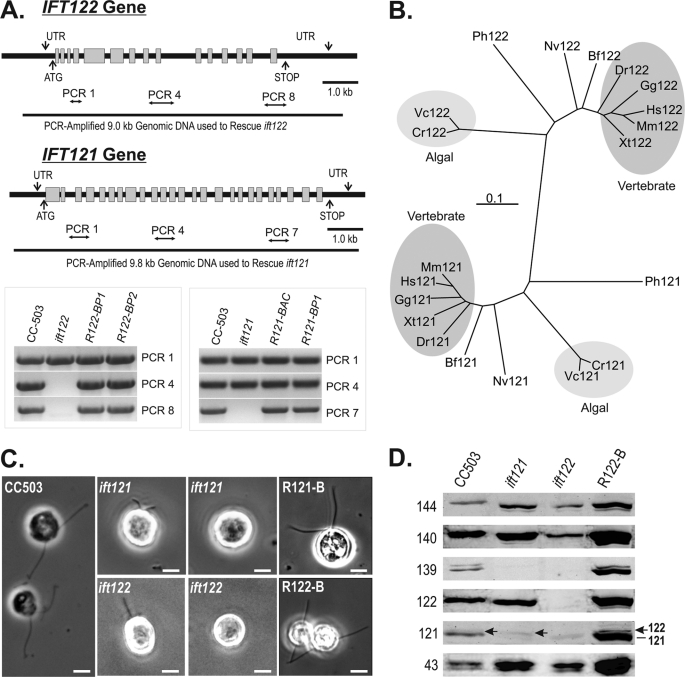
To identify functional aspects of IFT A, an insertional motility mutant library (52) was screened for disruptions in the IFT A genes. Two mutant strains were identified, ift121 and ift122, that carried significant interruptions in the IFT121 and IFT122 genes, respectively (Fig. 5A). PCR-based analysis of genomic DNA demonstrated that ∼⅓ of the 3′ end of the IFT121 gene was disrupted in the ift121 mutant, whereas greater than half of the IFT122 gene was disrupted in the ift122 mutant. Morphological examination of the parental (CC-503) and mutant strains showed a severe effect on flagellar assembly; most mutant cells were bald while a subset assembled short flagella (Fig. 5C). Immunostaining of ift121 and ift122 cells with anti-IFT172 (172.1) and anti-FLA10N revealed cytoplasmic pooling of kinesin-2 and IFT B near the base of the organelle similar to previous studies of wild-type cells (20) (supplemental Fig. S3). In the fraction of ift121 cells that generated short flagella, enrichment of IFT172 and, to a lesser extent FLA10, was frequently observed near the distal end of the organelle (supplemental Fig. S3). Similar modest enrichments were also observed near the tips of the ift122 flagella but for both mutant strains, the majority of the FLA10 and IFT172 staining was found in the cytoplasmic pools near the flagellar base.
FIGURE 5.
The loss of Chlamydomonas IFT121 or IFT122 results in distinctive disruptions to IFT complex A, both of which severely disrupt ciliogenesis. A, genomic maps and PCR analysis of genomic DNA isolated from wild-type, mutant, and rescued strains. R122-BP1, R122-BP2, and R121-BP1 represent mutant strains that were rescued with PCR-amplified genomic DNA, whereas R121-BAC represents an ift121 mutant strain that was rescued using intact BAC DNA. B, taxonomic relatedness of the IFT122 and IFT121 gene products. Cr, C. reinhardtii; Vc, Volvox carteri; Hs, Homo sapiens; Mm, Mus musculus; Gg, Gallus gallus; Xt, Xenopus tropicalis; Dr, Danio rerio; Bf, Branchiostoma floridae; Nv, Nematostella vectensis; Ph, Pediculus humanus corporis. C, phase-contrast images of ift121 and ift122 strains reveal cells with short or no flagella; >95% of ift121 and ift122 rescued with isolated BAC DNA containing IFT121 or IFT122 or PCR-amplified genomic fragments of the IFT121 and IFT122 genes, respectively, recovered assembly of full-length flagella. D, the levels of IFT A proteins extracted from 2 × 106 cells are measured using immunoblots. Soluble protein extracts from whole Chlamydomonas cells were resolved by SDS-PAGE on 10% polyacrylamide gels and transferred to nitrocellulose membrane before probing with antibodies against each of the six IFT A proteins. R122-B represents an ift122 mutant strain in which flagellar assembly was rescued using intact BAC DNA; R121-B represents an ift121 mutant strain rescued using PCR-amplified genomic IFT121.
To assign the flagellar assembly phenotype to disruptions in the IFT121 and IFT122 genes, the ift121 and ift122 mutant strains were complemented with BAC clones that included IFT121 or IFT122 genes, respectively. Because the BAC clones tend to be large genomic fragments that contain multiple genes, the specific IFT121 and IFT122 genes (plus 1–1.5 kb of 5′ and 3′ noncoding DNA) were PCR-amplified and transformed into the appropriate mutant strains and determined to be sufficient to rescue the flagellar assembly phenotype (Fig. 5C, R121-B and R122-B). PCR amplification of genomic DNA isolated from rescued strains verified that the IFT121 and IFT122 genes had been reintroduced into ift121 and ift122 mutant strains, respectively (Fig. 5A, gel inset). In light of the report that a second copy of the genomic IFT144 and IFT139 were capable of complementing the temperature-sensitive IFT A mutants, fla15 (ift139) and fla17 (ift144), respectively (72), we tested the ability of the IFT122-containing BAC to rescue the ift121 mutant and the ability of the IFT121-containing BAC to rescue the ift122 mutant; no complementation was achieved in this manner.
To determine how the loss of IFT121 and IFT122 affected IFT A protein levels, total protein from 2 × 106 whole cells were resolved by SDS-PAGE, transferred to nitrocellulose, and probed with IFT A subunit-specific antibodies (Fig. 5D). As expected, the parental strain (CC-503) contained all six complex A components. The ift121 mutant strain was deficient in IFT121 and IFT139 but contained normal levels of the remaining four IFT A proteins. The polyclonal anti-IFT121 detected a second band just above the IFT121 (Fig. 5D, arrow), which is believed to be IFT122 as the cross-reactive band disappeared in the ift122 mutant. The ift122 mutant, in contrast, was missing IFT121 and IFT139 in addition to the IFT122. The full set of IFT A proteins were restored in the complemented ift121 and ift122 mutant strains as shown for the BAC-rescued ift122 strain (R122-B) in Fig. 5D. Together these results indicate that the presence of IFT139 required IFT121, whereas the presence of IFT121 was dependent on IFT122.
IFT122, IFT140, and IFT144 Form a 12 S Subcomplex
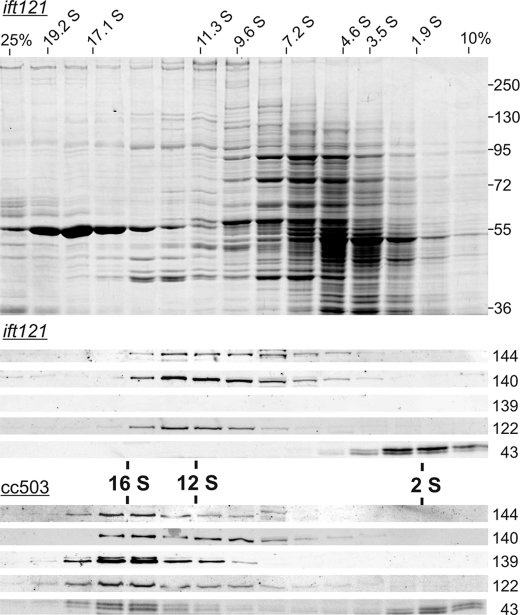
To determine how the loss of IFT121 and IFT139 affected the rest of the IFT A complex, soluble protein was isolated from the ift121 mutant and fractionated over a sucrose density gradient. In the absence of IFT121 (and IFT139), IFT144, IFT140, and IFT122 were found to co-sediment in a broad range that peaked at ∼12 S, whereas all of the IFT43 peaked near the top of the gradient at ∼2 S (Fig. 6). This is in contrast to the IFT A proteins isolated from the parental strain (CC-503) that also exhibited a fairly broad sedimentation, which clearly peaked at ∼16 S with approximately half of the IFT43 present in the 16 S fractions and the other half present near the top of the gradient. It is interesting to note that the 2 S and 16 S IFT43 ran on the SDS-PAGE as a doublet, which is consistent with a previous phosphoproteomic analysis that showed that a fraction of the flagellar form of Chlamydomonas IFT43 is phosphorylated (77). The presence of the IFT43 doublet in the ift121 mutant suggests that this phosphorylation is not dependent on the association of IFT43 with complex A. As expected, IFT121 (not shown) and IFT139 were not detected in the ift121 gradient.
FIGURE 6.
Co-sedimentation of an IFT A subcomplex extracted from ift121 cells. Whole cell extracts were fractionated on 5.0-ml 10–25% sucrose density gradients. Top, Coomassie-stained 8% polyacrylamide SDS-PAGE gel of ift121 fractionation with sedimentation standard S values shown ranging from 1.9 to 19.2 S. For reference, the prominent ∼55 kDa protein sedimenting at ∼19 S is the large subunit of RuBisCo. Middle, Western blots corresponding to the ift121 fractionation were probed with anti-IFT A antibodies. IFT144, IFT140, and IFT122 co-sediment with a peak of ∼12 S, whereas IFT43 peaks at ∼2 S; IFT121 (not shown) and IFT139 were absent from the extract. Bottom, corresponding Western blots of a sucrose density gradient fractionated whole cell extract from the parental strain, CC-503. Each of the IFT A subunits (IFT121 is not shown) co-sediment at 16–17 S; a fraction of IFT43, however, sedimented at ∼2 S.
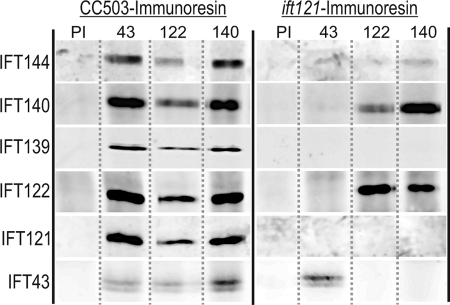
To determine whether the 12 S IFT A proteins observed in the ift121 gradient were, indeed, associated in a complex, whole cell extracts were immunoadsorbed using specific anti-IFT A antibody resins. Following extensive washing of the antibody resins, specifically bound protein was eluted with 1% SDS and then analyzed by SDS-PAGE and immunoblot analysis (Fig. 7). As expected, the parental strain, CC-503, contained the intact IFT A complex so that all six A components were pulled down using antibody resins specific for IFT43, IFT122, IFT139, and IFT140. This confirmed that at least a fraction of each of the six IFT A proteins participates in the formation of the IFT A complex. The control resin containing the combined antibodies made from a mixture of the anti-IFT A preimmune sera failed to bring down any of the complex A proteins. In agreement with the ift121 sucrose density gradient analysis, the anti-IFT122 and anti-IFT140 were both able to pull down a set of three IFT A proteins, IFT122, IFT140, and IFT144; no IFT139 or IFT121 was pulled down from these extracts. Although the IFT43 was present, it was only present on the anti-IFT43 resin, confirming the gradient observation that, in the absence of IFT121 (and IFT139), IFT43 is no longer associated with complex A. In a similar analysis of the ift122 mutant, the anti-IFT140 resin pulled down a small amount of IFT140 and IFT144, suggesting that in the absence of IFT122 (and IFT121 and IFT139), IFT140 and IFT144 are capable of forming a smaller, albeit apparently less stable, subcomplex (not shown).
FIGURE 7.
Antibody pull-downs of IFT A reveal formation of a subcomplex in ift121. Aliquots of soluble protein from whole cell extracts of CC-503 or ift121 were incubated with preimmune (PI) or subunit-specific antibody resins as indicated above each column. Immunoadsorbed proteins were resolved by SDS-PAGE and transferred to nitrocellulose and probed with anti-IFT A antibodies indicated to the left of each row.
DISCUSSION
Specific Interactions of IFT A and Formation of IFT A Core
We present here the first intensive analysis of the spatial organization and architecture of IFT complex A. With mass spectrometry, we verify the identity of all six IFT A gene products, including IFT43 (Fig. 1). We find that when extracted from either whole cells (Fig. 6) or cell bodies (data not shown), all six IFT A proteins co-sediment as a 16 S complex but that a fraction of IFT43 is unassociated with IFT A and is found to sediment at ∼2 S. This differs from flagellar isolations where essentially all of the IFT43 appears to be associated with IFT A. The cellular fraction of 2 S IFT43 also persists in the ift121 and ift122 mutants where complex A is severely compromised. The existence of this non-A-associated fraction of IFT43 suggests that this protein may play roles in IFT A-independent functions.
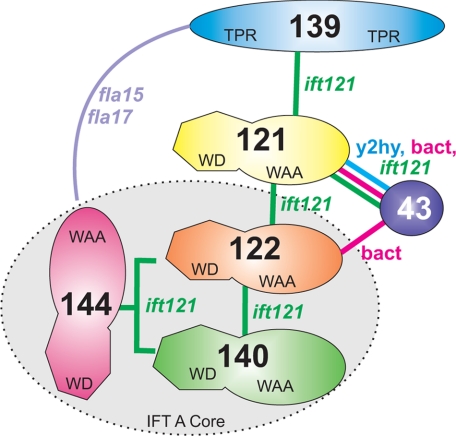
Using yeast-based two-hybrid analysis (Fig. 3) and recombinant bacterial co-expression (Fig. 4), we have established that the C-terminal half of IFT121 directly interacts with the C-terminal half of IFT43. Using the ift121 mutant, we have identified a stable, 12 S IFT A subcomplex containing IFT144, IFT140, and IFT122 (Fig. 6). Because these three IFT A components can associate without the remaining three subunits, we refer to this subcomplex as the IFT A core. The composition of the Chlamydomonas IFT A core is consistent with a similar subcomplex generated in mammalian cell cultures using selective RNAi knockdowns of IFT A gene expression (73), which suggests that the architecture of complex A is well conserved. The presence of a minor component of IFT144 and IFT140 in the ∼8 S fraction of ift121 extracts further suggests that IFT144 and IFT140 are capable of interacting directly in the absence of other IFT A components. Last, because IFT139 is essentially absent in the ift121 strains, we conclude that the presence of IFT121 is required to assemble IFT139 as well as IFT43 onto the A complex. Thus, IFT121 may interact directly with IFT139 as suggested in the IFT A interaction model diagrammed in Fig. 8. Included in this model is a complex interaction of IFT139 and IFT144 uncovered by genetic analysis (72). In brief, double gene dosage of IFT139 is capable of rescuing a temperature-sensitive mutation in IFT144, whereas a double gene dosage of IFT144 is capable of rescuing a temperature-sensitive mutation in the IFT139 gene. The molecular mechanism underlying this complementation is not understood but it should be noted that the temperature-sensitive phenotypes in fla15 and fla17–1 result from a single amino acid substitution and a minor in-frame deletion, respectively, which still allow protein expression. It is possible that null mutants in IFT144 and IFT139 would not be susceptible to this sort of complementation by another IFT A gene. In light of the Iomini et al. (72) result, we did attempt rescue of the ift121 and ift122 mutants with genomic fragments containing the IFT122 and IFT121 genes, respectively, but were unable to restore flagellar assembly. No attempts were made to complement these mutant strains using any other IFT A genes.
FIGURE 8.
Interaction model of interactions within the Chlamydomonas IFT A complex. The model shown summarizes the IFT A subunit interactions that have been directly visualized, such as IFT121-IFT43, or implied, such as IFT139-IFT121. Based on biochemical analysis of the ift121 mutant, we propose that IFT122, IFT140, and IFT144 assemble to create a heterotrimeric stable core complex. The interactions of IFT121 with both IFT139 and IFT43 are supported by the observation that loss of IFT121 completely removes IFT139 and IFT43 from complex A. Last, IFT139 and IFT144 demonstrate a complex genetic interaction that suggests a functional association (72).
IFT A Genes Are Ciliogenic but Functionally Distinct from IFT B Genes
Functional analyses of IFT A genes in various model organisms have shown that IFT A plays a role in ciliogenesis but disruptions in IFT A genes often allow for the assembly of flagella, albeit often shorter than wild-type organelles, whereas disruptions in IFT B genes often result in a more severe blocking of ciliogenesis that results in little or no assembly of the organelle (25, 27, 30, 31, 43, 44, 48, 68, 72, 250). These differences indicate that, although they both participate in intraflagellar transport, IFT A and B have functional responsibilities that do not completely overlap. One common feature of many IFT A mutant phenotypes is the accumulation of IFT B and other proteins at or near the distal ends of the cilia, which are often shorter than in wild-type cells. Because this sort of behavior suggests an unbalanced ratio of anterograde to retrograde transport, IFT A has been associated with retrograde transport of materials out of the organelle (28, 47, 48, 70–72, 79). Although the molecular basis for these disruptions in retrograde transport is not clear, elegant analysis of Chlamydomonas mutants with defective IFT A components, IFT144, and IFT139 revealed reduced rates of retrograde IFT, whereas anterograde IFT remains essentially normal; the unbalanced transport results in ciliary accumulations of IFT B components (27, 72). One functional role for IFT A could be to efficiently associate the IFT particles with the retrograde IFT dynein 1b/2. Alternatively, IFT A could be responsible for efficient turn around of materials at the ciliary tip. Another possibility, however, is that IFT A may be responsible for the anterograde transport of essential retrograde-associated components into the organelle. A recent example of an IFT A-mediated anterograde transport was demonstrated by Mukhopadhyay and colleagues (73) who showed that knockdown of IFT A in mammalian cells reduced the transport of TULP3 and associated G-protein receptors into the organelle. The idea of IFT A-specific and/or IFT B-specific entry into and exit out of the organelle is suggested by a recent study of the Chlamydomonas CEP290 gene (80, 81). In the absence of the transition zone protein, CEP290, IFT B accumulates in the flagella, whereas IFT A levels are reduced. The concomitant reduction of IFT A and other flagellar proteins, such as adenylate kinase and FAP12, is consistent with the idea that some of these proteins may be IFT A-specific cargos.
IFT A and Human Ciliopathies
Genes encoding five of the six IFT A proteins have recently been associated with human ciliopathies where many of the disease-causing mutations result in single amino acid substitutions or small deletions in IFT43, IFT121, IFT122, IFT139, or IFT144 (82–86). A complete loss of an IFT A subunit, such as that observed with a murine IFT122 knock-out strain shows that the loss of the IFT A complex is likely to be embryonic lethal in humans (49, 87). Based on our results and the recent mammalian IFT A studies (77), it is likely that complex A is severely disrupted in the IFT122 knock-out strain; such a disruption would explain why the cilia in these mice are considerably shorter than normal (incomplete assembly) and engorged (defective in retrograde transport). Thus, mutations resulting in more subtle phenotypes are likely required to generate disease states that allow full gestation and live birth. The mutations in the IFT A genes in some patients suffering from Sensenbrenner syndrome, for example, result in single amino acid substitutions in conserved residues of IFT122 and IFT121 (79, 82). Although these mutations appear to affect ciliary function, it seems unlikely that they block assembly of complex A (88). A mutation in the initiation codon of IFT43 in one family of Sensenbrenner patients results in aberrant translation of two IFT43 isoforms that are missing the N-terminal 21 amino acids and differ from one another due to splice variation (84). According to our interaction results, these changes in the mutant IFT43 proteins should not prevent it from interacting with IFT121. Last, it is interesting to note that we show a close physical association of IFT121 with IFT43 and IFT122, which have all been associated with Sensenbrenner syndrome.
Future Studies
There are very compelling reasons to pursue structural and functional analyses of IFT A. First, functional studies to date indicate that IFT A is responsible for tasks that are separate from IFT B; a complete understanding of this complex transport process will require a thorough elucidation of the functions attributable to both A and B. Second, mutations in no less than four of the six IFT A genes give rise to human disease states but the molecular mechanism(s) underlying these disease states remains a mystery. Last, as has been proven for so many complex molecular machines, such as the ribosome or the pyruvate dehydrogenase complex, understanding the architectural structure of the machine and the molecular structure of the individual components is essential to understanding the mechanism(s) by which the machine works. Thus, it is now critically important to solve high resolution structures and determine specific functions of the IFT A components.
Supplementary Material
Acknowledgments
We are grateful to Dr. John Leszyk (University of Massachusetts Medical School) for expert analysis using MALDI-TOF mass spectrometry Edman protein sequencing. We are grateful to Ann Norton and the University of Idaho Imaging Center for assistance with microscopic imaging. We thank all members of the Cole laboratory who have provided critical feedback and technical assistance. Last, we are especially grateful to Dr. Joel Rosenbaum for providing guidance, encouragement, and facilities during the early stages of our proteomic analysis.
This work was supported, in whole or in part, by National Institutes of Health Grant R01-GM61920 (to D. G. C.) and INBRE Program of the National Center for Research Resources Grant P20-RR016454.

This article contains supplemental Table S1 and Figs. S1–S3.
- IFT
- intraflagellar transport
- AD
- GAL4 activation domain
- BD
- GAL4 DNA binding domain
- MBP
- maltose-binding protein
- PI
- protease inhibitor
- MCS
- multicloning site.
REFERENCES
- 1. Afzelius B. A. (1995) Role of cilia in human health. Cell Motil. Cytoskeleton. 32, 95–97 [DOI] [PubMed] [Google Scholar]
- 2. Vincensini L., Blisnick T., Bastin P. (2011) 1001 model organisms to study cilia and flagella. Biol. Cell 103, 109–130 [DOI] [PubMed] [Google Scholar]
- 3. Corbit K. C., Aanstad P., Singla V., Norman A. R., Stainier D. Y., Reiter J. F. (2005) Vertebrate Smoothened functions at the primary cilium. Nature 437, 1018–1021 [DOI] [PubMed] [Google Scholar]
- 4. Eggenschwiler J. T., Anderson K. V. (2007) Cilia and developmental signaling. Annu. Rev. Cell Dev. Biol. 23, 345–373 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Berbari N. F., O'Connor A. K., Haycraft C. J., Yoder B. K. (2009) The primary cilium as a complex signaling center. Curr. Biol. 19, R526–535 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Badano J. L., Mitsuma N., Beales P. L., Katsanis N. (2006) The ciliopathies. An emerging class of human genetic disorders. Annu. Rev. Genomics Hum. Genet. 7, 125–148 [DOI] [PubMed] [Google Scholar]
- 7. Marshall W. F. (2008) The cell biological basis of ciliary disease. J. Cell Biol. 180, 17–21 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Cardenas-Rodriguez M., Badano J. L. (2009) Ciliary biology: understanding the cellular and genetic basis of human ciliopathies. Am. J. Med. Genet. C Semin. Med. Genet. 151C, 263–280 [DOI] [PubMed] [Google Scholar]
- 9. Baker K., Beales P. L. (2009) Making sense of cilia in disease: the human ciliopathies. Am. J. Med. Genet. C Semin. Med. Genet. 151C, 281–295 [DOI] [PubMed] [Google Scholar]
- 10. Brugmann S. A., Cordero D. R., Helms J. A. (2010) Craniofacial ciliopathies: a new classification for craniofacial disorders. Am. J. Med. Genet. A 152A, 2995–3006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Hildebrandt F., Benzing T., Katsanis N. (2011) Ciliopathies. N. Engl. J. Med. 364, 1533–1543 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Lee L. (2011) Mechanisms of mammalian ciliary motility. Insights from primary ciliary dyskinesia genetics. Gene 473, 57–66 [DOI] [PubMed] [Google Scholar]
- 13. Rosenbaum J. L., Witman G. B. (2002) Intraflagellar transport. Nat. Rev. Mol. Cell Biol. 3, 813–825 [DOI] [PubMed] [Google Scholar]
- 14. Scholey J. M. (2003) Intraflagellar transport. Annu. Rev. Cell Dev. Biol. 19, 423–443 [DOI] [PubMed] [Google Scholar]
- 15. Cole D. G., Snell W. J. (2009) SnapShot. Intraflagellar transport. Cell 137, 784–784.e1 [DOI] [PubMed] [Google Scholar]
- 16. Kozminski K. G., Johnson K. A., Forscher P., Rosenbaum J. L. (1993) A motility in the eukaryotic flagellum unrelated to flagellar beating. Proc. Natl. Acad. Sci. U.S.A. 90, 5519–5523 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Kozminski K. G., Beech P. L., Rosenbaum J. L. (1995) The Chlamydomonas kinesin-like protein FLA10 is involved in motility associated with the flagellar membrane. J. Cell Biol. 131, 1517–1527 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Shakir M. A., Fukushige T., Yasuda H., Miwa J., Siddiqui S. S. (1993) C. elegans osm-3 gene mediating osmotic avoidance behaviour encodes a kinesin-like protein. Neuroreport 4, 891–894 [DOI] [PubMed] [Google Scholar]
- 19. Walther Z., Vashishtha M., Hall J. L. (1994) The Chlamydomonas FLA10 gene encodes a novel kinesin-homologous protein. J. Cell Biol. 126, 175–188 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Cole D. G., Diener D. R., Himelblau A. L., Beech P. L., Fuster J. C., Rosenbaum J. L. (1998) Chlamydomonas kinesin II-dependent intraflagellar transport (IFT). IFT particles contain proteins required for ciliary assembly in Caenorhabditis elegans sensory neurons. J. Cell Biol. 141, 993–1008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Signor D., Wedaman K. P., Rose L. S., Scholey J. M. (1999) Two heteromeric kinesin complexes in chemosensory neurons and sensory cilia of Caenorhabditis elegans. Mol. Biol. Cell 10, 345–360 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Pazour G. J., Dickert B. L., Witman G. B. (1999) The DHC1b (DHC2) isoform of cytoplasmic dynein is required for flagellar assembly. J. Cell Biol. 144, 473–481 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Porter M. E., Bower R., Knott J. A., Byrd P., Dentler W. (1999) Cytoplasmic dynein heavy chain 1b is required for flagellar assembly in Chlamydomonas. Mol. Biol. Cell 10, 693–712 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Piperno G., Mead K. (1997) Transport of a novel complex in the cytoplasmic matrix of Chlamydomonas flagella. Proc. Natl. Acad. Sci. U.S.A. 94, 4457–4462 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Piperno G., Siuda E., Henderson S., Segil M., Vaananen H., Sassaroli M. (1998) Distinct mutants of retrograde intraflagellar transport (IFT) share similar morphological and molecular defects. J. Cell Biol. 143, 1591–1601 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Cole D. G. (2009) Intraflagellar Transport. The Chlamydomonas Sourcebook: Cell Motility and Behavior, 2nd Ed., Vol. 3, Chap. 4, pp. 71–112, Academic Press, San Diego, CA [Google Scholar]
- 27. Iomini C., Babaev-Khaimov V., Sassaroli M., Piperno G. (2001) Protein particles in Chlamydomonas flagella undergo a transport cycle consisting of four phases. J. Cell Biol. 153, 13–24 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Qin H., Rosenbaum J. L., Barr M. M. (2001) An autosomal recessive polycystic kidney disease gene homolog is involved in intraflagellar transport in C. elegans ciliated sensory neurons. Curr. Biol. 11, 457–461 [DOI] [PubMed] [Google Scholar]
- 29. Wang Z., Fan Z. C., Williamson S. M., Qin H. (2009) Intraflagellar transport (IFT) protein IFT25 is a phosphoprotein component of IFT complex B and physically interacts with IFT27 in Chlamydomonas. PLoS ONE 4, e5384. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Pazour G. J., Dickert B. L., Vucica Y., Seeley E. S., Rosenbaum J. L., Witman G. B., Cole D. G. (2000) Chlamydomonas IFT88 and its mouse homologue, polycystic kidney disease gene tg737, are required for assembly of cilia and flagella. J. Cell Biol. 151, 709–718 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Brazelton W. J., Amundsen C. D., Silflow C. D., Lefebvre P. A. (2001) The bld1 mutation identifies the Chlamydomonas osm-6 homolog as a gene required for flagellar assembly. Curr. Biol. 11, 1591–1594 [DOI] [PubMed] [Google Scholar]
- 32. Pazour G. J., Baker S. A., Deane J. A., Cole D. G., Dickert B. L., Rosenbaum J. L., Witman G. B., Besharse J. C. (2002) The intraflagellar transport protein, IFT88, is essential for vertebrate photoreceptor assembly and maintenance. J. Cell Biol. 157, 103–113 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Qin H., Diener D. R., Geimer S., Cole D. G., Rosenbaum J. L. (2004) Intraflagellar transport (IFT) cargo. IFT transports flagellar precursors to the tip and turnover products to the cell body. J. Cell Biol. 164, 255–266 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Lucker B. F., Behal R. H., Qin H., Siron L. C., Taggart W. D., Rosenbaum J. L., Cole D. G. (2005) Characterization of the intraflagellar transport complex B core. Direct interaction of the IFT81 and IFT74/72 subunits. J. Biol. Chem. 280, 27688–27696 [DOI] [PubMed] [Google Scholar]
- 35. Pedersen L. B., Miller M. S., Geimer S., Leitch J. M., Rosenbaum J. L., Cole D. G. (2005) Chlamydomonas IFT172 is encoded by FLA11, interacts with CrEB1, and regulates IFT at the flagellar tip. Curr. Biol. 15, 262–266 [DOI] [PubMed] [Google Scholar]
- 36. Hou Y., Qin H., Follit J. A., Pazour G. J., Rosenbaum J. L., Witman G. B. (2007) Functional analysis of an individual IFT protein. IFT46 is required for transport of outer dynein arms into flagella. J. Cell Biol. 176, 653–665 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Qin H., Wang Z., Diener D., Rosenbaum J. (2007) Intraflagellar transport protein 27 is a small G protein involved in cell-cycle control. Curr. Biol. 17, 193–202 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Lechtreck K. F., Luro S., Awata J., Witman G. B. (2009) HA-tagging of putative flagellar proteins in Chlamydomonas reinhardtii identifies a novel protein of intraflagellar transport complex B. Cell Motil. Cytoskeleton 66, 469–482 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Fan Z. C., Behal R. H., Geimer S., Wang Z., Williamson S. M., Zhang H., Cole D. G., Qin H. (2010) Chlamydomonas IFT70/CrDYF-1 is a core component of IFT particle complex B and is required for flagellar assembly. Mol. Biol. Cell 21, 2696–2706 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Pedersen L. B., Rosenbaum J. L. (2008) Intraflagellar transport (IFT) role in ciliary assembly, resorption, and signalling. Curr. Top. Dev. Biol. 85, 23–61 [DOI] [PubMed] [Google Scholar]
- 41. Blacque O. E., Cevik S., Kaplan O. I. (2008) Intraflagellar transport. From molecular characterization to mechanism. Front. Biosci. 13, 2633–2652 [DOI] [PubMed] [Google Scholar]
- 42. Silverman M. A., Leroux M. R. (2009) Intraflagellar transport and the generation of dynamic, structurally, and functionally diverse cilia. Trends Cell Biol. 19, 306–316 [DOI] [PubMed] [Google Scholar]
- 43. Perkins L. A., Hedgecock E. M., Thomson J. N., Culotti J. G. (1986) Mutant sensory cilia in the nematode Caenorhabditis elegans. Dev. Biol. 117, 456–487 [DOI] [PubMed] [Google Scholar]
- 44. Collet J., Spike C. A., Lundquist E. A., Shaw J. E., Herman R. K. (1998) Analysis of osm-6, a gene that affects sensory cilium structure and sensory neuron function in Caenorhabditis elegans. Genetics 148, 187–200 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Haycraft C. J., Swoboda P., Taulman P. D., Thomas J. H., Yoder B. K. (2001) The C. elegans homolog of the murine cystic kidney disease gene Tg737 functions in a ciliogenic pathway and is disrupted in osm-5 mutant worms. Development 128, 1493–1505 [DOI] [PubMed] [Google Scholar]
- 46. Avidor-Reiss T., Maer A. M., Koundakjian E., Polyanovsky A., Keil T., Subramaniam S., Zuker C. S. (2004) Decoding cilia function. Defining specialized genes required for compartmentalized cilia biogenesis. Cell. 117, 527–539 [DOI] [PubMed] [Google Scholar]
- 47. Absalon S., Blisnick T., Kohl L., Toutirais G., Doré G., Julkowska D., Tavenet A., Bastin P. (2008) Intraflagellar transport and functional analysis of genes required for flagellum formation in trypanosomes. Mol. Biol. Cell 19, 929–944 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Tsao C. C., Gorovsky M. A. (2008) Tetrahymena IFT122A is not essential for cilia assembly but plays a role in returning IFT proteins from the ciliary tip to the cell body. J. Cell Sci. 121, 428–436 [DOI] [PubMed] [Google Scholar]
- 49. Cortellino S., Wang C., Wang B., Bassi M. R., Caretti E., Champeval D., Calmont A., Jarnik M., Burch J., Zaret K. S., Larue L., Bellacosa A. (2009) Defective ciliogenesis, embryonic lethality, and severe impairment of the Sonic Hedgehog pathway caused by inactivation of the mouse complex A intraflagellar transport gene Ift122/Wdr10, partially overlapping with the DNA repair gene Med1/Mbd4. Dev. Biol. 325, 225–237 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Pigino G., Geimer S., Lanzavecchia S., Paccagnini E., Cantele F., Diener D. R., Rosenbaum J. L., Lupetti P. (2009) Electron-tomographic analysis of intraflagellar transport particle trains in situ. J. Cell Biol. 187, 135–148 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Baker S. A., Freeman K., Luby-Phelps K., Pazour G. J., Besharse J. C. (2003) IFT20 links kinesin II with a mammalian intraflagellar transport complex that is conserved in motile flagella and sensory cilia. J. Biol. Chem. 278, 34211–34218 [DOI] [PubMed] [Google Scholar]
- 52. Lucker B. F., Miller M. S., Dziedzic S. A., Blackmarr P. T., Cole D. G. (2010) Direct interactions of intraflagellar transport complex B proteins IFT88, IFT52, and IFT46. J. Biol. Chem. 285, 21508–21518 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Taschner M., Bhogaraju S., Vetter M., Morawetz M., Lorentzen E. (2011) Biochemical mapping of interactions within the intraflagellar transport (IFT) B core complex. IFT52 binds directly to four other IFT-B subunits. J. Biol. Chem. 286, 26344–26352 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Zhao C., Malicki J. (2011) Nephrocystins and MKS proteins interact with IFT particle and facilitate transport of selected ciliary cargos. EMBO J. 30, 2532–2544 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Gorman D. S., Levine R. P. (1965) Cytochrome f and plastocyanin. Their sequence in the photosynthetic electron transport chain of Chlamydomonas reinhardi. Proc. Natl. Acad. Sci. U.S.A. 54, 1665–1669 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Pazour G. J., Witman G. B. (2000) Forward and reverse genetic analysis of microtubule motors in Chlamydomonas. Methods 22, 285–298 [DOI] [PubMed] [Google Scholar]
- 57. Sizova I., Fuhrmann M., Hegemann P. (2001) A Streptomyces rimosus aphVIII gene coding for a new type phosphotransferase provides stable antibiotic resistance to Chlamydomonas reinhardtii. Gene 277, 221–229 [DOI] [PubMed] [Google Scholar]
- 58. Berthold P., Schmitt R., Mages W. (2002) An engineered Streptomyces hygroscopicus aph7" gene mediates dominant resistance against hygromycin B in Chlamydomonas reinhardtii. Protist 153, 401–412 [DOI] [PubMed] [Google Scholar]
- 59. Witman G. B., Carlson K., Berliner J., Rosenbaum J. L. (1972) Chlamydomonas flagella. I. Isolation and electrophoretic analysis of microtubules, matrix, membranes, and mastigonemes. J. Cell Biol. 54, 507–539 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60. Asamizu E., Nakamura Y., Sato S., Fukuzawa H., Tabata S. (1999) A large scale structural analysis of cDNAs in a unicellular green alga, Chlamydomonas reinhardtii. I. Generation of 3433 non-redundant expressed sequence tags. DNA Res. 6, 369–373 [DOI] [PubMed] [Google Scholar]
- 61. Asamizu E., Miura K., Kucho K., Inoue Y., Fukuzawa H., Ohyama K., Nakamura Y., Tabata S. (2000) Generation of expressed sequence tags from low-CO2 and high-CO2 adapted cells of Chlamydomonas reinhardtii. DNA Res. 7, 305–307 [DOI] [PubMed] [Google Scholar]
- 62. Wilkerson C. G., King S. M., Witman G. B. (1994) Molecular analysis of the γ-heavy chain of Chlamydomonas flagellar outer-arm dynein. J. Cell Sci. 107, 497–506 [DOI] [PubMed] [Google Scholar]
- 63. Sambrook J., Fritsch E. F., Maniatis T. (1989) Molecular Cloning: A Laboratory Manual, Wiley-Interscience, New York [Google Scholar]
- 64. Schuler G. D., Altschul S. F., Lipman D. J. (1991) A workbench for multiple alignment construction and analysis. Proteins 9, 180–190 [DOI] [PubMed] [Google Scholar]
- 65. Kathir P., LaVoie M., Brazelton W. J., Haas N. A., Lefebvre P. A., Silflow C. D. (2003) Molecular map of the Chlamydomonas reinhardtii nuclear genome. Eukaryot. Cell. 2, 362–379 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66. Towbin H., Staehelin T., Gordon J. (1979) Electrophoretic transfer of proteins from polyacrylamide gels to nitrocellulose sheets. Procedure and some applications. Proc. Natl. Acad. Sci. U.S.A. 76, 4350–4354 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67. Pazour G. J., Agrin N., Leszyk J., Witman G. B. (2005) Proteomic analysis of a eukaryotic cilium. J. Cell Biol. 170, 103–113 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68. Efimenko E., Blacque O. E., Ou G., Haycraft C. J., Yoder B. K., Scholey J. M., Leroux M. R., Swoboda P. (2006) Caenorhabditis elegans DYF-2, an orthologue of human WDR19, is a component of the intraflagellar transport machinery in sensory cilia. Mol. Biol. Cell 17, 4801–4811 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69. Blacque O. E., Li C., Inglis P. N., Esmail M. A., Ou G., Mah A. K., Baillie D. L., Scholey J. M., Leroux M. R. (2006) The WD repeat-containing protein IFTA-1 is required for retrograde intraflagellar transport. Mol. Biol. Cell 17, 5053–5062 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70. Tran P. V., Haycraft C. J., Besschetnova T. Y., Turbe-Doan A., Stottmann R. W., Herron B. J., Chesebro A. L., Qiu H., Scherz P. J., Shah J. V., Yoder B. K., Beier D. R. (2008) THM1 negatively modulates mouse sonic hedgehog signal transduction and affects retrograde intraflagellar transport in cilia. Nat. Genet. 40, 403–410 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71. Lee E., Sivan-Loukianova E., Eberl D. F., Kernan M. J. (2008) An IFT-A protein is required to delimit functionally distinct zones in mechanosensory cilia. Curr. Biol. 18, 1899–1906 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72. Iomini C., Li L., Esparza J. M., Dutcher S. K. (2009) Retrograde intraflagellar transport mutants identify complex A proteins with multiple genetic interactions in Chlamydomonas reinhardtii. Genetics 183, 885–896 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73. Mukhopadhyay S., Wen X., Chih B., Nelson C. D., Lane W. S., Scales S. J., Jackson P. K. (2010) TULP3 bridges the IFT-A complex and membrane phosphoinositides to promote trafficking of G protein-coupled receptors into primary cilia. Genes Dev. 24, 2180–2193 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74. Bell L. R., Stone S., Yochem J., Shaw J. E., Herman R. K. (2006) The molecular identities of the Caenorhabditis elegans intraflagellar transport genes dyf-6, daf-10, and osm-1. Genetics 173, 1275–1286 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75. Das A. K., Cohen P. W., Barford D. (1998) The structure of the tetratricopeptide repeats of protein phosphatase 5. Implications for TPR-mediated protein-protein interactions. EMBO J. 17, 1192–1199 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76. Xue B., Dunbrack R. L., Williams R. W., Dunker A. K., Uversky V. N. (2010) PONDR-FIT. A meta-predictor of intrinsically disordered amino acids. Biochim. Biophys. Acta 1804, 996–1010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77. Boesger J., Wagner V., Weisheit W., Mittag M. (2009) Analysis of flagellar phosphoproteins from Chlamydomonas reinhardtii. Eukaryot. Cell 8, 922–932 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78. Haycraft C. J., Schafer J. C., Zhang Q., Taulman P. D., Yoder B. K. (2003) Identification of CHE-13. A novel intraflagellar transport protein required for cilia formation. Exp. Cell Res. 284, 251–263 [DOI] [PubMed] [Google Scholar]
- 79. Inglis P. N., Blacque O. E., Leroux M. R. (2009) Functional genomics of intraflagellar transport-associated proteins in C. elegans. Methods Cell Biol. 93, 267–304 [DOI] [PubMed] [Google Scholar]
- 80. Craige B., Tsao C. C., Diener D. R., Hou Y., Lechtreck K. F., Rosenbaum J. L., Witman G. B. (2010) CEP290 tethers flagellar transition zone microtubules to the membrane and regulates flagellar protein content. J. Cell Biol. 190, 927–940 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81. Betleja E., Cole D. G. (2010) Ciliary trafficking. CEP290 guards a gated community. Curr. Biol. 20, R928–931 [DOI] [PubMed] [Google Scholar]
- 82. Gilissen C., Arts H. H., Hoischen A., Spruijt L., Mans D. A., Arts P., van Lier B., Steehouwer M., van Reeuwijk J., Kant S. G., Roepman R., Knoers N. V., Veltman J. A., Brunner H. G. (2010) Exome sequencing identifies WDR35 variants involved in Sensenbrenner syndrome. Am. J. Hum. Genet. 87, 418–423 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83. Walczak-Sztulpa J., Eggenschwiler J., Osborn D., Brown D. A., Emma F., Klingenberg C., Hennekam R. C., Torre G., Garshasbi M., Tzschach A., Szczepanska M., Krawczynski M., Zachwieja J., Zwolinska D., Beales P. L., Ropers H. H., Latos-Bielenska A., Kuss A. W. (2010) Cranioectodermal dysplasia, Sensenbrenner syndrome, is a ciliopathy caused by mutations in the IFT122 gene. Am. J. Hum. Genet. 86, 949–956 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84. Davis E. E., Zhang Q., Liu Q., Diplas B. H., Davey L. M., Hartley J., Stoetzel C., Szymanska K., Ramaswami G., Logan C. V., Muzny D. M., Young A. C., Wheeler D. A., Cruz P., Morgan M., Lewis L. R., Cherukuri P., Maskeri B., Hansen N. F., Mullikin J. C., Blakesley R. W., Bouffard G. G., NISC Comparative Sequencing Program, Gyapay G., Rieger S., Tönshoff B., Kern I., Soliman N. A., Neuhaus T. J., Swoboda K. J., Kayserili H., Gallagher T. E., Lewis R. A., Bergmann C., Otto E. A., Saunier S., Scambler P. J., Beales P. L., Gleeson J. G., Maher E. R., Attié-Bitach T., Dollfus H., Johnson C. A., Green E. D., Gibbs R. A., Hildebrandt F., Pierce E. A., Katsanis N. (2011) TTC21B contributes both causal and modifying alleles across the ciliopathy spectrum. Nat. Genet. 43, 189–196 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85. Arts H. H., Bongers E. M., Mans D. A., van Beersum S. E., Oud M. M., Bolat E., Spruijt L., Cornelissen E. A., Schuurs-Hoeijmakers J. H., de Leeuw N., Cormier-Daire V., Brunner H. G., Knoers N. V., Roepman R. (2011) C14ORF179 encoding IFT43 is mutated in Sensenbrenner syndrome. J. Med. Genet. 48, 390–395 [DOI] [PubMed] [Google Scholar]
- 86. Bredrup C., Saunier S., Oud M. M., Fiskerstrand T., Hoischen A., Brackman D., Leh S. M., Midtbø M., Filhol E., Bole-Feysot C., Nitschké P., Gilissen C., Haugen O. H., Sanders J. S., Stolte-Dijkstra I., Mans D. A., Steenbergen E. J., Hamel B. C., Matignon M., Pfundt R., Jeanpierre C., Boman H., Rødahl E., Veltman J. A., Knappskog P. M., Knoers N. V., Roepman R., Arts H. H. (2011) Ciliopathies with skeletal anomalies and renal insufficiency due to mutations in the IFT-A gene WDR19. Am. J. Hum. Genet. 89, 634–643 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87. Qin J., Lin Y., Norman R. X., Ko H. W., Eggenschwiler J. T. (2011) Intraflagellar transport protein 122 antagonizes Sonic Hedgehog signaling and controls ciliary localization of pathway components. Proc. Natl. Acad. Sci. U.S.A. 108, 1456–1461 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88. Mill P., Lockhart P. J., Fitzpatrick E., Mountford H. S., Hall E. A., Reijns M. A., Keighren M., Bahlo M., Bromhead C. J., Budd P., Aftimos S., Delatycki M. B., Savarirayan R., Jackson I. J., Amor D. J. (2011) Human and mouse mutations in WDR35 cause short-rib polydactyly syndromes due to abnormal ciliogenesis. Am. J. Hum. Genet. 88, 508–515 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.