Background: Linker histones are functionally heterogeneous.
Results: Using a novel FRAP approach, the N-terminal domain modulates binding affinities of H10 and H1c; the C-terminal domain influences the nucleosomal orientation of the globular domain.
Conclusion: The variable terminal domains have distinct roles in the chromatin binding characteristics of linker histones.
Significance: Evidence for a structural basis for the functional heterogeneity of linker histones is presented.
Keywords: Chromatin, Chromatin Structure, Gene Regulation, Histones, Molecular Dynamics, Molecular Imaging, Nucleosome, Fluorescence Recovery after Photobleaching (FRAP), Linker Histones
Abstract
Eukaryotic linker or H1 histones modulate DNA compaction and gene expression in vivo. In mammals, these proteins exist as multiple isotypes with distinct properties, suggesting a functional significance to the heterogeneity. Linker histones typically have a tripartite structure composed of a conserved central globular domain flanked by a highly variable short N-terminal domain and a longer highly basic C-terminal domain. We hypothesized that the variable terminal domains of individual subtypes contribute to their functional heterogeneity by influencing chromatin binding interactions. We developed a novel dual color fluorescence recovery after photobleaching assay system in which two H1 proteins fused to spectrally separable fluorescent proteins can be co-expressed and their independent binding kinetics simultaneously monitored in a single cell. This approach was combined with domain swap and point mutagenesis to determine the roles of the terminal domains in the differential binding characteristics of the linker histone isotypes, mouse H10 and H1c. Exchanging the N-terminal domains between H10 and H1c changed their overall binding affinity to that of the other variant. In contrast, switching the C-terminal domains altered the chromatin interaction surface of the globular domain. These results indicate that linker histone subtypes bind to chromatin in an intrinsically specific manner and that the highly variable terminal domains contribute to differences between subtypes. The methods developed in this study will have broad applications in studying dynamic properties of additional histone subtypes and other mobile proteins.
Introduction
Eukaryotic DNA is compacted into a nucleoprotein complex called chromatin (1). The dynamic nature of chromatin facilitates structural and functional plasticity required for DNA-dependent cellular processes (2). The nucleosome is the basic repeating unit of chromatin composed of ∼147 bp of DNA wrapped around an octamer of core histones plus linker DNA and associated proteins (3, 4). At the nucleosomal level, post-translational modifications of core histones and nucleosome remodeling modulate chromatin structure (5). Linker histones are a family of lysine-rich proteins that bind at or near the point at which DNA enters and exits the nucleosomal core and organize an additional ∼20 bp of linker DNA, to form the chromatosome (6–8). Although binding of linker histones further stabilizes chromatin into higher order structures (9–12), these proteins interact with chromatin in a dynamic manner as components of a highly intricate and active network of chromatin-binding proteins (13–16).
Eleven nonallelic primary sequence isotypes or variants of histone H1 have been identified in mammals (17–19). Several studies have reported functional heterogeneity between H1 isotypes, which includes their ability to activate or repress expression of specific genes (20–23). Thus, differences in the primary structures of linker histones, post-translational modification patterns, and competition with other dynamic DNA-binding proteins could determine their distinct chromatin binding properties (24, 25).
Linker histones have a tripartite domain structure that includes short N-terminal and longer C-terminal domains, which are basic and relatively unstructured in solution (26). These domains flank a central globular domain, which is a three-helix bundle forming a winged helix motif similar to that found in DNA-binding transcription factors (27, 28). Previous studies from our laboratory identified the interaction surface and positioning of the globular domain of linker histone H10 onto the chromatosome (7). In an analogous study, we showed that the globular domain of variant H1c interacts with the nucleosome with a binding orientation that is distinct than that of H10 (29). It is plausible that differential orientation of the globular domain could impart functional specificity to the linker histone isotypes. Thus, it is important to comprehensively elucidate the structural properties of H1 isotypes that dictate their binding characteristics.
The globular domain sequence shows a high degree of conservation across individual linker histones, whereas those of the terminal domains are quite divergent within the same species (17–19), suggesting that the functional heterogeneity among the isotypes may arise because of the differences in their terminal domains. Although both the globular and C-terminal domains of H1 are required for nucleosomal binding, only the C-terminal domain stabilizes chromatin higher order folding (16, 30). The role of the N-terminal domain is unknown and is proposed to be involved in proper positioning and anchoring of the globular domain onto chromatin.
FRAP2 is a powerful system for measuring protein dynamics in living cells (31, 32). However, it does not account for biological and experimental variations leading to discrepancies in quantitative interexperimental conclusions and is prone to several drawbacks that can lead to faulty inferences (33). In this manuscript, we report a novel dual color FRAP assay approach, which permits simultaneous determination of the in vivo binding kinetics of two distinct forms of linker histone proteins within a subnuclear region of a single cell. In this system, sequences encoding two linker histones are separately fused with sequences encoding spectrally separable fluorescent proteins and are co-expressed from a bicistronic vector. Thus, an extremely stringent and precise measurement and comparison of the binding behavior of the two proteins is possible. Using this system, combined with a domain swap and a mutagenesis-based experimental approach, we determined that the N- and C-terminal domains have distinct roles in the chromatin binding characteristics of linker histone isotypes H10 and H1c. We conclude that the N-terminal domain contributes to overall chromatin binding affinity, whereas the C-terminal domain directs differential nucleosomal orientation of the H10 and H1c globular domains.
EXPERIMENTAL PROCEDURES
Expression Vectors and Cell Lines
Plasmids containing H1 isotypes tagged at the C terminus with enhanced GFP have been described previously (16). Plasmids containing mouse H1 isotypes tagged with mCherry (ChFP) were constructed by inserting the coding sequence of monomeric mCherry (a kind gift from R. Tsein) after the last lysine residue of the H1 isotypes. Plasmids containing H1 isotypes tagged at the N terminus, separated by a 20-amino acid linker, were also constructed. Domain shift hybrids were generated in multiple steps using PCR-based and silent mutagenesis. Point mutations were introduced using the QuikChange mutagenesis kit (Stratagene) or by introduction of annealed oligonucleotides between restriction sites. Details of the constructs used are presented in Fig. 1 and supplemental Fig. S1. Bicistronic constructs were transfected using Lipofectamine 2000 reagent (Invitrogen) as per the manufacturer's instruction into mouse BALB/c 3T3 fibroblast cells cultured in DMEM (Invitrogen) supplemented with 10% fetal calf serum. Multiple stable clonal cell lines were selected for each construct using 500 μg/ml G418 sulfate (Clontech).
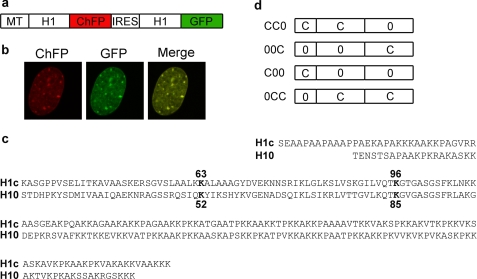
FIGURE 1.
Simultaneous expression of two linker histones in the nucleus of a single cell. a, bicistronic expression vectors contain the mouse metallothionein I promoter (MT) driving expression of a mRNA containing an ORF encoding H1-ChFP, an internal ribosome entry site (IRES) sequence, and a second ORF encoding H1-GFP. b, images acquired by live cell confocal microscopy of a cell expressing H10-ChFP and H10-GFP. c, sequence alignment of H1c and H10. Top two lines, N-terminal domains. Second two lines, central globular domains. Last four lines, C-terminal domains. Residues in bold type were mutagenized to alanine for constructs used in Fig. 5. d, schematic of domain switch mutants. Construct CC0 consists of amino acids 1–109 of H1c and amino acids 98–193 of H10. Construct 00C consists of amino acids 1–97 of H10 and amino acids 110–211 of H1c. Construct C00 consists of amino acids 1–32 of H1c and amino acids 21–193 of H10. Construct 0CC consists of amino acids 1–20 of H10 and amino acids 32–211 of H1c.
Dual Color FRAP Microscopy
The cells were plated at 50% confluency and imaged on Lab-Tek II chambered coverglass (Nalgene Nunc) in growth DMEM in the absence of inducer. The Zeiss 510 LSM META confocal microscope with a Plan Apochromat ×63/1.4 oil-immersion objective lens was used for imaging studies. The cells were imaged at 37 °C using a heat-regulated stage (Heating insert P) and an objective heater (Carl Zeiss Microimaging, Inc.). For excitation of GFP, the 488-nm line of an argon laser (nominal output, 30 milliwatt) operating at 75% output laser power was used. For excitation of ChFP, the 543-nm line of 10 milliwatt helium-neon lasers was used. Fluorescence emission of GFP and ChFP was detected by using the BP 505–530- and LP 585-nm filters, respectively. The detection was achieved using the multitracking mode at 4–5% laser intensity in the linear range of detection with maximal gain of 650. Scanning was bidirectional at the highest possible rate using a 4× zoom and image format 512 × 512 pixels with the pinhole of 3 Airy units. Control experiments with cells expressing only H1-mCherry or H1-GFP confirmed absence of bleed through of fluorescence signals between channels (supplemental Fig. S2). For all FRAP experiments, three prebleach scans of the cells were acquired. Bleaching of a random heterochromatic spot in the nucleus was performed simultaneously for GFP and mCherry with a single pass with all laser lines between 488 and 543 nm set to maximal power. The bleach area was set to be a spot of 2 μm, and bleaching was achieved using 10 consecutive bleach scans of 49-ms duration. Fluorescence recovery was monitored for 150 s after bleaching by scanning the cell at 2-s intervals. Identical settings were used for the pre- and postbleach images.
Analysis of Dual Color FRAP Measurements
Image analysis was carried out using MBF ImageJ software. Each cell image was split into two separate channel windows for ChFP and GFP, and image alignment was performed for each with the TurboReg Plugin (34). Double normalization was carried out to normalize the raw fluorescence recovery data to the average prebleach signal while correcting for acquisition bleaching and background subtraction to obtain the relative fluorescence intensity as described (35). The normalized recovery curves were processed in separate channels for mCherry and GFP for each cell. The normalization measurements were carried out in the same regions for mCherry and GFP in a single cell, by applying the defined region of interest for the bleached region, total cell nucleus, and a random region outside the cell for background subtraction, saved using ImageJ ROI manager to both channels.
Curve Fitting
The fluorescence recovery data for GFP- and ChFP-tagged H1 proteins were obtained by plotting the normalized relative fluorescence intensity versus time. The t50 values for both were obtained by fitting their normalized recovery curves to a single exponential equation (Origin 6.1; OriginLab) as described previously (35), and the relative ratios of the t50 values were calculated. The data sets from at least eight random cells from a pool of three independent cell lines were averaged to obtain the final quantitative values.
RESULTS
Quantification of Histone H1 Binding Dynamics Using Dual Color FRAP Analysis
A bicistronic expression vector containing an internal ribosome entry site was constructed to co-express two linker histone-fluorescent protein hybrids (Fig. 1a). The spectrally distinct fluorescent markers ChFP and GFP can be monitored independently and simultaneously in a single cell (Fig. 1b). Stable cell lines expressing linker histone proteins encoded by the bicistronic vector were established in murine BALB/c 3T3 fibroblast cells. These proteins are expressed at similar levels and comprise less than 5% of the total H1 population (supplemental Fig. S3). Dual color FRAP analysis was performed, and the binding affinity of individual proteins was estimated from the t50 value obtained by fitting their normalized recovery curves to a single exponential equation (35). We limited the analysis to the first 150 s of recovery to focus on the highly mobile fraction of H1 that constitutes more than 75% of the total linker histone pool (16, 36).
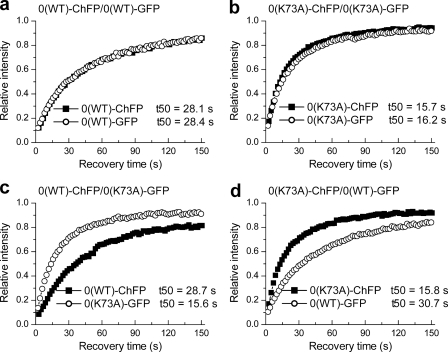
To validate the dual color FRAP approach, proof-of-principle constructs were made in which we systematically switched the position of expression of WT and mutant linker histone proteins and their fluorescent tags in the vector (Fig. 2). The mutant proteins contained a K73A mutation within the globular domain of histone H10. This residue is critical for binding, and the mutation to alanine results in faster recovery kinetics of H10 (7). Dual color FRAP experiments with WT H10-ChFP co-expressed with WT H10-GFP displayed overlapping recovery kinetics with similar t50 values of ∼ 28 s (Fig. 2a and Table 1). Similarly, mutant proteins H10(K73A)-ChFP and H10(K73A)-GFP had nearly identical t50 values of ∼16 s (Fig. 2b and Table 1). Thus, identical forms of H1 proteins fused with GFP or ChFP showed similar t50 values, indicating that the binding kinetics of these proteins is not appreciably affected by the type of fluorescent tag. Simultaneous FRAP analysis of H10(K73A)-GFP and WT H10-ChFP (Fig. 2c and Table 1) or of H10(K73A)-ChFP and WT H10-GFP (Fig. 2d and Table 1) displayed recovery kinetics for the mutant and WT proteins similar to that obtained with vectors expressing only mutant or only WT proteins. Collectively, these results indicate that neither the position of the protein within the vector nor the fluorescent tag significantly affects the binding behavior. The results also indicate that expression of a mutant protein at the low levels used in these assays does not affect the binding of the WT protein (37). Analysis of the data reveals some distinct advantages of the dual color FRAP approach. The t50 values obtained for any given protein vary considerably as noted by the standard deviations. However, the ratios of the values obtained for two proteins in a single cell display much less variability (supplemental Fig. S4) as is shown by the coefficients of variability, which are much lower for the ratios. Thus, the experimental system provides stringent internal controls for biological and instrumentation variability inherent in quantitative FRAP experiments. Additionally, the design allows the application of paired Student's t tests for statistical analysis that can reduce the number of replicates needed for accurate determination of significant differences.
FIGURE 2.
Validation of the dual color FRAP approach. a–d, quantitative dual color FRAP analysis of stable transfectants co-expressing WT H10-ChFP and WT H10-GFP (a), H10(K73A)-ChFP and H10(K73A)-GFP (b), WT H10-ChFP and H10(K73A)-GFP (c), and H10(K73A)-ChFP and WT H10-GFP (d). The values for the half-time of recovery (t50) were determined as previously described (35) and represent the means ± S.D. of at least eight independent measurements from a pool of three stable cell lines. The error bars are omitted from the plots for clarity. Table 1 provides the corresponding statistical analyses.
TABLE 1.
Quantitative analyses validating the dual color FRAP approach
| Construct |
t50a |
t50 ratio of GFP/ChFP | p valueb | |
|---|---|---|---|---|
| ChFP | GFP | |||
| s | ||||
| 0(WT)ChFP/0(WT)GFP | 28.1 ± 7.2 (0.26) | 28.4 ± 6.5 (0.23) | 0.99 ± 0.08 (0.08) | 0.4115 |
| 0(K73A)ChFP/0(K73A)GFP | 15.7 ± 2.3 (0.15) | 16.2 ± 2.5 (0.15) | 0.97 ± 0.06 (0.06) | 0.2758 |
| 0(WT)ChFP/0(K73A)GFP | 28.7 ± 4.6 (0.16) | 15.6 ± 2.8 (0.15) | 0.54 ± 0.02 (0.04) | 0.0001 |
| 0(K73A)ChFP/0(WT)GFP | 15.8 ± 3.8 (0.26) | 30.7 ± 6.3 (0.2) | 0.52 ± 0.02 (0.04)c | 0.0001 |
a The values for t50 were determined as previously described (35). The values are the means ± S.D. from at least eight independent measurements. The values in parentheses denote the calculated coefficients of variation.
b Paired Student's t test for H1-GFP versus H1-ChFP.
c The ratio of GFP/ChFP was inverted to ChFP/GFP for comparison.
N-terminal Domain Determines Specific and Differential Chromatin Binding Affinities of Linker Histones H10 and H1c
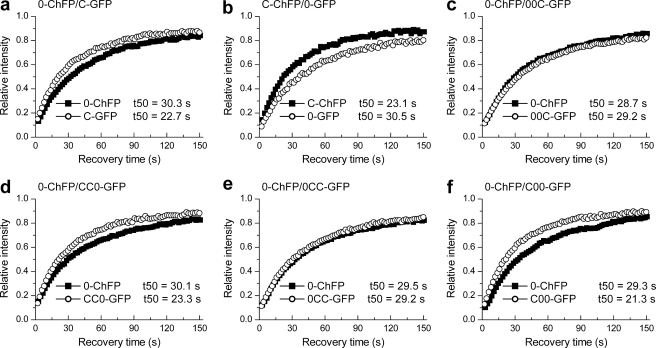
We utilized the dual color FRAP approach to determine the relative quantitative differences in the chromatin binding affinity of two distinct isotypes: H10 and H1c. Cells co-expressing WT H10-ChFP and WT H1c-GFP displayed significant differences in their recovery curves (Fig. 3a and Table 2). WT H10-ChFP recovered with a t50 of ∼30 s, whereas WT H1c-GFP had a t50 of ∼23 s. The reciprocal construct generated essentially identical results (Fig. 3b and Table 2). These results are consistent with our previous observations in independent cell lines (29).
FIGURE 3.
Quantitative analysis of the relative binding kinetics of isotypes H10 and H1c and effects of swapping their terminal domains. a and b, FRAP analysis with cells co-expressing WT H10-ChFP and WT H1c-GFP (a) or WT H10-GFP and WT H1c-ChFP (b) shows faster recovery kinetics for H1c than H10. c–f, simultaneous recovery curves of the C-terminal switch mutants 00C-GFP relative to WT H10-ChFP (c) and CC0-GFP relative to WT H10-ChFP (d) and of the N-terminal switch mutants 0CC-GFP relative to WT H10-ChFP (e) and C00-GFP relative to WT H10-ChFP (f). The values for the half-time of recovery (t50) were determined as previously described (35) and represent the means ± S.D. of at least eight independent measurements from a pool of three stable cell lines. The error bars are omitted from the plots for clarity. Table 2 provides the corresponding statistical analyses.
TABLE 2.
Quantitative analysis of H10 versus H1c and domain swap mutants
| Construct |
t50a |
t50 ratio of GFP/ChFP | p valueb | |
|---|---|---|---|---|
| ChFP | GFP | |||
| s | ||||
| 0-ChFP/C-GFP | 30.3 ± 1.9 (0.06) | 22.7 ± 1.6 (0.07) | 0.75 ± 0.03 (0.03) | 0.0001 |
| C-ChFP/0-GFP | 23.1 ± 4.6 (0.2) | 30.5 ± 7.0 (0.23) | 0.77 ± 0.05 (0.07) | 0.0002 |
| 0-ChFP/00C-GFP | 28.7 ± 3.8 (0.13) | 29.2 ± 4.2 (0.14) | 1.02 ± 0.06 (0.06) | 0.4066 |
| 0-ChFP/CC0-GFP | 30.1 ± 3.9 (0.13) | 23.3 ± 3.8 (0.16) | 0.77 ± 0.04 (0.06) | 0.0001 |
| 0-ChFP/0CC-GFP | 29.5 ± 5.0 (0.17) | 29.2 ± 4.7 (0.16) | 0.99 ± 0.05 (0.05) | 0.3383 |
| 0-ChFP/C00-GFP | 29.3 ± 3.7 (0.13) | 21.3 ± 3.0 (0.14) | 0.72 ± 0.04 (0.05) | 0.0004 |
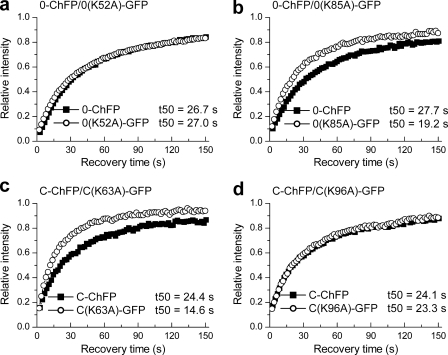
| 0-ChFP/0(K52A)-GFP | 26.7 ± 4.8 (0.18) | 27.0 ± 5.3 (0.2) | 1.01 ± 0.07 (0.07) | 0.6349 |
| 0-ChFP/0(K85A)-GFP | 27.7 ± 2.8 (0.1) | 19.2 ± 2.9 (0.15) | 0.69 ± 0.06 (0.08) | 0.0001 |
| C-ChFP/C(K63A)-GFP | 24.4 ± 3.1 (0.13) | 14.6 ± 2.1 (0.07) | 0.60 ± 0.07 (0.08) | 0.0001 |
| C-ChFP/C(K96A)-GFP | 24.1 ± 6.5 (0.27) | 23.3 ± 6.7 (0.29) | 0.96 ± 0.06 (0.06) | 0.1316 |
| 00C-ChFP/00C-GFP | 27.0 ± 3.6 (0.13) | 27.1 ± 3.8 (0.12) | 1.01 ± 0.06 (0.06) | 0.7508 |
| 00C-ChFP/00(K52A)C-GFP | 28.4 ± 6.2 (0.22) | 14.9 ± 3.3 (0.22) | 0.53 ± 0.06 (0.11) | 0.0001 |
| 00C-ChFP/00(K85A)C-GFP | 30.2 ± 4.7 (0.15) | 29.5 ± 6.6 (0.22) | 0.96 ± 0.10 (0.10) | 0.3926 |
| CC0-ChFP/CC0-GFP | 22.3 ± 4.4 (0.19) | 22.1 ± 4.1 (0.19) | 1.00 ± 0.06 (0.06) | 0.7552 |
| CC0-ChFP/CC(K63A)0-GFP | 21.6 ± 2.9 (0.14) | 22.8 ± 2.5 (0.11) | 1.06 ± 0.05 (0.05) | 0.206 |
| CC0-ChFP/CC(K96A)0-GFP | 21.6 ± 3.7 (0.17) | 14.3 ± 1.7 (0.17) | 0.67 ± 0.06 (0.09) | 0.0001 |
| 0CC-ChFP/0CC-GFP | 25.2 ± 4.3 (0.17) | 26.1 ± 3.9 (0.15) | 1.04 ± 0.05 (0.05) | 0.6677 |
| 0CC-ChFP/0C(K63A)C-GFP | 25.0 ± 2.6 (0.14) | 13.7 ± 1.2 (0.09) | 0.55 ± 0.03 (0.06) | 0.0001 |
| 0CC-ChFP/0C(K96A)C-GFP | 25.7 ± 4.2 (0.16) | 26.8 ± 6.5 (0.24) | 1.04 ± 0.10 (0.10) | 0.2123 |
| C00-ChFP/C00-GFP | 22.9 ± 3.1 (0.14) | 23.6 ± 3.6 (0.15) | 1.03 ± 0.07 (0.07) | 0.6832 |
| C00-ChFP/C0(K85A)0-GFP | 23.0 ± 2.9 (0.13) | 15.6 ± 2.2 (0.14) | 0.69 ± 0.06 (0.10) | 0.0001 |
| C00-ChFP/C0(K52A)0-GFP | 24.3 ± 1.7 (0.07) | 16.0 ± 2.0 (0.13) | 0.66 ± 0.06 (0.09) | 0.0001 |
a The values for t50 were determined as previously described (35). The values are the means ± S.D. from at least eight independent measurements. The values in parentheses denote the calculated coefficient of variation.
b Paired Student's t test for H1-GFP versus H1-ChFP.
To characterize the contribution of the terminal domains of linker histones H10 and H1c to their differential chromatin binding affinities, we generated domain switch constructs in which either of the terminal domains was replaced by that from the other variant. These individual mutants, tagged with GFP, were cloned into the bicistronic vector and co-expressed with WT H10-ChFP (Fig. 1, c and d).
We first switched the C-terminal domains of H10 and H1c to determine whether they contribute to their differential in vivo binding affinities. The mutant protein 00C-GFP, consisting of the N-terminal and globular domains of H10 and the C-terminal domain of H1c, showed recovery kinetics (t50 = ∼29 s) similar to that observed for the co-expressed WT H10-ChFP (Fig. 3c and Table 2). Thus, exchanging the C terminus of H10 with that of H1c does not change its binding affinity. The reciprocal domain swap mutant CC0-GFP, generated by replacing the C-terminal domain of H1c with that of H10, showed faster recovery (t50 = 23.3 s) compared with that of WT H10-ChFP (t50 = 30.1 s) (Fig. 3d and Table 2). Thus, the CC0 mutant displays recovery kinetics similar to that of H1c. This result was confirmed by co-expressing the CC0-GFP mutant protein with WT H1c-ChFP (supplemental Fig. S5). These proteins showed overlapping recovery curves with identical t50 values of ∼23 s. These results indicate that the C-terminal domain does not contribute to the differential chromatin binding affinities of linker histones H10 and H1c.
To determine whether the N-terminal domain contributes to the differential in vivo binding affinities of H10 and H1c, we swapped the N-terminal domain of H10 with that of H1c and vice versa. The binding affinities of these domain swap mutants were compared with the simultaneously expressed WT H10. The domain swap protein 0CC-GFP, consisting of the globular and C-terminal domains of H1c and the N-terminal domain of H10, showed recovery kinetics nearly identical to that of WT H10-ChFP with t50 values of ∼29 s (Fig. 3e and Table 2). Thus, swapping the N-terminal domain of H1c with that of H10 changes its chromatin residence time to that of WT H10. The reciprocal mutant C00-GFP, consisting of the globular and C-terminal domains of H10 and the N-terminal domain of H1c, displayed a significantly faster recovery (t50 = 21.3 s) than that of WT H10-ChFP (t50 = 29.3 s) (Fig. 3f and Table 2). These results indicate that exchanging the N-terminal domain of H10 with that of H1c changes its chromatin binding affinity to that of WT H1c. We thus conclude that swapping of the N-terminal domains between H10 and H1c changes their overall binding affinity to that of the variant from which the N-terminal domain was derived.
It is possible that the location of the fluorescent tag at the C terminus might compromise or obscure influences of this domain on the binding affinity. We therefore repeated this analysis with constructs in which the tag was fused to the N terminus and obtained nearly identical results (supplemental Fig. S6 and Table S1).
C-terminal Domain Influences Nucleosomal Geometry of Globular Domain
We previously showed that the nucleosomal interaction surfaces of the globular domains of linker histone isotypes H10 and H1c are distinct and proposed that they may bind to chromatin with different orientations (7, 29). However, the role of H10 and H1c terminal domains in determining the distinct nucleosomal interaction surface and the orientation of their respective globular domains onto chromatin are unknown. We previously identified key conserved residues that occupy similar positions in the globular domains of H10 and H1c (Fig. 1c) and yet display differential contributions toward their in vivo binding (7, 29). Residues Lys-52 and Lys-85 within the H10 globular domain and their respective homologous residues Lys-63 and Lys-96 within the H1c globular domain are conserved in all the major somatic H1 isotypes. We confirmed the contribution of these residues to H1 binding using the dual color FRAP system. A K52A point mutation did not significantly affect binding of H10 (Fig. 4a), but a K63A mutation significantly impaired binding of H1c (Fig. 4c). Conversely, a K85A mutation significantly impaired binding of H10 (Fig. 4b), but a K96A mutation did not affect H1c binding (Fig. 4d). We then asked whether the terminal domains influence the differential contribution of these homologous residues in H10 and H1c toward the nucleosomal binding of their respective globular domains.
FIGURE 4.
Identification of conserved residues in the globular domains of H10 and H1c showing differential binding behavior. a and b, plots showing recovery kinetics of H10 mutants H10(K52A)-GFP (a) and H10(K85A)-GFP (b), relative to co-expressed H10-ChFP. c and d, plots showing recovery kinetics of H1c mutants H1c(K63A)-GFP (c) and H1c(K96A)-GFP (d), relative to co-expressed H1c-ChFP. The values for the half-time of recovery (t50) were determined as previously described (35) and represent the means ± S.D. of at least eight independent measurements from a pool of three stable cell lines. The error bars are omitted from the plots for clarity. Table 2 provides the corresponding statistical analyses.
Although molecular models have been proposed but not confirmed (7, 29), we nevertheless consider that the affect of mutations at the Lys-52/63 and Lys-85/96 sites to be a reasonable indicator of differential binding geometry of these two isotypes. Point mutations K52A or K85A in the H10 globular domain and K63A or K96A in H1c globular domain were introduced in the previously studied N- and C-terminal domain swap constructs. The bicistronic vector thus had the domain swap constructs harboring mutated globular domains tagged with GFP in one ORF, whereas the domain swap construct with nonmutated globular domain served as the endogenous control. Thus, we could determine the role of these individual residues in the binding of globular domains of H10 and H1c in mutant proteins where their terminal domains are derived from the other variant. The domain swap constructs without any mutations in their globular domains showed identical recovery kinetics (Fig. 5, a and d, and Table 2).
FIGURE 5.
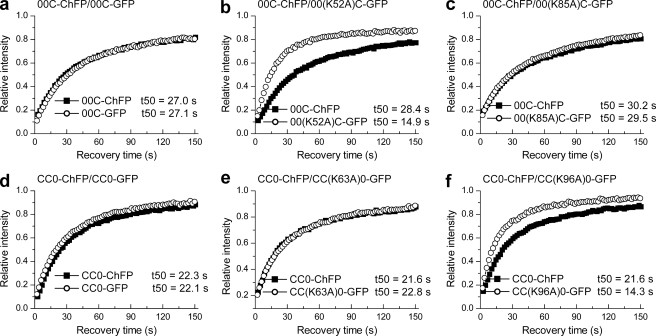
Quantitative FRAP analysis on cells expressing C-terminal domain swap bearing mutations in the globular domain. a–f, dual color FRAP analysis with cells co-expressing 00C-GFP and 00C-ChFP (a), 00(K52A)C-GFP and 00C-ChFP (b), 00(K85A)C-GFP and 00C-ChFP (c), CC0-GFP and CC0-ChFP (d), CC(K63A)0-GFP and CC0-ChFP (e), and CC(K96A)0-GFP and CC0-ChFP (f). The values for the half-time of recovery (t50) were determined as previously described (35) and represent the means ± S.D. of at least eight independent measurements from a pool of three stable cell lines. The error bars are omitted from the plots for clarity. Table 2 provides the corresponding statistical analyses.
Dual color FRAP analysis was conducted on cells co-expressing the 00(K52A)C-GFP and 00C-ChFP domain swap proteins. Surprisingly, the 00(K52A)C-GFP protein showed faster recovery kinetics (t50 = 14.9 s) relative to that of the co-expressed 00C-ChFP (t50 = 28.4 s) (Fig. 5b and Table 2), indicating that in the presence of the C-terminal domain of H1c, the globular domain of H10 in the 00C mutant requires the Lys-52 residue for in vivo chromatin binding in contrast to its role as a nonbinder in WT H10 (Fig. 4a). We then compared the binding kinetics of the 00(K85A)C-GFP domain swap mutant with that of the co-expressed 00C-ChFP. The 00(K85A)C-GFP mutant showed a t50 of 29.5 s similar to that of the 00C-ChFP (t50 = 30.2 s) (Fig. 5c and Table 2). Thus, the Lys-85 residue, critical for binding of the globular domain of WT H10 to chromatin (Fig. 4b), is dispensable in the 00C mutant. These results suggest that under the influence of the C-terminal domain of linker histone H1c, the globular domain of H10 switches its nucleosomal interaction surface to that of H1c. To further confirm that the chromatin interaction surface of globular domain is influenced by the C-terminal domain, the contribution of residues Lys-63 and Lys-96 in the H1c globular domain was examined in domain swap mutant CC0; which is composed of the N-terminal and globular domains of H1c but the C-terminal domain of H10. The CC(K63A)0-GFP protein recovered with kinetics similar to the simultaneously analyzed CC0-ChFP with observed t50 values of 22.8 and 21.6 s, respectively (Fig. 5e and Table 2). Thus, in the presence of C-terminal domain of H10, residue Lys-63 in the globular domain of H1c is not critical for binding. Conversely, the K96A domain swap mutant, CC(K96A)0-GFP, showed faster recovery kinetics (t50 = 14.3 s) relative to CC0-ChFP (t50 = 21.6 s) (Fig. 5f and Table 2). Thus, in the domain swap mutant CC0, the Lys-96 residue contributes to binding of the H1c globular domain. The results indicate a switch in the binding contribution of residues Lys-63 and Lys-96 from that observed in WT H1c (Fig. 4, c and d). From these results, we conclude that the C-terminal domain may influence the binding geometry of the globular domain.
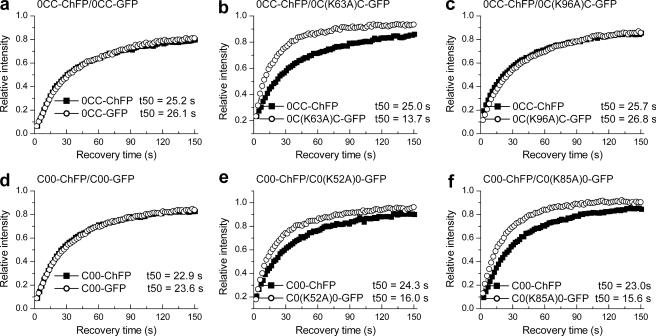
The N-terminal domain swap mutants, with one exception, showed no change in the contribution of these resides to the binding of their respective globular domains (Fig. 6 and Table 2). The exception is that the K52A mutation in the C00 hybrid compromises binding to some extent. This may be due to its proximity to the junction point. Collectively, the data indicate that the N-terminal domain does not contribute to the differential in vivo interaction surface of the globular domains of histones H10 and H1c or conversely, consistent with the above results, the binding properties of these residues were dictated by its native C terminus that remained unchanged in these mutants.
FIGURE 6.
Quantitative FRAP analysis on cells expressing N-terminal domain swap bearing mutations in the globular domain. a–f, dual color FRAP analysis with cells co-expressing 0CC-GFP and 0CC-ChFP (a), 0C(K63A)C-GFP and 0CC-ChFP (b), 0C(K96A)C-GFP and 0CC-ChFP (c), C00-GFP and C00-ChFP (d), C0(K52A)0-GFP and C00-ChFP (e), and C0(K85A)0-GFP and C00-ChFP (f). The values for the half-time of recovery (t50) were determined as previously described (35) and represent the means ± S.D. of at least eight independent measurements from a pool of three stable cell lines. The error bars are omitted from the plots for clarity. Table 2 provides the corresponding statistical analyses.
DISCUSSION
The variation in expression and intracellular localization patterns among members of the heterogenous family of H1 proteins is reflected in divergence at the sequence and structural level (18, 19). This divergence is most striking within the N- and C-terminal domains. Furthermore, individual isotypes show a higher degree of sequence conservation with orthologs from related species than with paralogs within the same species, suggesting a functionally relevant role for this variability. Several recent studies have provided evidence that individual histone isotypes have unique functions not only in regulating specific genes but also in other DNA-dependent processes (18). Thus, differences in their in vivo chromatin binding properties may form a mechanistic basis for this functional heterogeneity, and the structural differences among individual histone isotypes need to be considered as important determinants of their dynamic interactions with chromatin.
Several studies have reported contradictory evidence in defining the relative differences among individual isotypes in their chromatin binding affinities and in their ability to promote chromatin condensation (17–19). H1 isotypes isolated from rat brains were classified as possessing high (H1e, H10, and H1d), intermediate (H1c and H1b), and low binding affinities (H1a) (38). No appreciable differences in the binding affinities between human H1 isotypes H1–0 and H1.2 (the paralog of mouse H1c) were found in in vitro assays (39), although these isotypes were reported to have different binding characteristics by in vivo experiments (40).
In the present study, we utilized a dual color FRAP-based approach and show that H1c binds chromatin with a relatively lower affinity than that observed for H10. We have previously shown that overexpression of H10 in mouse fibroblasts leads to a global reduction in gene expression and causes a delay in cell cycle progression, whereas that of H1c results in induction of specific genes (22). Microarray analyses on fibroblasts overexpressing these isotypes also showed variant-specific differences in gene expression (41). Additionally, shRNA knockdown of H1.2 in human breast cancer cells results in G1 phase arrest and repressed expression of certain cell cycle genes, whereas depletion of H10 resulted in normal cell growth (21). Furthermore, knock-out studies showed that H10 is essential for terminal differentiation of dendritic cells (42), whereas H1.2 is involved with double-strand break-induced apoptosis (43). We recently reported that the nucleosome interaction surface of the globular domain of H1c differs from that of H10 and presented evidence that these isotypes will bind to the nucleosome with distinct orientations (29). Collectively, these observations suggest that there are significant functional differences between linker histone isotypes H10 and H1c. In the present study, we investigated the structural basis for differences in the in vivo chromatin binding properties of the mouse H10 and H1c isotypes.
Both the globular and the C-terminal domains have been demonstrated to be critical for H1 binding to the nucleosome (16, 40). The detailed molecular mechanism governing the differential chromatin binding properties of H1 isotypes is not very clear. Replacing the C-terminal domains of the lowest affinity human variant, H1.1, with that of higher affinity isotypes H1.4 and H1.5 resulted in an intermediate binding phenotype for the chimeric proteins. The authors suggested that the relative differences in lengths of the C-terminal domains of H1 isotypes determine their differential chromatin binding affinities (40). However, the chimeric protein did not show recovery kinetics identical to the variant that contributed the C-terminal domain, suggesting that the C-terminal domain may not be the sole determinant of differences in their binding affinities. Also, the mouse H10 and human H1–0 isotypes display the highest binding affinities among the isotypes yet have the shortest C-terminal domains. The role of the N-terminal domain in H1 binding is yet unknown.
In our study, we did not observe any change in the binding affinity of H10 and H1c upon swapping their C-terminal domains. Our results showed that exchanging the N-terminal domains between H10 and H1c resulted in the domain swap mutant protein exhibiting binding kinetics similar to the variant that donated the N-terminal domain. Hence, we report that the N-terminal domain plays a critical role in determining the specific and differential chromatin binding affinity of linker histone isotypes H10 and H1c.
The N-terminal domain of H1 comprises of two distinct subregions; the region proximal to the globular domain is highly basic, whereas the distal half is devoid of any basic residues (44). Subtle differences in the inherent secondary structure, post-translational modifications, and/or selective protein-binding partners within the N-terminal domain could form the molecular basis for their contribution to differential chromatin binding affinities of individual isotypes. Consistent with this hypothesis, using circular dichroism, high resolution NMR, and IR spectroscopy, the N-terminal domain of H10 was shown to contain a single nonamphipathic α-helical element, whereas that of H1e contains two amphipathic α-helices separated by a Gly-Gly motif (45, 46). This structural flexibility could allow interaction with distinct protein binding partners as suggested in the case of Msx1 and HP1 (47, 48). Recent studies have also mapped distinct post-translational modifications within the N-terminal domains of H1 (49, 50). The N terminus of H10 differs from that of H1c in length (20 versus 32 residues) with shorter basic and distal subregions. Notably, the H10 N terminus contains five Ser/Thr residues, whereas the H1c N terminus has a single N-terminal Ser residue. The Ser/Thr residues in H10 may be targets for phosphorylation, although this has not been demonstrated. Although our study was limited to H10 and H1c, future studies with other H1 isotypes will provide additional insight to the contribution of the N-terminal domain to histone H1 binding.
In a recent study we mapped the nucleosome interaction surface of the globular domain of H1c (29) and compared it with that previously determined for H10 (7). This was done by mutagenizing individual basic residues to alanine and determining the effect of these mutations on the in vivo binding of the mutants to chromatin by FRAP analysis. We identified two highly conserved lysine residues on each variant that displayed very different contributions to binding. Mutation of Lys-63 in H1c severely diminished nucleosome binding, whereas mutation of H10 at the corresponding Lys-52 residue did not affect binding. Conversely, mutation of Lys-96 in H1c had no effect on binding, whereas mutation at Lys-85 in H10 significantly compromised binding. Molecular modeling to accommodate these differences suggested that these isotypes bind to the nucleosome with distinct orientations. Although we do not know the details of the different orientations, our approach considered the analysis of effects of mutagenesis at the Lys-52/63 and Lys-85/96 residues to be a reasonable indicator of the different orientations. Surprisingly, we find that exchanging the C terminus of one variant with that of the other completely changes the contribution of these key residues toward H1 binding. For example, in the CC0 hybrid, containing the N-terminal and globular domains of H1c and the C-terminal domain of H10, the K96A mutation reduces binding affinity, whereas the K63A mutation has no effect. From these results we propose that the C-terminal domain in some way influences the orientation of the globular domain and the interface between the globular domain and the nucleosome. Our results are consistent with a recent study that showed cooperativity between the DNA-binding regions of the H10 globular and its C-terminal domain, which initiates H1 binding (51).
We hypothesize that the C-terminal domain initializes H1 binding and leads to a conformational change in the DNA, thereby re-orienting it. This change in turn facilitates organization of the DNA-binding targets of the globular domain, allowing its proper positioning. Our hypothesis is supported by molecular modeling experiments that have previously shown that the C-terminal domain is responsible for bending and altering the path of DNA and may adopt a high mobility group box-like fold upon binding to the nucleosome, leading to chromatin compaction (52). This change in DNA conformation may be aided by acquiring of an α-helical secondary structure by the C-terminal domain upon interacting with DNA (53). Deletion or phosphorylation of (S/T)PXK motifs within the C-terminal domain changed its secondary structure and DNA condensing properties (54, 55). The numbers and distribution of these motifs differ among isotypes and can result in differences in the secondary structures of their C termini. Differences in the secondary structure of C-terminal domains of individual isotypes could thus allow subtle differences in DNA conformation, contributing to differential positioning of the globular domains onto the nucleosome.
Thus our results show that the N-terminal domain contributes toward the differential chromatin binding affinity, whereas the C-terminal domain contributes toward distinct nucleosomal interface of isotypes H10 and H1c. These specific contributions of the variable terminal domains perhaps led to distinct modes of chromatin interactions for individual isotypes, thereby enabling them to fine-tune their differential roles in DNA dependent processes while maintaining their global role as stabilizers of the higher order of chromatin organization.
Supplementary Material
Acknowledgments
We thank Thomas Flanagan, Eric George, Stephen Anderson, Thomas Smith, and Stan Smith for technical assistance and Pratik Shah for critical reading of the manuscript. FRAP studies were performed at the Mississippi Functional Genomics Network Imaging Facility of the University of Southern Mississippi, and we are grateful to G. Shearer and B. Kang for assistance.
This work was supported by the Mississippi Idea Networks of Biomedical Research Excellence funded by grants from the National Center for Research Resources (5P20RR016476-11) and NIGMS (8 P20 GM103476-11) from the National Institutes of Health. This work was also supported by Grant MCB0235800 from the National Science Foundation (to D. T. B.).

This article contains supplemental Table S1 and Figs. S1–S6.
- FRAP
- fluorescence recovery after photobleaching
- ChFP
- mCherry fluorescent protein.
REFERENCES
- 1. van Holde K. (1989) Chromatin, Springer-Verlag, New York [Google Scholar]
- 2. Luger K., Hansen J. C. (2005) Nucleosome and chromatin fiber dynamics. Curr. Opin. Struct. Biol. 15, 188–196 [DOI] [PubMed] [Google Scholar]
- 3. Luger K., Mäder A. W., Richmond R. K., Sargent D. F., Richmond T. J. (1997) Crystal structure of the nucleosome core particle at 2.8 A resolution. Nature 389, 251–260 [DOI] [PubMed] [Google Scholar]
- 4. Woodcock C. L. (2006) Chromatin architecture. Curr. Opin. Struct. Biol. 16, 213–220 [DOI] [PubMed] [Google Scholar]
- 5. Hansen J. C., Nyborg J. K., Luger K., Stargell L. A. (2010) Histone chaperones, histone acetylation, and the fluidity of the chromogenome. J. Cell Physiol. 224, 289–299 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Zhou Y. B., Gerchman S. E., Ramakrishnan V., Travers A., Muyldermans S. (1998) Position and orientation of the globular domain of linker histone H5 on the nucleosome. Nature 395, 402–405 [DOI] [PubMed] [Google Scholar]
- 7. Brown D. T., Izard T., Misteli T. (2006) Mapping the interaction surface of linker histone H10 with the nucleosome of native chromatin in vivo. Nat. Struct. Mol. Biol. 13, 250–255 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Simpson R. T. (1978) Structure of the chromatosome, a chromatin particle containing 160 base pairs of DNA and all the histones. Biochemistry 17, 5524–5531 [DOI] [PubMed] [Google Scholar]
- 9. Thoma F., Koller T., Klug A. (1979) Involvement of histone H1 in the organization of the nucleosome and of the salt-dependent superstructures of chromatin. J. Cell Biol. 83, 403–427 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Bednar J., Horowitz R. A., Grigoryev S. A., Carruthers L. M., Hansen J. C., Koster A. J., Woodcock C. L. (1998) Nucleosomes, linker DNA, and linker histone form a unique structural motif that directs the higher-order folding and compaction of chromatin. Proc. Natl. Acad. Sci. U.S.A. 95, 14173–14178 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Carruthers L. M., Bednar J., Woodcock C. L., Hansen J. C. (1998) Linker histones stabilize the intrinsic salt-dependent folding of nucleosomal arrays. Mechanistic ramifications for higher-order chromatin folding. Biochemistry 37, 14776–14787 [DOI] [PubMed] [Google Scholar]
- 12. Robinson P. J., Rhodes D. (2006) Structure of the ‘30 nm’ chromatin fibre. A key role for the linker histone. Curr. Opin. Struct. Biol. 16, 336–343 [DOI] [PubMed] [Google Scholar]
- 13. Catez F., Yang H., Tracey K. J., Reeves R., Misteli T., Bustin M. (2004) Network of dynamic interactions between histone H1 and high-mobility-group proteins in chromatin. Mol. Cell Biol. 24, 4321–4328 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Phair R. D., Scaffidi P., Elbi C., Vecerová J., Dey A., Ozato K., Brown D. T., Hager G., Bustin M., Misteli T. (2004) Global nature of dynamic protein-chromatin interactions in vivo. Three-dimensional genome scanning and dynamic interaction networks of chromatin proteins. Mol. Cell Biol. 24, 6393–6402 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Lever M. A., Th'ng J. P., Sun X., Hendzel M. J. (2000) Rapid exchange of histone H1.1 on chromatin in living human cells. Nature 408, 873–876 [DOI] [PubMed] [Google Scholar]
- 16. Misteli T., Gunjan A., Hock R., Bustin M., Brown D. T. (2000) Dynamic binding of histone H1 to chromatin in living cells. Nature 408, 877–881 [DOI] [PubMed] [Google Scholar]
- 17. Parseghian M. H., Hamkalo B. A. (2001) A compendium of the histone H1 family of somatic subtypes. An elusive cast of characters and their characteristics. Biochem. Cell Biol. 79, 289–304 [PubMed] [Google Scholar]
- 18. Izzo A., Kamieniarz K., Schneider R. (2008) The histone H1 family. Specific members, specific functions? Biol. Chem. 389, 333–343 [DOI] [PubMed] [Google Scholar]
- 19. Happel N., Doenecke D. (2009) Histone H1 and its isoforms. Contribution to chromatin structure and function. Gene 431, 1–12 [DOI] [PubMed] [Google Scholar]
- 20. Alami R., Fan Y., Pack S., Sonbuchner T. M., Besse A., Lin Q., Greally J. M., Skoultchi A. I., Bouhassira E. E. (2003) Mammalian linker-histone subtypes differentially affect gene expression in vivo. Proc. Natl. Acad. Sci. U.S.A. 100, 5920–5925 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Sancho M., Diani E., Beato M., Jordan A. (2008) Depletion of human histone H1 variants uncovers specific roles in gene expression and cell growth. PLoS Genet. 4, e1000227. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Brown D. T., Alexander B. T., Sittman D. B. (1996) Differential effect of H1 variant overexpression on cell cycle progression and gene expression. Nucleic Acids Res. 24, 486–493 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Fan Y., Nikitina T., Zhao J., Fleury T. J., Bhattacharyya R., Bouhassira E. E., Stein A., Woodcock C. L., Skoultchi A. I. (2005) Histone H1 depletion in mammals alters global chromatin structure but causes specific changes in gene regulation. Cell 123, 1199–1212 [DOI] [PubMed] [Google Scholar]
- 24. Catez F., Ueda T., Bustin M. (2006) Determinants of histone H1 mobility and chromatin binding in living cells. Nat. Struct. Mol. Biol. 13, 305–310 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Brown D. T. (2003) Histone H1 and the dynamic regulation of chromatin function. Biochem. Cell Biol. 81, 221–227 [DOI] [PubMed] [Google Scholar]
- 26. Hartman P. G., Chapman G. E., Moss T., Bradbury E. M. (1977) Studies on the role and mode of operation of the very-lysine-rich histone H1 in eukaryote chromatin. The three structural regions of the histone H1 molecule. Eur. J. Biochem. 77, 45–51 [DOI] [PubMed] [Google Scholar]
- 27. Cerf C., Lippens G., Ramakrishnan V., Muyldermans S., Segers A., Wyns L., Wodak S. J., Hallenga K. (1994) Homo- and heteronuclear two-dimensional NMR studies of the globular domain of histone H1. Full assignment, tertiary structure, and comparison with the globular domain of histone H5. Biochemistry 33, 11079–11086 [DOI] [PubMed] [Google Scholar]
- 28. Ramakrishnan V., Finch J. T., Graziano V., Lee P. L., Sweet R. M. (1993) Crystal structure of globular domain of histone H5 and its implications for nucleosome binding. Nature 362, 219–223 [DOI] [PubMed] [Google Scholar]
- 29. George E. M., Izard T., Anderson S. D., Brown D. T. (2010) Nucleosome interaction surface of linker histone H1c is distinct from that of H10. J. Biol. Chem. 285, 20891–20896 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Allan J., Mitchell T., Harborne N., Bohm L., Crane-Robinson C. (1986) Roles of H1 domains in determining higher order chromatin structure and H1 location. J. Mol. Biol. 187, 591–601 [DOI] [PubMed] [Google Scholar]
- 31. Phair R. D., Misteli T. (2000) High mobility of proteins in the mammalian cell nucleus. Nature 404, 604–609 [DOI] [PubMed] [Google Scholar]
- 32. Mueller F., Wach P., McNally J. G. (2008) Evidence for a common mode of transcription factor interaction with chromatin as revealed by improved quantitative fluorescence recovery after photobleaching. Biophys J 94, 3323–3339 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Mueller F., Mazza D., Stasevich T. J., McNally J. G. (2010) FRAP and kinetic modeling in the analysis of nuclear protein dynamics. What do we really know? Curr. Opin. Cell Biol. 22, 403–411 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Thévenaz P., Ruttimann U. E., Unser M. (1998) A pyramid approach to subpixel registration based on intensity. IEEE Trans. Image Process 7, 27–41 [DOI] [PubMed] [Google Scholar]
- 35. Phair R. D., Gorski S. A., Misteli T. (2004) Measurement of dynamic protein binding to chromatin in vivo, using photobleaching microscopy. Methods Enzymol. 375, 393–414 [DOI] [PubMed] [Google Scholar]
- 36. Raghuram N., Carrero G., Stasevich T. J., McNally J. G., Th'ng J., Hendzel M. J. (2010) Core histone hyperacetylation impacts cooperative behavior and high-affinity binding of histone H1 to chromatin. Biochemistry 49, 4420–4431 [DOI] [PubMed] [Google Scholar]
- 37. Yellajoshyula D., Brown D. T. (2006) Global modulation of chromatin dynamics mediated by dephosphorylation of linker histone H1 is necessary for erythroid differentiation. Proc. Natl. Acad. Sci. U.S.A. 103, 18568–18573 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Orrego M., Ponte I., Roque A., Buschati N., Mora X., Suau P. (2007) Differential affinity of mammalian histone H1 somatic subtypes for DNA and chromatin. BMC Biol. 5, 22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Clausell J., Happel N., Hale T. K., Doenecke D., Beato M. (2009) Histone H1 subtypes differentially modulate chromatin condensation without preventing ATP-dependent remodeling by SWI/SNF or NURF. PLoS One 4, e0007243. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Th'ng J. P., Sung R., Ye M., Hendzel M. J. (2005) H1 family histones in the nucleus. Control of binding and localization by the C-terminal domain. J. Biol. Chem. 280, 27809–27814 [DOI] [PubMed] [Google Scholar]
- 41. Bhan S., May W., Warren S. L., Sittman D. B. (2008) Global gene expression analysis reveals specific and redundant roles for H1 variants, H1c and H10, in gene expression regulation. Gene 414, 10–18 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Gabrilovich D. I., Cheng P., Fan Y., Yu B., Nikitina E., Sirotkin A., Shurin M., Oyama T., Adachi Y., Nadaf S., Carbone D. P., Skoultchi A. I. (2002) H10 histone and differentiation of dendritic cells. A molecular target for tumor-derived factors. J. Leukocyte Biol. 72, 285–296 [PubMed] [Google Scholar]
- 43. Konishi A., Shimizu S., Hirota J., Takao T., Fan Y., Matsuoka Y., Zhang L., Yoneda Y., Fujii Y., Skoultchi A. I., Tsujimoto Y. (2003) Involvement of histone H1.2 in apoptosis induced by DNA double-strand breaks. Cell 114, 673–688 [DOI] [PubMed] [Google Scholar]
- 44. Böhm L., Mitchell T. C. (1985) Sequence conservation in the N-terminal domain of histone H1. FEBS Lett. 193, 1–4 [DOI] [PubMed] [Google Scholar]
- 45. Vila R., Ponte I., Collado M., Arrondo J. L., Jiménez M. A., Rico M., Suau P. (2001) DNA-induced α-helical structure in the NH2-terminal domain of histone H1. J. Biol. Chem. 276, 46429–46435 [DOI] [PubMed] [Google Scholar]
- 46. Vila R., Ponte I., Jiménez M. A., Rico M., Suau P. (2002) An inducible helix-Gly-Gly-helix motif in the N-terminal domain of histone H1e. A CD and NMR study. Protein Sci. 11, 214–220 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Daujat S., Zeissler U., Waldmann T., Happel N., Schneider R. (2005) HP1 binds specifically to Lys26-methylated histone H1.4, whereas simultaneous Ser27 phosphorylation blocks HP1 binding. J. Biol. Chem. 280, 38090–38095 [DOI] [PubMed] [Google Scholar]
- 48. Lee H., Habas R., Abate-Shen C. (2004) MSX1 cooperates with histone H1b for inhibition of transcription and myogenesis. Science 304, 1675–1678 [DOI] [PubMed] [Google Scholar]
- 49. Garcia B. A., Busby S. A., Barber C. M., Shabanowitz J., Allis C. D., Hunt D. F. (2004) Characterization of phosphorylation sites on histone H1 isoforms by tandem mass spectrometry. J. Proteome Res. 3, 1219–1227 [DOI] [PubMed] [Google Scholar]
- 50. Wisniewski J. R., Zougman A., Krüger S., Mann M. (2007) Mass spectrometric mapping of linker histone H1 variants reveals multiple acetylations, methylations, and phosphorylation as well as differences between cell culture and tissue. Mol. Cell Proteomics 6, 72–87 [DOI] [PubMed] [Google Scholar]
- 51. Stasevich T. J., Mueller F., Brown D. T., McNally J. G. (2010) Dissecting the binding mechanism of the linker histone in live cells. An integrated FRAP analysis. EMBO J. 29, 1225–1234 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Bharath M. M., Chandra N. R., Rao M. R. (2003) Molecular modeling of the chromatosome particle. Nucleic Acids Res. 31, 4264–4274 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Roque A., Iloro I., Ponte I., Arrondo J. L., Suau P. (2005) DNA-induced secondary structure of the carboxyl-terminal domain of histone H1. J. Biol. Chem. 280, 32141–32147 [DOI] [PubMed] [Google Scholar]
- 54. Roque A., Ponte I., Arrondo J. L., Suau P. (2008) Phosphorylation of the carboxy-terminal domain of histone H1. Effects on secondary structure and DNA condensation. Nucleic Acids Res. 36, 4719–4726 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Bharath M. M., Ramesh S., Chandra N. R., Rao M. R. (2002) Identification of a 34 amino acid stretch within the C-terminus of histone H1 as the DNA-condensing domain by site-directed mutagenesis. Biochemistry 41, 7617–7627 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.