Background: In quiescent endothelial cells, the transcription factor Erg regulates cell homeostasis by repressing expression of proinflammatory genes.
Results: Erg represses NF-κB p65-dependent transcription of ICAM-1, partly by inhibiting p65 binding to DNA.
Conclusion: Erg acts as a gatekeeper in quiescent endothelial cells to inhibit basal NF-κB activity.
Significance: Novel pathway controlling endothelial cell activation and inflammation.
Keywords: Endothelium, Ets Family Transcription Factor, NF-κB Transcription Factor, Transcription Repressor, Vascular Biology, Erg Transcription Factor, ICAM-1, Endothelial Homeostasis, Endothelial Quiescence
Abstract
The interaction of transcription factors with specific DNA sequences is critical for activation of gene expression programs. In endothelial cells (EC), the transcription factor NF-κB is important in the switch from quiescence to activation, and is tightly controlled to avoid excessive inflammation and organ damage. Here we describe a novel mechanism that controls the activation of NF-κB in EC. The transcription factor Erg, the most highly expressed ETS member in resting EC, controls quiescence by repressing proinflammatory gene expression. Focusing on intercellular adhesion molecule 1(ICAM)-1 as a model, we identify two ETS binding sites (EBS −118 and −181) within the ICAM-1 promoter required for Erg-mediated repression. We show that Erg binds to both EBS −118 and EBS −181, the latter located within the NF-κB binding site. Interestingly, inhibition of Erg expression in quiescent EC results in increased NF-κB-dependent ICAM-1 expression, indicating that Erg represses basal NF-κB activity. Erg prevents NF-κB p65 from binding to the ICAM-1 promoter, suggesting a direct mechanism of interference. Gene set enrichment analysis of transcriptome profiles of Erg and NF-κB-dependent genes, together with chromatin immunoprecipitation (ChIP) studies, reveals that this mechanism is common to other proinflammatory genes, including cIAP-2 and IL-8. These results identify a role for Erg as a gatekeeper controlling vascular inflammation, thus providing an important barrier to protect against inappropriate endothelial activation.
Introduction
Members of the transcription factor family nuclear factor (NF)-κB are important mediators of proinflammatory responses in the vasculature. Activation of the NF-κB pathway leads to the expression of multiple genes involved in inflammation, including cytokines and chemokines, adhesion molecules and growth factors. NF-κB family members include RelA (p65), RelB, c-Rel, NF-κB1 (p50), and NF-κB2 (p52), which bind as homodimers or heterodimers to the NF-κB binding sites on the promoters of target genes (1). In resting endothelial cells (EC),2 NF-κB is sequestered in the cytoplasm by proteins of the inhibitor of NF-κB (IκB) family. Activation by stimuli including tumor necrosis factor (TNF)-α, interleukin (IL)-1, and lipopolysaccharide (LPS) triggers a signaling cascade that leads to the degradation of IκBα and the phosphorylation and translocation of NF-κB to the nucleus (2). NF-κB activity is tightly regulated through feedback repressive mechanisms (3, 4). Control of NF-κB activity is essential to maintain quiescence and for the resolution of the inflammatory response; in chronic inflammation the tight control on NF-κB is lost, leading to vascular diseases such as atherosclerosis (5). The identification of key mediators to regain control of EC homeostasis has great potential for the development of novel therapeutics.
There is growing evidence on the importance of transcription factor Erg in maintaining EC homeostasis. Erg is a member of the ETS family of transcription factors, characterized by a conserved ETS DNA binding domain that binds to a core consensus motif of GGA(A/T) (6). Erg is the most highly expressed ETS factor in resting EC (7), where it acts as an activator of genes involved in homeostasis, including endoglin (8), ICAM-2 (9), vascular endothelial-cadherin (10), and heme oxygenase-1 (11). Recently, we and others have shown that Erg represses endothelial expression of proinflammatory molecules ICAM-1 and IL-8 in quiescent cells, and that inhibition of Erg induces leukocyte adhesion to unstimulated human umbilical vein endothelial cells (HUVEC) (12, 13), suggesting an important role for Erg in maintaining EC homeostasis by repressing basal expression of proinflammatory genes.
The clinical relevance of Erg in repressing EC activation is supported by its pattern of expression in atherosclerotic plaques: Erg is expressed in the healthy endothelium of human coronary artery but is absent from the activated endothelium over inflammatory infiltrate in the plaque shoulder (12). This is likely to be the result of endothelial activation by proinflammatory stimuli, because Erg levels have been shown to decrease upon LPS or TNF-α stimulation (9, 13). These data suggest that Erg may be important in maintaining endothelial quiescence and in the termination of the inflammatory response, by preventing the induction of proinflammatory gene expression.
In this study we investigate the mechanisms used by Erg to repress inflammatory gene expression in quiescent EC, focusing on ICAM-1 as a model gene. We show that in quiescent EC Erg prevents NF-κB p65 binding to DNA, suggesting that Erg may compete with p65 for DNA binding. We demonstrate that Erg binds to two ETS binding sites (EBS) in the ICAM-1 promoter and we show that both EBS and NF-κB consensus sites are required for the repressive activity of Erg. Using bioinformatic analysis of transcriptome profiling datasets and validation by ChIP, we show that this mechanism is common to other proinflammatory genes and we identify a specific subset of NF-κB target genes repressed by Erg in quiescent endothelial cells.
EXPERIMENTAL PROCEDURES
Cells
HUVEC were isolated and cultured in supplemented M199 media as previously described (10).
Erg and ETS Factor Inhibition
Erg expression was inhibited using either Genebloc (Silence Therapeutics AG, Berlin, Germany) as previously described (10), or by RNA interference with short interfering RNA (siRNA). siRNA treatment to inhibit Erg, Fli1, and Ets2 expression was carried out using Hs_ERG_7, Hs_FLI1_7, Hs_ETS2_7, and Hs_GAPBα_10 FlexiTube siRNA (Qiagen), respectively, and AllStars Negative Control siRNA (Qiagen). HUVEC were seeded onto 1% gelatin-coated plates and grown in EGM-2 medium (Lonza, Wokingham, United Kingdom). The following day, siRNA (10 nm) was mixed with AtuFect01 lipid (1 μg/ml, Silence Therapeutics) at 5 times concentration in Opti-MEM (Invitrogen), then added to cells for 24 or 48 h.
Transduction of HUVEC with Erg Adenovirus
Erg overexpression was carried out using a V5-tagged Erg-3 adenovirus (AdErg), as described previously (12). Briefly, HUVEC (3 × 104 cells/well) seeded onto 1% gelatin-coated 24-well plates in EGM2 were transduced with 50 multiplicity of infection of AdErg or β-galactosidase adenovirus (AdLacZ).
Transduction of HUVEC with IκBα Super Repressor Adenovirus
HUVEC (1 × 105 cells/well) were seeded onto 1% gelatin-coated 6-well plates in supplemented M199 media. The following day, cells were transduced with 100 multiplicity of infection of IκBα Super Repressor Adenovirus (AdIκBαSR) (14) or AdLacZ in serum-free M199 medium for 2 h before replacing with complete M199 medium. After 24 h, cells were transfected with Erg or control siRNA as described above. Alternatively, after 42 h following adenovirus transduction, cells were treated with 10 ng/ml of TNF-α for 6 h. ICAM-1 mRNA levels were assessed by quantitative RT-PCR, normalized to GAPDH. Oligonucleotides used in this study are described in supplemental Table S1.
Inhibition of NF-κB in HUVEC with BAY 11-7085
HUVEC (1 × 105 cells/well) were seeded onto 1% gelatin-coated 6-well plates in EGM-2. After 39 h, cells were treated with BAY 11-7085 (5 μm, Sigma) diluted in dimethyl sulfoxide and incubated for a further 24 h. Cells were treated with 10 ng/ml of TNF-α for the final 6 h. ICAM-1 protein levels were measured by Western blot using an anti-ICAM-1 (clone 15.2, kind gift of Prof. Nancy Hogg) antibody, and normalized to GAPDH (MAB374 Millipore).
Chromatin Immunoprecipitation
Chromatin immunoprecipitation (ChIP) was performed using ChIP-IT express (Active Motif, Rixensart, Belgium) as previously described (10). Briefly, sheared chromatin from HUVEC untreated, or treated with Erg or control siRNA, was fixed in 1% formaldehyde for 10 min before shearing chromatin using a Bioruptor (Diagenode, Liege, Belgium). Chromatin was immunoprecipitated with 2 μg antibody to Erg (sc-353, Santa Cruz Biotechnology, Inc), NF-κB p65 (ab7970, AbCam, Cambridge, United Kingdom), or negative control rabbit IgG (PP64, Chemicon, Millipore). Immunoprecipitated DNA was then used as template for quantitative PCR using primers specific for the ICAM-1 promoter, cIAP2 promoter, IL-8 promoter, and the negative control gene GAPDH. Oligonucleotide sequences are listed in supplemental Table S1.
Site-directed Mutagenesis of ICAM-1 Promoter Constructs
The ICAM-1 promoter construct pGL4 ICAM-1 1.3 containing the first 1.3 kb upstream from the transcription start site as described in Ref. 12 was mutated within ETS binding sites (EBS), or within the NF-κB binding site as previously shown (15), using the QuikChange® lightning multi site-directed mutagenesis kit (Agilent), all primers were designed using the QuikChange® Primer Design Program (Agilent). Briefly, pGL4 ICAM-1 1.3 plasmid was amplified using a primer designed to mutate a specific EBS from GGAA to CCAA or the NF-κB site from TTGGAAATTCC to TTCTAGATTAG. Sequencing was carried out to confirm mutation of the desired EBS. Mutations within more than one EBS were carried out sequentially. Primers for site-directed mutagenesis are listed in supplemental Table S1.
Reporter Gene Assays
HUVEC were seeded at 3 × 104 per well in EGM-2 medium (Lonza) on a gelatin-coated 24-well plate, and the following day either transduced with AdErg or AdLacZ (50 multiplicity of infection), or transfected with 100 nm Erg or control Genebloc and maintained in EGM-2 medium. After 24 h, co-transfection of HUVEC was performed with Genejuice transfection reagent (Merck Chemicals), according to the manufacturer's instructions. Briefly, cells were incubated with 1.5 μl of GeneJuice, 250 ng of pGL4 ICAM-1 1.3, pGL4ICAM1-EBS mutants, or pGL4.10 empty vector and 250 ng of pGL4.73 Rluc/TK Renilla luciferase (Promega) for 24 h. Luciferase activity was measured using the Dual Luciferase® Reporter Assay System (Promega) on a Syngergy HT microplate reader (BioTek, Winooski, VT) to calculate the dual luciferase ratio.
Electromobility Shift Assay (EMSA)
Nuclear lysates from a confluent monolayer of HUVEC were extracted using the Nuclear extract kit (Active Motif) following the manufacturer's instructions. DNA protein interactions were then investigated using the Lightshift chemiluminescent EMSA kit (Pierce). Briefly, biotinylated double-stranded oligonucleotides containing the ICAM-1 promoter sequence surrounding and including EBS −118 and −181 were incubated with 2 μg of HUVEC nuclear lysate with or without excess unbiotinylated oligonucleotides. Components of the protein-DNA complexes were investigated by the addition of anti-Erg (sc-353, Santa Cruz), anti-NF-κB p65 (ab7970, AbCam), or IgG (PP64, Millipore) control antibodies. EMSA reactions were run on a 0.5× Tris borate-EDTA (TBE) polyacrylamide gel and transferred to Hybond N+ nylon membrane (GE Healthcare, Amersham Biosciences) before detection. See supplemental Table S1 for oligonucleotide sequences.
Gene Set Enrichment Analysis
Gene set enrichment analysis (GSEA) overlap studies were carried out using GSEA software version 2.0 (Broad Institute). The query dataset were the 1138 genes identified as being up-regulated following a 48-h Erg inhibition in HUVEC (16), which were then compared against all the studies in the C2 curated gene sets. GSEA correlation of microarray data from Erg GeneBloc-treated HUVEC from the following published studies: TNF-α-treated pancreatic cancer cells (17), TNF-α-treated endothelial cells (18), NF-κB p65 ChIP seq data (19), was performed using Gene Set Enrichment Analysis version 2.0 software using 1000 data permutations.
RESULTS
Erg Repression of ICAM-1 Expression Is Not Shared by Other Constitutively Expressed ETS Factors
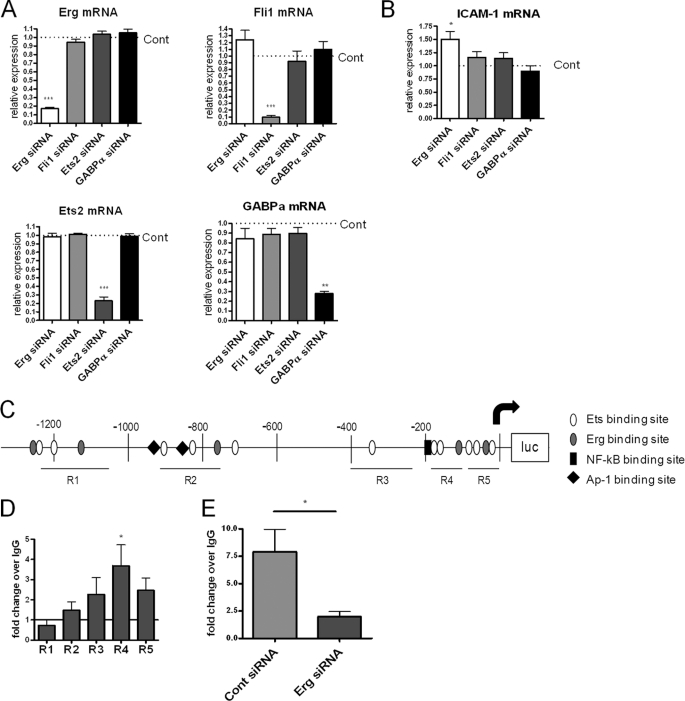
Recently we have shown that inhibition of Erg expression using antisense (Genebloc or GB) up-regulates basal ICAM-1 mRNA and protein levels (12). Because a number of ETS factors are constitutively expressed in resting EC (7), and all share the DNA consensus motif GGA(A/T), we investigated whether other ETS factors can repress ICAM-1 expression in EC, using siRNA. We selected the following ETS factors, all expressed in resting HUVEC, although at lower levels than Erg (supplemental Fig. S1 and Ref. 7): Fli-1, the ETS factor with the closest homology to Erg; Ets-2, a more distantly related ETS factor known to interact with Erg (20); and GABPα, which can act as an activator or repressor of transcription (21, 22). siRNA to Fli-1 inhibited expression of Fli-1 but not Erg, Ets-2, or GAPBα; similarly, siRNA to Ets-2 affected only Ets-2 expression, not Erg, Fli-1, or GAPBα expression; GABP-α siRNA only affected GABP-α expression and not Erg, Fli-1, or Ets-2; and finally Erg siRNA did not affect expression of the other three ETS factors (Fig. 1A). Only siRNA to Erg caused a significant increase in ICAM-1 mRNA levels (Fig. 1B), indicating that repression of ICAM-1 expression is not a shared property of constitutive ETS factors.
FIGURE 1.
Erg represses ICAM-1 and binds to the ICAM-1 promoter region 4 in resting EC. A and B, HUVEC were treated with siRNA for Erg, Fli-1, Ets-2, or GABPα for 24 h; relative expression of Erg, Fli-1, Ets-2, and GABPα mRNA (A) or ICAM-1 mRNA (B) was quantified by RT-PCR and expressed as relative to control siRNA treatment (Cont). C, analysis of 1.3 kb of the ICAM-1 promoter identified putative EBS, Erg consensus sites, AP1/ETS sites, and an NF-κB site as indicated. Location of quantitative PCR amplicons covering R1, R2, R3, R4, and R5 are indicated by black lines below. D and E, ChIP was carried out using an anti-Erg or control IgG antibody on sheared chromatin from confluent resting HUVEC (D) or HUVEC treated with Erg or control siRNA (E). Immunoprecipitated DNA was analyzed by qPCR for primers covering ICAM-1 promoter regions 1–5 (D) or region 4 only (E) and negative control region. Results are expressed as fold-change compared with IgG normalized to input and negative control region. n = 6 (B), n = 5 (C), *, p < 0.05; **, p < 0.01; ***, p < 0.001.
Erg Binds to ICAM-1 Promoter in a Region Proximal to the Transcription Start Site
We have previously shown that Erg binds to the ICAM-1 promoter (12). The first 1.3 kb upstream of the ICAM-1 transcription start site are required for ICAM-1 basal expression and regulation by inflammatory stimuli (23). Bioinformatic analysis using MatInspector shows that this region contains 16 EBS consensus sequences (Fig. 1C). A number of these EBS have been shown to play a role in ICAM-1 expression, including two AP1-EBS repeats involved in ICAM-1 induction after H2O2 stimulation (24), and two EBS that are involved in ICAM-1 expression in non-endothelial cells (25–27). Analysis of oligonucleotide screening and ChIP sequencing (ChIP Seq) data has suggested a possible specific Erg consensus motif of (A/C)GGAA(G/A) (28) or AGGA(A/T)(G/A) (29). This consensus sequence appears in the ICAM-1 promoter five times, at −1239, −1136, −773, −118, and −75 relative to the transcription start site (Fig. 1C). To investigate whether Erg binding to the ICAM-1 promoter is localized around these sites, ChIP was carried out by scanning the EBS in the first 1.3 kb of the ICAM-1 promoter, using quantitative PCR (qPCR) with primers for regions (R) named 1, 2, 3, 4, and 5 (Fig. 1C). Erg enrichment at R4, which includes EBS −118 and EBS −181, was greater than at other regions containing EBS (Fig. 1D). To confirm the specificity for Erg binding at this site, ChIP was carried out on chromatin from HUVEC treated with Erg siRNA. This showed a decrease in the amount of Erg bound to R4 after Erg inhibition, compared with control siRNA treatment (Fig. 1E). These data indicate that Erg binds to the ICAM-1 promoter in a region detected by the R4 primers, ∼188 to 103 bp upstream of the transcription start site.
Erg Repression of ICAM-1 Is Dependent on Two EBS −181 and −118 Upstream of the Transcription Start Site
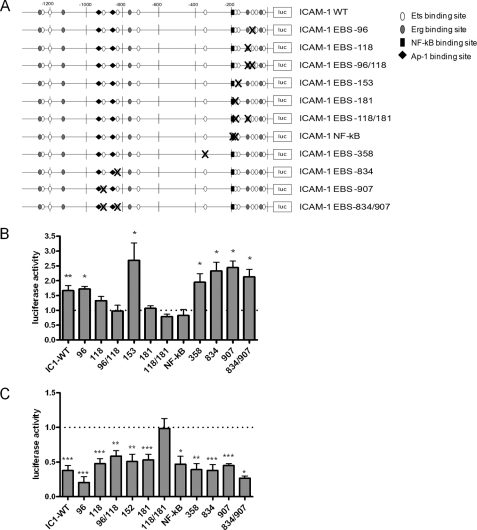
Previously, using an ICAM-1 promoter luciferase construct containing the first 1.3 kb upstream of the ICAM-1 transcription start site, we have shown that Erg represses ICAM-1 promoter activity in resting EC (12). Inhibition of Erg expression increased the activity of the ICAM-1 promoter luciferase construct; conversely, overexpression of Erg by adenovirus (AdErg) repressed basal promoter activity. To identify which EBS are involved in Erg-mediated repression, we generated ICAM-1 promoter constructs with mutations within single EBS, or two EBS together, if they had previously been shown to have a cooperative role (24, 25). Additionally, we mutated the NF-κB site at −188, previously identified as important for cytokine-mediated up-regulation of ICAM-1 (15) (Fig. 2A). In agreement with previous data (12), inhibition of Erg expression by Genebloc antisense significantly increased wild type (WT) ICAM-1 promoter activity by 1.66-fold, compared with control (Fig. 2B). The ICAM-1 promoter EBS mutants −96, −153, −358, −834, and −907 also showed significantly increased promoter activity after Erg inhibition. However, the EBS mutant constructs −118 and −181, and the combined −118/−181 mutant, showed no significant increase in promoter activity after Erg Genebloc treatment compared with control Genebloc, indicating involvement of these sites in Erg-mediated repression of ICAM-1 promoter activity. The double mutant EBS −96/−118, shown previously to have a role in both transactivation and repression of ICAM-1 by ETS factors in rabbit kidney RK13 cells and HEK 293 cells (25–27), also showed no increase in promoter activity after Erg inhibition; however, this is likely to occur through EBS −118, as mutation of EBS −96 alone was responsive to Erg inhibition. Finally, mutation of the NF-κB site at −188 resulted in loss of ICAM-1 up-regulation following Erg Genebloc, indicating that NF-κB is involved in the Erg-dependent repression. In conclusion, this set of experiments indicates that Erg-mediated repression of ICAM-1 involves EBS −118 and EBS −181, which is located within the NF-κB consensus sequence, and requires a functional NF-κB binding site.
FIGURE 2.
Identification of the DNA binding site involved in Erg-mediated repression of ICAM-1. A, schematic diagram of ICAM-1 promoter mutant constructs. pGL4 ICAM-1 1.3 was mutated in either single or double EBS or in the NF-κB binding site. EBS were mutated from GGAA to CCAA. NF-κB was mutated as previously shown (15). B, ICAM-1 promoter activity of EBS mutants after Erg Genebloc treatment. Results are expressed as luciferase activity relative to control Genebloc-treated cells. C, ICAM-1 promoter activity of EBS mutants after AdErg treatment, expressed as luciferase activity relative to AdLacZ (n = 3–7). *, p < 0.05; **, p < 0.01; ***, p < 0.001.
To consolidate these findings, we used the opposite approach. ICAM-1 promoter activity is repressed after Erg overexpression in HUVEC (12); therefore we investigated whether this repression was lost after mutation of EBS in the ICAM-1 promoter. In agreement with previous data, transduction with AdErg repressed ICAM-1 promoter activity to 38% of treatment with AdLacZ (Fig. 2C). Only one construct, containing mutation of the combined EBS −118 and −181, was not repressed by AdErg transduction. However, Erg overexpression repressed the EBS −118 and −181 constructs when these EBS were mutated individually (Fig. 2C). Taken together, these data suggest that Erg repression involves two EBS, 118 and 181 bp upstream of the transcription start site and either of these sites is sufficient for Erg-mediated repression. One of these sites (EBS −181) is located within the consensus binding site for NF-κB.
Erg Binds to EBS −118 and EBS −181 in the ICAM-1 Promoter
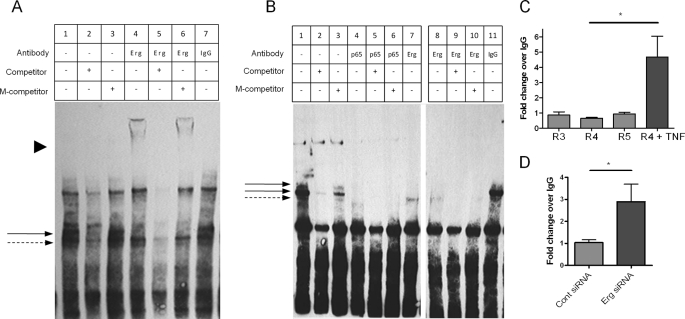
The above data suggest that Erg is repressing ICAM-1 through a mechanism that involves EBS −118 and EBS −181. To investigate whether Erg is binding directly to EBS −118 or −181, we studied the interaction between Erg and these sequences by electrophoretic mobility shift assay (EMSA). Nuclear extracts from resting HUVEC were incubated with biotinylated double-stranded oligonucleotides containing either the EBS −118 or −181 sequence. The oligonucleotide containing EBS −118 formed a number of complexes with nuclear proteins from resting HUVEC (Fig. 3A, lane 1, arrows), two of which were competed by excess unlabeled probe but not by a probe containing a mutation in the EBS −118 site, indicating specificity for this site. Addition of an anti-Erg antibody resulted in the appearance of a supershift (Fig. 3A, lane 4, arrowhead) and a decrease in the intensity of a complex (Fig. 3A, solid arrow), indicating that Erg binds to the EBS −118 site. The supershift was specific, as it was competed off by an excess of unlabeled probe, but not by unlabeled probe containing a mutation of the EBS −118 (Fig. 3A, lanes 5 and 6, respectively). Addition of IgG antibodies had no effect on the band pattern (Fig. 3A, lane 7), confirming that Erg specifically interacts with the oligonucleotide containing EBS −118.
FIGURE 3.
Erg binds to ICAM-1 promoter through EBS −118 and −181, and NF-κB p65 binds to EBS −181. A, and B, biotinylated oligonucleotides containing sequences from the ICAM-1 promoter EBS −118 (A) or the EBS −181 (B) were incubated with nuclear lysate from resting HUVEC. Antibodies to Erg, p65, or IgG were incubated with nuclear extract-oligonucleotide complexes as indicated. Specificity was measured by addition of saturating amounts of competing oligonucleotide (competitor) or competing oligonucleotide with a mutation in the EBS −118 (A) or −181 (B) (M-competitor). Shifted protein-oligonucleotide complexes are indicated by an arrow and super-shifted complexes are indicated by arrowhead. Images are a single representation of at least 3 separate experiments. C and D, ChIP was carried out on sheared chromatin from confluent resting HUVEC ± TNF (10 ng/ml for 30 min) (C), or HUVEC treated with Erg or control siRNA (D) using an anti-NF-κB p65 or control IgG antibody. Immunoprecipitated DNA was analyzed by qPCR for primers covering ICAM-1 promoter regions (R) 3–5 (C) or 4 (D) and negative control region. Results are expressed as fold-change compared with IgG normalized to input and negative control region. n = 4 (C), n = 6 (D); *, p < 0.05.
EMSA carried out with the EBS −181 oligonucleotide showed the formation of three specific complexes with nuclear proteins from resting HUVEC (Fig. 3B, lane 1, arrows), which were competed by excess unlabeled probe but not by a probe containing a mutation in the EBS −181 site (Fig. 3B, lanes 2 and 3). The addition of an anti-Erg antibody resulted in a decrease in the intensity of the two upper bands, suggesting that Erg binds to this complex (Fig. 3B, lanes 7–10); however, contrary to what was observed for the −118 EMSA, no supershift was detectable. A similar pattern was observed with a different anti-Erg antibody (data not shown), suggesting that the absence of a supershift is not due to a technical issue related to a single antibody but possibly because the new complex is too large to be identified by this method. In support of the specificity of the interaction of Erg with this site, addition of control IgG antibodies had no effect on the band pattern (Fig. 3B, lane 11 compared with lane 1). In conclusion, the EMSA experiments indicate that Erg binds to both EBS −118 and −181 in the ICAM-1 promoter.
NF-κB p65 Binds to EBS −181 in the ICAM-1 Promoter
EBS −181 is part of an NF-κB consensus sequence that is bound by NF-κB p65 after TNF-α stimulation in HUVEC (15, 30). Although the majority of NF-κB p65 in resting EC is located in the cytoplasm, low levels of p65 are also found in the nucleus (12, 31, 32). In quiescent EC, nuclear-localized NF-κB p65 appears to have a role in constitutive expression of genes, such as ICAM-2 and P-selectin, as mutation of the NF-κB binding sites within the promoters of these genes results in a decrease in basal activity (9, 33). Using EMSA, we investigated whether, in resting HUVEC, p65 interacts with the NF-κB site at −188, which overlaps with EBS −181. Addition of an anti-NF-κB p65 antibody caused a marked decrease in the intensity of all three bands identified with this oligonucleotide (Fig. 3B, lanes 4–6, arrows), suggesting that the anti-NF-κB p65 antibody binds to the protein-oligonucleotide complex at this site. No supershifted band was visible after the addition of the anti NF-κB p65 antibody. However, loss of the major complex after addition of NF-κB p65 antibody suggests that, at least in vitro, nuclear NF-κB p65 is able to bind to the EBS −181 oligonucleotide in resting HUVEC as well as in activated HUVEC.
Erg Inhibits NF-κB p65 Binding to the ICAM-1 Promoter
As shown above, Erg repression of the ICAM-1 promoter requires EBS −181, which is part of the NF-κB consensus sequence. Because EMSA studies have shown that Erg can bind to this site, we investigated the possibility that, in resting EC, Erg might block NF-κB p65 binding to the ICAM-1 promoter at EBS −181. We first tested whether NF-κB p65 could bind to the region containing the NF-κB site and EBS −181 within the native chromatin structure of resting HUVEC, using ChIP. ChIP was carried out on a confluent monolayer of quiescent or TNF-α-treated HUVEC. Chromatin was enriched with an NF-κB p65 antibody and binding sites were analyzed by quantitative PCR, using the ICAM-1 promoter primers described above (see Fig. 1D). No enrichment for NF-κB p65 was found in R4, which contains the NF-κB binding sites, EBS −181 and −118, or in flanking R3 and R5 (Fig. 3C); however, NF-κB p65 was enriched in R4 after treatment with TNF-α, as expected (34) (Fig. 3C). Thus, these results show that although p65 can bind to the NF-κB site within this region of the ICAM-1 promoter, when presented as an oligonucleotide, such as in the EMSA (Fig. 3B), in fact p65 does not bind to this same sequence in native chromatin from unstimulated HUVEC. Therefore we speculated that there may be an inhibitory element in the native chromatin structure of the ICAM-1 promoter that inhibits the binding of p65 to this site. Because Erg also binds to this region, we investigated whether the presence of Erg inhibits the binding of NF-κB p65 to DNA, by performing ChIP for NF-κB p65 on HUVEC treated with Erg siRNA. As expected, inhibition of Erg expression by siRNA decreased the amount of Erg binding to the ICAM-1 promoter (see Fig. 1E). Interestingly, this resulted in a significant increase in NF-κB p65 binding to R4 of the ICAM-1 promoter (Fig. 3D). These results suggest that in quiescent EC Erg prevents endogenous nuclear NF-κB p65 from binding to the ICAM-1 promoter.
Erg Represses NF-κB-mediated ICAM-1 Basal Expression
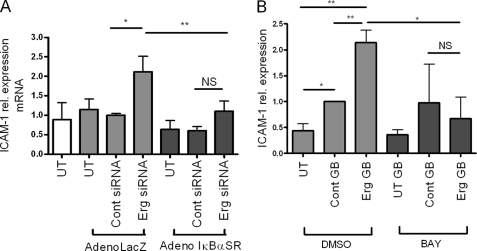
The above data suggest that Erg is repressing ICAM-1 expression by inhibiting NF-κB p65 binding to the ICAM-1 promoter. We therefore investigated whether NF-κB is responsible for the up-regulation of ICAM-1 expression after Erg inhibition. NF-κB activity was inhibited using an adenovirus expressing a mutant super repressor version of IκBα (AdIκBαSR), which cannot be phosphorylated and degraded, resulting in sequestration of NF-κB in the cytoplasm (14). After transduction with AdIκBαSR, the increase in ICAM-1 mRNA expression following Erg inhibition was lost, compared with cells transduced with AdlacZ (Fig. 4A). The inhibitory activity of AdIκBαSR on NF-κB was confirmed on TNF-α-stimulated HUVEC (supplemental Fig. 2A). Similar results were observed using the BAY-117085 inhibitor compound, which blocks IκBα degradation (35): the up-regulation of ICAM-1 protein levels after Erg inhibition was lost in cells treated with BAY-117085 compared with dimethyl sulfoxide control (Fig. 4B). The ability of BAY-117085 to repress the NF-κB pathway was confirmed in TNF-α treated HUVEC (supplemental Fig. 2B). Thus, using two separate inhibitors of NF-κB, we have demonstrated that the up-regulation of ICAM-1 after Erg inhibition is mediated by the activity of NF-κB.
FIGURE 4.
NF-κB induces ICAM-1 expression after Erg inhibition. A, Erg siRNA or control siRNA-treated HUVEC were transduced with AdIκBαSR or AdLacZ. ICAM-1 mRNA levels were measured by quantitative RT-PCR and results are expressed as fold-change compared with control siRNA AdLacZ-treated. n = 5. B, Erg or control Genebloc-transfected HUVEC were treated with BAY-11-7085 or dimethyl sulfoxide (DMSO) control. ICAM-1 protein levels were measured by Western blot using an anti-ICAM-1 antibody and normalized to levels of GAPDH (n = 3). *, p < 0.05; **, p < 0.01.
Genome-wide Analysis Shows That Erg Represses Multiple NF-κB Target Genes
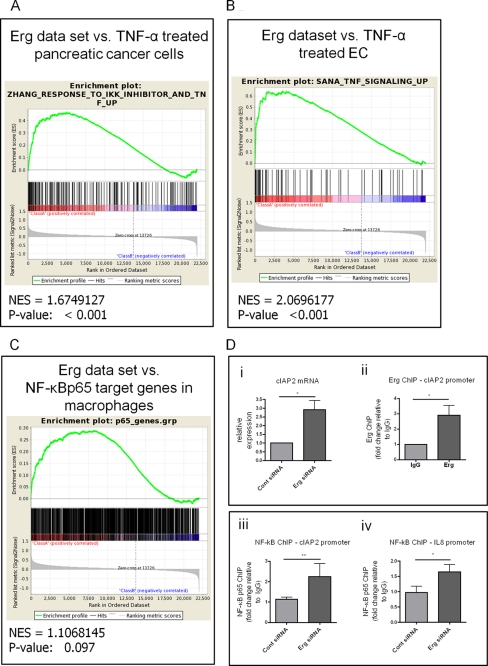
Because Erg inhibits ICAM-1 expression by repressing NF-κB activity, we hypothesized that Erg may also inhibit the expression of other NF-κB target genes in resting endothelium. We have previously investigated the genome-wide targets of Erg by transcription profiling of HUVEC treated with Erg Genebloc (16), and found that Erg represses a number of genes involved in inflammatory processes. To expand this analysis to the entire Erg dataset and find patterns that could support the biological data, we used GSEA (36–38), and compared the Erg dataset with other relevant gene sets, to identify common biological functions. We carried out an initial screen to identify any overlap between genes up-regulated following Erg inhibition and published datasets held in the Broad Institute curated GSEA MSigDB database. Of the top 9 studies with the most significant overlaps in regulated genes, two involved the response to treatment with the NF-κB activating cytokine TNF-α. The first study, in pancreatic cancer cells, investigated genes up-regulated (at least 1.5-fold) by TNF-α treatment that were inhibited by an IKK inhibitor, therefore regulated by NF-κB (17). The second study investigated the response of microvascular and macrovascular endothelial cells to TNF-α, interferon γ, or IL-4 (18). The sets of genes regulated by TNF-α in both studies significantly overlapped with genes up-regulated by Erg inhibition, suggesting a specific alignment of Erg with NF-κB regulatory pathways. We then carried out a separate GSEA between the whole Erg dataset and these two studies. Comparison between the Erg dataset and the genes up-regulated in pancreatic cancer cells after TNF-α treatment gave a significant normalized enrichment score of 1.67 (Fig. 5A), indicating a correlation between NF-κB target genes in pancreatic cancer cells and genes up-regulated by Erg inhibition in HUVEC. Comparison with genes up-regulated in TNF-α-treated endothelial cells gave a significant normalized enrichment score of 2.06 (Fig. 5B). This indicates a correlation between NF-κB pathway target genes in endothelial cells and genes up-regulated by Erg inhibition in HUVEC. In addition, we also used GSEA to compare the Erg-regulated genes with the data from a recent ChIP seq analysis, which identified regulatory sequences bound by NF-κB p65 in LPS-treated macrophages (19). This comparison gave a normalized enrichment score of 1.11 with a p value of 0.097 (Fig. 5C), indicating a trend toward a correlation. Analysis of the three studies together suggests a strong correlation, although not complete overlap, between genes repressed by Erg and genes transactivated by NF-κB. Table 1 lists the shared genes enriched in more than one study. This analysis adds power to our detailed study of repression of ICAM-1 by Erg and suggests that constitutive repression of NF-κB target genes by Erg is a common mechanism to maintain endothelial quiescence.
FIGURE 5.
GSEA demonstrates highly significant enrichment of genes repressed by Erg with genes up-regulated by NF-κB activating cytokine TNF-α. A–C, GSEA was carried out using standard settings. The graphical outputs show enrichment (green curve) of genes up-regulated in TNF-α-treated pancreatic cancer cells (A), endothelial cells (B), or genes bound by NF-κB p65 in macrophages after LPS stimulation (C), along a ranked list of genes up- or down-regulated by 48 h of Erg inhibition. The normalized enrichment score (NES) reflects the degree to which a gene set is overrepresented at the top or bottom of a ranked list, normalized for differences in gene set size and in correlations between gene sets and the expression dataset (D, panel i) mRNA from HUVEC treated with Erg or control siRNA was analyzed by quantitative RT-PCR with primers specific for cIAP2 and normalized to GAPDH, expressed as relative to control siRNA-treated cells (n = 3), *, p < 0.05. D, panel ii, ChIP was carried out on sheared chromatin from confluent resting HUVEC, using an anti-Erg or control IgG antibody. Immunoprecipitated DNA was analyzed by qPCR for primers covering EBS in cIAP2 (see supplemental Fig. S3) and negative control GAPDH promoter region. Data are expressed as fold-change compared with IgG normalized to input and control regions (n = 3), *, p < 0.05. (D, panels iii and iv) ChIP was carried out on sheared chromatin from HUVEC treated with control or Erg siRNA, using an anti NF-κB p65 or control IgG antibody. Immunoprecipitated DNA was analyzed by qPCR for primers covering the NF-κB site and EBS in cIAP2 (D, panel iii), or IL-8 (panel iv) (see supplemental Fig. S3) and a negative control GAPDH promoter region. Data are expressed as fold-change compared with IgG normalized to input and control region (n = 8). *, p < 0.05; **, p < 0.01.
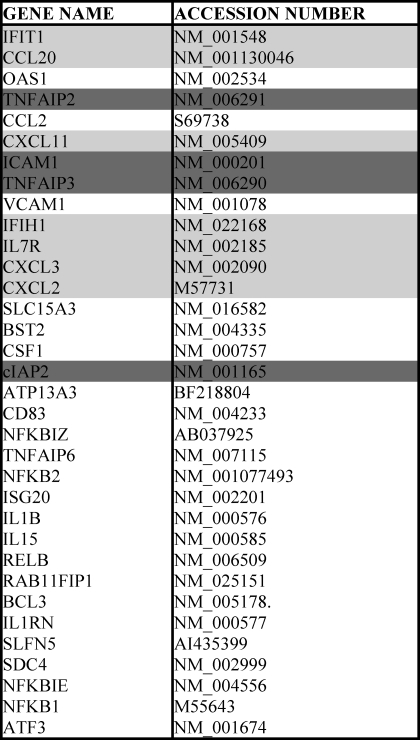
TABLE 1.
Common gene hits and accession numbers from GSEA analysis
List of shared genes enriched in more than one study. Common genes enriched for Erg GSEA among all three studies are in dark grey. Common genes enriched for Erg GSEA with TNF-α-treated pancreatic cancer cells and TNF-α-treated EC studies are in light grey. The remaining genes were commonly enriched between genes bound by NF-κB p65 after LPS treatment, and either TNF-α-treated pancreatic cancer cells or TNF-α-treated EC.
To confirm that Erg repression of NF-κB activation is a common mechanism in resting EC, we focused on two genes: cellular inhibitor of apoptosis (cIAP)-2 and IL-8. cIAP-2 was identified in all three studies used for the GSEA comparisons, whereas IL-8 was identified in the GSEA comparison with the study on TNF-α-treated EC (18) and was previously shown to be repressed by Erg (13). Levels of cIAP2 and IL-8 are low in quiescent EC and are up-regulated by proinflammatory cytokines such as TNF-α (39, 40). Therefore both genes are candidates for regulation by both Erg and NF-κB. First, we showed that Erg inhibition in HUVEC significantly increased the mRNA levels of cIAP2 (Fig. 5D, panel i), confirming that cIAP2 is repressed by Erg in resting HUVEC. We then carried out ChIP to test whether Erg binds to the promoter of cIAP2 in resting HUVEC, using primers designed around the Erg consensus sequence near NF-κB sites (Fig. 5D, panel ii, and supplemental Fig. S3). This showed that Erg binds to the promoter of cIAP2. Previous studies have demonstrated that Erg binds to an EBS in the IL-8 promoter (13). Analysis of the IL-8 promoter showed that the EBS identified as the binding site of Erg is located within the functional NF-κB binding site, similarly to what was observed for ICAM-1 and cIAP2 (supplemental Fig. S3).
Our data shows that Erg inhibits NF-κB p65 binding to the ICAM-1 promoter in quiescent EC. To investigate whether the same applies to the other target genes investigated here, we tested whether Erg inhibits binding of NF-κB p65 to the IL-8 and cIAP2 promoters. ChIP on resting HUVEC treated with control or Erg siRNA showed that NF-κB p65 binding to the IL-8 and cIAP2 promoters increases significantly after Erg inhibition (Fig. 5D, panels iii and iv, respectively). These results indicate that, as with ICAM-1, Erg inhibits constitutive binding of nuclear NF-κB p65 to the promoters of IL-8 and cIAP2. Therefore Erg represses expression of multiple NF-κB target genes in resting EC. In summary, the data in this study describe a novel mechanism controlling endothelial homeostasis, based on the transcription factor Erg, and suggest a novel pathway of inhibition of NF-κB activity in resting endothelium.
DISCUSSION
Healthy EC maintain homeostasis through a dynamic balance between expression of protective genes and repression of proinflammatory genes. In this study we identify a novel mechanism that controls endothelial quiescence through inhibition of NF-κB activity. The ETS transcription factor Erg is known to drive the expression of genes that promote EC homeostasis (8, 10, 11); here we demonstrate that Erg also maintains EC homeostasis through the repression of NF-κB p65 activity. We show that Erg binds to the promoters of a number of NF-κB target genes, repressing their basal expression. We also show that Erg blocks NF-κB p65 binding to the promoters of ICAM-1, IL-8, and cIAP2 in resting HUVEC and that inhibition of Erg results in NF-κB-mediated induction of the expression of these genes. We suggest that in quiescent EC, Erg acts as a gatekeeper, to inhibit the activity of low constitutive levels of nuclear NF-κB p65 and protect the endothelium from the up-regulation of proinflammatory genes, thus maintaining quiescence. Tight control of NF-κB activation is crucial to cellular homeostasis, and several mechanisms, both at the transcriptional and non-transcriptional levels, have been identified. We propose that Erg may provide a checkpoint to pass before induction of NF-κB target genes occurs in EC.
In this study, Erg was shown to interact with two EBS in the ICAM-1 promoter, and both were found to be required for Erg repression of ICAM-1 expression. One of the sites is within the NF-κB consensus binding site, and our data suggest that Erg may directly compete with NF-κB p65 for binding at this site. The role of EBS −118 in Erg repression of ICAM-1 activity, however, is less clear. In the ICAM-1 promoter, the EBS −118 and the NF-κB site are 70 bp apart, which suggest they are approximately half a nucleosome apart. This may allow Erg-mediated repression through modification of the nucleosomal structure of the ICAM-1 promoter. Analysis of the promoter sequences of other genes highlighted in the GSEA identified putative Erg binding sites within a distance equivalent to 1–2 nucleosomes away from an NF-κB site.3 The involvement of distant sites in transcriptional repression has been well described, and may involve recruitment of co-repressors and/or modification of chromatin structure (41). A recent example is provided by BCL6, which was shown to repress a subset of NF-κB target genes in macrophages by binding the promoters at a distance equivalent to approximately one nucleosome or 200 bp away from NF-κB binding sites (19). In the absence of inflammatory stimuli, BCL6 recruited HDAC3 to promoters of NF-κB target genes and repressed their transcription. Erg may similarly repress NF-κB p65 binding by recruiting co-repressors to the promoters of NF-κB target genes. Erg interacts with the histone-3 lysine-9 specific-methyltranferase Erg-associated protein with a SET domain (ESET), and ESET binds co-repressors HDAC-1 and -2, and mSin3A and -B (42, 43). Whether Erg represses NF-κB target genes through recruiting these or other co-repressors is being investigated.
ETS factors regulate specific target genes through combinatorial promoter motifs and interaction with other transcription factors. The enhancers of endothelial specific genes are synergistically activated by FOX and ETS transcription factors containing FOXO:ETS motifs (44). Also, analysis of the genome-wide binding sites of 10 key regulators of blood stem/progenitor cells identified a combinatorial interaction between a heptad of transcription factors including Erg (29). Other ETS combinatorial transcription factor motifs include the serum response elements that bind ternary complex ETS factors and the serum response factor (45), and ETS:AP1 binding motifs (46). Our data highlights for the first time a mechanism of combinatorial repression involving Erg and NF-κB, controlling basal repression of ICAM-1 and other proinflammatory endothelial genes.
Inhibition of Erg in resting HUVEC stimulates leukocyte recruitment (13). This suggests that Erg is required to protect EC against a low level of inflammatory stimuli that may otherwise cause chronic activation and inflammation. Erg levels are inhibited by TNF-α stimulation (9, 13), and recently we have shown that overexpression of Erg inhibits TNF-α-induced NF-κB activity. Moreover we showed that overexpression of Erg inhibited acute inflammation in mouse paws induced by TNF-α (12). This, in conjunction with our present study of the basal repression of proinflammatory genes, suggests that inhibition of Erg is required to induce a NF-κB-mediated inflammatory response in the endothelium, and that Erg can control NF-κB activity at multiple levels.
Interestingly, not all the genes identified by the GSEA comparison of Erg and NF-κB-dependent datasets were repressed through the mechanisms identified here (data not shown), suggesting that other pathways may be involved in Erg-mediated repression. It is possible that some of the genes may be indirect targets of Erg; genomic ChIP sequencing of Erg DNA binding profiles will address this question.
One of the EBS (−118) involved in Erg-mediated repression of ICAM-1 expression was previously shown to be required for the regulation of ICAM-1 in non-endothelial cells. Ets-1, Ets-2, and ERM overexpression in RK13 cells increased ICAM-1 promoter activity through EBS −118 (25). This site is also responsible for the repression of ICAM-1 by the ETS factor FEV. FEV expression is restricted to prostate and small intestine tissue (47) and Dami megakaryocytic cells (27). FEV contains an ETS DNA binding domain, which is highly homologous to Erg and Fli-1 (48); however, its restricted pattern of expression suggests that FEV is not likely to play a role in transcriptional repression in endothelial cells. EBS −834 and −907, suggested to have a role in H2O2-mediated activation of the ICAM-1 promoter (24), do not appear to have a role in Erg-mediated repression.
The role of Erg as a repressor of inflammation is in contrast to that of other ETS factors, which have previously been shown to act synergistically with NF-κB in promoting inflammatory gene expression. Overlapping ETS and NF-κB binding sites have been identified in the regulatory regions of inflammatory genes such as IL-3, IL-12, IL-2, IL-2 receptor-α, and granulocyte-macrophage colony stimulating factor (49–52). Chimeric decoy oligonucleotides containing a combination of EBS and NF-κB consensus sequences were able to inhibit inflammation in abdominal aortic aneurysms to a greater level than either site alone (53). In EC, Erg expression is down-regulated by inflammatory stimuli (9, 13); this is in contrast with ETS factors including Ets-1, Ets-2, and ESE-1, whose expression is increased after treatment with agents such as IL-1β, TNF-α, angiotensin II, or thrombin (9, 17, 54–59). These ETS factors have been shown, in endothelial cells, to drive the expression of proinflammatory genes such as VCAM-1, inducible nitric-oxide synthase, cyclooxygenase-2, and MCP1. In this study we showed that Ets-2, Fli-1, and GABPα, although constitutively expressed in EC, do not repress ICAM-1 expression. The different expression levels and activities of ETS factors highlights their important role in regulating inflammation.
In conclusion, this study provides evidence of the crucial role Erg plays in maintaining a quiescent state in endothelial cells. By inhibiting the activity of constitutive low levels of nuclear NF-κB, Erg represses the transactivation of a set of proinflammatory NF-κB target genes. Thus Erg provides a checkpoint to protect endothelial cells against inappropriate activation, and may prevent the onset of chronic inflammatory vascular diseases, such as atherosclerosis.
Supplementary Material
Acknowledgments
We thank Dr. Odile Dumont, Dr. Richard Starke, Professor Guido Franzoso, Dr. Paul Evans, Professor Malcolm Parker, and Dr. Irina Udalova (Imperial College London) for discussions. Professor Nancy Hogg (Cancer Research UK) for the kind gift of the anti-ICAM-1 antibody, clone 15.5.
This work was supported by grants from the British Heart Foundation (to N. H. D., S. A. M., G. M. B., S. T. K., and A. S.) and Leukemia and Lymphoma Research and the National Institute for Health Research Cambridge Biomedical Research Centre (to R. L. H. and B. G.).

This article contains supplemental Table S1 and Figs. S1–S3.
N. H. Dryden and A. M. Randi, unpublished data.
- EC
- endothelial cell
- ETS
- E-twenty six
- Erg
- ETS-related gene
- HUVEC
- human umbilical vein endothelial cell
- GSEA
- gene set enrichment analysis
- qPCR
- quantitative PCR
- AdErg
- Erg adenovirus
- R
- region
- EGM-2
- endothelial growth medium-2
- ICAM-1
- intercellular adhesion molecule 1
- EBS
- ETS binding site.
REFERENCES
- 1. Oeckinghaus A., Ghosh S. (2009) The NF-κB family of transcription factors and its regulation. Cold Spring Harbor Perspect. Biol. 1, a000034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Li Q., Verma I. M. (2002) NF-κB regulation in the immune system. Nat. Rev. Immunol. 2, 725–734 [DOI] [PubMed] [Google Scholar]
- 3. Hoffmann A., Levchenko A., Scott M. L., Baltimore D. (2002) The IκB-NF-κB signaling module. Temporal control and selective gene activation. Science 298, 1241–1245 [DOI] [PubMed] [Google Scholar]
- 4. Sun S. C., Ganchi P. A., Ballard D. W., Greene W. C. (1993) NF-κB controls expression of inhibitor IκBα. Evidence for an inducible autoregulatory pathway. Science 259, 1912–1915 [DOI] [PubMed] [Google Scholar]
- 5. Rocha V. Z., Libby P. (2009) Obesity, inflammation, and atherosclerosis. Nat. Rev. Cardiol. 6, 399–409 [DOI] [PubMed] [Google Scholar]
- 6. Karim F. D., Urness L. D., Thummel C. S., Klemsz M. J., McKercher S. R., Celada A., Van Beveren C., Maki R. A., Gunther C. V., Nye J. A. (1990) The ETS domain. A new DNA-binding motif that recognizes a purine-rich core DNA sequence. Genes Dev. 4, 1451–1453 [DOI] [PubMed] [Google Scholar]
- 7. Hollenhorst P. C., Jones D. A., Graves B. J. (2004) Expression profiles frame the promoter specificity dilemma of the ETS family of transcription factors. Nucleic Acids Res. 32, 5693–5702 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Pimanda J. E., Chan W. Y., Donaldson I. J., Bowen M., Green A. R., Göttgens B. (2006) Endoglin expression in the endothelium is regulated by Fli-1, Erg, and Elf-1 acting on the promoter and a −8-kb enhancer. Blood 107, 4737–4745 [DOI] [PubMed] [Google Scholar]
- 9. McLaughlin F., Ludbrook V. J., Kola I., Campbell C. J., Randi A. M. (1999) Characterization of the tumor necrosis factor (TNF)-α response elements in the human ICAM-2 promoter. J. Cell Sci. 112, 4695–4703 [DOI] [PubMed] [Google Scholar]
- 10. Birdsey G. M., Dryden N. H., Amsellem V., Gebhardt F., Sahnan K., Haskard D. O., Dejana E., Mason J. C., Randi A. M. (2008) Transcription factor Erg regulates angiogenesis and endothelial apoptosis through VE-cadherin. Blood 111, 3498–3506 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Deramaudt B. M., Remy P., Abraham N. G. (1999) Up-regulation of human heme oxygenase gene expression by Ets family proteins. J. Cell. Biochem. 72, 311–321 [PubMed] [Google Scholar]
- 12. Sperone A., Dryden N. H., Birdsey G. M., Madden L., Johns M., Evans P. C., Mason J. C., Haskard D. O., Boyle J. J., Paleolog E. M., Randi A. M. (2011) The transcription factor Erg inhibits vascular inflammation by repressing NF-κB activation and proinflammatory gene expression in endothelial cells. Arterioscler. Thromb. Vasc. Biol. 31, 142–150 [DOI] [PubMed] [Google Scholar]
- 13. Yuan L., Nikolova-Krstevski V., Zhan Y., Kondo M., Bhasin M., Varghese L., Yano K., Carman C. V., Aird W. C., Oettgen P. (2009) Anti-inflammatory effects of the ETS factor ERG in endothelial cells are mediated through transcriptional repression of the interleukin-8 gene. Circ. Res. 104, 1049–1057 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Brockman J. A., Scherer D. C., McKinsey T. A., Hall S. M., Qi X., Lee W. Y., Ballard D. W. (1995) Coupling of a signal response domain in IκBα to multiple pathways for NF-κB activation. Mol. Cell. Biol. 15, 2809–2818 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Ledebur H. C., Parks T. P. (1995) Transcriptional regulation of the intercellular adhesion molecule-1 gene by inflammatory cytokines in human endothelial cells. Essential roles of a variant NF-κB site and p65 homodimers. J. Biol. Chem. 270, 933–943 [DOI] [PubMed] [Google Scholar]
- 16. Birdsey G. M., Dryden N. H., Shah A. V., Hannah R., Hall M. D., Haskard D. O., Parsons M., Mason J. C., Zvelebil M., Gottgens B., Ridley A. J., Randi A. M. (2012) The transcription factor Erg regulates expression of histone deacetylase 6 and multiple pathways involved in endothelial cell migration and angiogenesis. Blood 119, 894–903 [DOI] [PubMed] [Google Scholar]
- 17. Zhang Y., Gavriil M., Lucas J., Mandiyan S., Follettie M., Diesl V., Sum F. W., Powell D., Haney S., Abraham R., Arndt K. (2008) IκBα kinase inhibitor IKI-1 conferred tumor necrosis factor α sensitivity to pancreatic cancer cells and a xenograft tumor model. Cancer Res. 68, 9519–9524 [DOI] [PubMed] [Google Scholar]
- 18. Sana T. R., Janatpour M. J., Sathe M., McEvoy L. M., McClanahan T. K. (2005) Microarray analysis of primary endothelial cells challenged with different inflammatory and immune cytokines. Cytokine 29, 256–269 [DOI] [PubMed] [Google Scholar]
- 19. Barish G. D., Yu R. T., Karunasiri M., Ocampo C. B., Dixon J., Benner C., Dent A. L., Tangirala R. K., Evans R. M. (2010) Bcl-6 and NF-κB cistromes mediate opposing regulation of the innate immune response. Genes Dev. 24, 2760–2765 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Prasad D. D., Rao V. N., Reddy E. S. (1992) Structure and expression of human Fli-1 gene. Cancer Res. 52, 5833–5837 [PubMed] [Google Scholar]
- 21. Jeong B. C., Kim M. Y., Lee J. H., Kee H. J., Kho D. H., Han K. E., Qian Y. R., Kim J. K., Kim K. K. (2006) Brain-specific angiogenesis inhibitor 2 regulates VEGF through GABP that acts as a transcriptional repressor. FEBS Lett. 580, 669–676 [DOI] [PubMed] [Google Scholar]
- 22. Okada Y., Yano K., Jin E., Funahashi N., Kitayama M., Doi T., Spokes K., Beeler D. L., Shih S. C., Okada H., Danilov T. A., Maynard E., Minami T., Oettgen P., Aird W. C. (2007) A 3-kb fragment of the human Robo4 promoter directs cell type-specific expression in endothelium. Circ. Res. 100, 1712–1722 [DOI] [PubMed] [Google Scholar]
- 23. Voraberger G., Schäfer R., Stratowa C. (1991) Cloning of the human gene for intercellular adhesion molecule 1 and analysis of its 5′-regulatory region. Induction by cytokines and phorbol ester. J. Immunol. 147, 2777–2786 [PubMed] [Google Scholar]
- 24. Roebuck K. A., Rahman A., Lakshminarayanan V., Janakidevi K., Malik A. B. (1995) H2O2 and tumor necrosis factor-α activate intercellular adhesion molecule 1 (ICAM-1) gene transcription through distinct cis-regulatory elements within the ICAM-1 promoter. J. Biol. Chem. 270, 18966–18974 [DOI] [PubMed] [Google Scholar]
- 25. de Launoit Y., Audette M., Pelczar H., Plaza S., Baert J. L. (1998) The transcription of the intercellular adhesion molecule-1 is regulated by Ets transcription factors. Oncogene 16, 2065–2073 [DOI] [PubMed] [Google Scholar]
- 26. Yockell-Lelièvre J., Spriet C., Cantin P., Malenfant P., Heliot L., de Launoit Y., Audette M. (2009) Functional cooperation between Stat-1 and ets-1 to optimize icam-1 gene transcription. Biochem. Cell Biol. 87, 905–918 [DOI] [PubMed] [Google Scholar]
- 27. Maurer P., T'Sas F., Coutte L., Callens N., Brenner C., Van Lint C., de Launoit Y., Baert J. L. (2003) FEV acts as a transcriptional repressor through its DNA-binding ETS domain and alanine-rich domain. Oncogene 22, 3319–3329 [DOI] [PubMed] [Google Scholar]
- 28. Wei G. H., Badis G., Berger M. F., Kivioja T., Palin K., Enge M., Bonke M., Jolma A., Varjosalo M., Gehrke A. R., Yan J., Talukder S., Turunen M., Taipale M., Stunnenberg H. G., Ukkonen E., Hughes T. R., Bulyk M. L., Taipale J. (2010) Genome-wide analysis of ETS family DNA-binding in vitro and in vivo. EMBO J. 29, 2147–2160 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Wilson N. K., Foster S. D., Wang X., Knezevic K., Schütte J., Kaimakis P., Chilarska P. M., Kinston S., Ouwehand W. H., Dzierzak E., Pimanda J. E., de Bruijn M. F., Göttgens B. (2010) Combinatorial transcriptional control in blood stem/progenitor cells. Genome-wide analysis of 10 major transcriptional regulators. Cell Stem Cell 7, 532–544 [DOI] [PubMed] [Google Scholar]
- 30. Hou J., Baichwal V., Cao Z. (1994) Regulatory elements and transcription factors controlling basal and cytokine-induced expression of the gene encoding intercellular adhesion molecule 1. Proc. Natl. Acad. Sci. U.S.A. 91, 11641–11645 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Chen W., Bacanamwo M., Harrison D. G. (2008) Activation of p300 histone acetyltransferase activity is an early endothelial response to laminar shear stress and is essential for stimulation of endothelial nitric-oxide synthase mRNA transcription. J. Biol. Chem. 283, 16293–16298 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Wang J. G., Mahmud S. A., Thompson J. A., Geng J. G., Key N. S., Slungaard A. (2006) The principal eosinophil peroxidase product, HOSCN, is a uniquely potent phagocyte oxidant inducer of endothelial cell tissue factor activity. A potential mechanism for thrombosis in eosinophilic inflammatory states. Blood 107, 558–565 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Pan J., McEver R. P. (1995) Regulation of the human P-selectin promoter by Bcl-3 and specific homodimeric members of the NF-κB/Rel family. J. Biol. Chem. 270, 23077–23083 [DOI] [PubMed] [Google Scholar]
- 34. Xue J., Thippegowda P. B., Hu G., Bachmaier K., Christman J. W., Malik A. B., Tiruppathi C. (2009) NF-κB regulates thrombin-induced ICAM-1 gene expression in cooperation with NFAT by binding to the intronic NF-κB site in the ICAM-1 gene. Physiol. Genomics 38, 42–53 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Pierce J. W., Schoenleber R., Jesmok G., Best J., Moore S. A., Collins T., Gerritsen M. E. (1997) Novel inhibitors of cytokine-induced IκBα phosphorylation and endothelial cell adhesion molecule expression show anti-inflammatory effects in vivo. J. Biol. Chem. 272, 21096–21103 [DOI] [PubMed] [Google Scholar]
- 36. Subramanian A., Tamayo P., Mootha V. K., Mukherjee S., Ebert B. L., Gillette M. A., Paulovich A., Pomeroy S. L., Golub T. R., Lander E. S., Mesirov J. P. (2005) Gene set enrichment analysis. A knowledge-based approach for interpreting genome-wide expression profiles. Proc. Natl. Acad. Sci. U.S.A. 102, 15545–15550 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Subramanian A., Kuehn H., Gould J., Tamayo P., Mesirov J. P. (2007) GSEA-P, a desktop application for Gene Set Enrichment Analysis. Bioinformatics 23, 3251–3253 [DOI] [PubMed] [Google Scholar]
- 38. Mootha V. K., Lindgren C. M., Eriksson K. F., Subramanian A., Sihag S., Lehar J., Puigserver P., Carlsson E., Ridderstråle M., Laurila E., Houstis N., Daly M. J., Patterson N., Mesirov J. P., Golub T. R., Tamayo P., Spiegelman B., Lander E. S., Hirschhorn J. N., Altshuler D., Groop L. C. (2003) PGC-1α-responsive genes involved in oxidative phosphorylation are coordinately down-regulated in human diabetes. Nat. Genet. 34, 267–273 [DOI] [PubMed] [Google Scholar]
- 39. Furusu A., Nakayama K., Xu Q., Konta T., Sugiyama H., Kitamura M. (2001) Expression, regulation, and function of inhibitor of apoptosis family genes in rat mesangial cells. Kidney Int. 60, 579–586 [DOI] [PubMed] [Google Scholar]
- 40. Strieter R. M., Kunkel S. L., Showell H. J., Remick D. G., Phan S. H., Ward P. A., Marks R. M. (1989) Endothelial cell gene expression of a neutrophil chemotactic factor by TNF-α, LPS, and IL-1α. Science 243, 1467–1469 [DOI] [PubMed] [Google Scholar]
- 41. Payankaulam S., Li L. M., Arnosti D. N. (2010) Transcriptional repression. Conserved and evolved features. Curr. Biol. 20, R764-R771 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Yang L., Xia L., Wu D. Y., Wang H., Chansky H. A., Schubach W. H., Hickstein D. D., Zhang Y. (2002) Molecular cloning of ESET, a novel histone H3-specific methyltransferase that interacts with ERG transcription factor. Oncogene 21, 148–152 [DOI] [PubMed] [Google Scholar]
- 43. Yang L., Mei Q., Zielinska-Kwiatkowska A., Matsui Y., Blackburn M. L., Benedetti D., Krumm A. A., Taborsky G. J., Jr., Chansky H. A. (2003) An ERG (ets-related gene)-associated histone methyltransferase interacts with histone deacetylases 1/2 and transcription co-repressors mSin3A/B. Biochem. J. 369, 651–657 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. De Val S., Chi N. C., Meadows S. M., Minovitsky S., Anderson J. P., Harris I. S., Ehlers M. L., Agarwal P., Visel A., Xu S. M., Pennacchio L. A., Dubchak I., Krieg P. A., Stainier D. Y., Black B. L. (2008) Combinatorial regulation of endothelial gene expression by Ets and Forkhead transcription factors. Cell 135, 1053–1064 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Buchwalter G., Gross C., Wasylyk B. (2004) Ets ternary complex transcription factors. Gene 324, 1–14 [DOI] [PubMed] [Google Scholar]
- 46. Wasylyk B., Hagman J., Gutierrez-Hartmann A. (1998) Ets transcription factors. Nuclear effectors of the Ras-MAP-kinase signaling pathway. Trends Biochem. Sci. 23, 213–216 [DOI] [PubMed] [Google Scholar]
- 47. Peter M., Couturier J., Pacquement H., Michon J., Thomas G., Magdelenat H., Delattre O. (1997) A new member of the ETS family fused to EWS in Ewing tumors. Oncogene 14, 1159–1164 [DOI] [PubMed] [Google Scholar]
- 48. Carrère S., Verger A., Flourens A., Stehelin D., Duterque-Coquillaud M. (1998) Erg proteins, transcription factors of the Ets family, form homo-, heterodimers, and ternary complexes via two distinct domains. Oncogene 16, 3261–3268 [DOI] [PubMed] [Google Scholar]
- 49. Gottschalk L. R., Giannola D. M., Emerson S. G. (1993) Molecular regulation of the human IL-3 gene. Inducible T cell-restricted expression requires intact AP-1 and Elf-1 nuclear protein binding sites. J. Exp. Med. 178, 1681–1692 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Thomas R. S., Tymms M. J., McKinlay L. H., Shannon M. F., Seth A., Kola I. (1997) ETS1, NFκB, and AP1 synergistically transactivate the human GM-CSF promoter. Oncogene 14, 2845–2855 [DOI] [PubMed] [Google Scholar]
- 51. Gri G., Savio D., Trinchieri G., Ma X. (1998) Synergistic regulation of the human interleukin-12 p40 promoter by NFκB and Ets transcription factors in Epstein-Barr virus-transformed B cells and macrophages. J. Biol. Chem. 273, 6431–6438 [DOI] [PubMed] [Google Scholar]
- 52. John S., Reeves R. B., Lin J. X., Child R., Leiden J. M., Thompson C. B., Leonard W. J. (1995) Regulation of cell type-specific interleukin-2 receptor α-chain gene expression. Potential role of physical interactions between Elf-1, HMG-I(Y), and NF-κB family proteins. Mol. Cell. Biol. 15, 1786–1796 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Shiraya S., Miwa K., Aoki M., Miyake T., Oishi M., Kataoka K., Ohgi S., Ogihara T., Kaneda Y., Morishita R. (2006) Hypertension accelerated experimental abdominal aortic aneurysm through up-regulation of nuclear factor κB and Ets. Hypertension 48, 628–636 [DOI] [PubMed] [Google Scholar]
- 54. Goetze S., Kintscher U., Kaneshiro K., Meehan W. P., Collins A., Fleck E., Hsueh W. A., Law R. E. (2001) TNFα induces expression of transcription factors c-fos, Egr-1, and Ets-1 in vascular lesions through extracellular signal-regulated kinases 1/2. Atherosclerosis 159, 93–101 [DOI] [PubMed] [Google Scholar]
- 55. Hultgårdh-Nilsson A., Cercek B., Wang J. W., Naito S., Lövdahl C., Sharifi B., Forrester J. S., Fagin J. A. (1996) Regulated expression of the ets-1 transcription factor in vascular smooth muscle cells in vivo and in vitro. Circ. Res. 78, 589–595 [DOI] [PubMed] [Google Scholar]
- 56. Cheng C., Tempel D., Den Dekker W. K., Haasdijk R., Chrifi I., Bos F. L., Wagtmans K., van de Kamp E. H., Blonden L., Biessen E. A., Moll F., Pasterkamp G., Serruys P. W., Schulte-Merker S., Duckers H. J. (2011) Ets2 determines the inflammatory state of endothelial cells in advanced atherosclerotic lesions. Circ. Res. 109, 382-395 [DOI] [PubMed] [Google Scholar]
- 57. Redlich K., Kiener H. P., Schett G., Tohidast-Akrad M., Selzer E., Radda I., Stummvoll G. H., Steiner C. W., Gröger M., Bitzan P., Zenz P., Smolen J. S., Steiner G. (2001) Overexpression of transcription factor Ets-1 in rheumatoid arthritis synovial membrane. Regulation of expression and activation by interleukin-1 and tumor necrosis factor α. Arthritis Rheum. 44, 266–274 [DOI] [PubMed] [Google Scholar]
- 58. Rudders S., Gaspar J., Madore R., Voland C., Grall F., Patel A., Pellacani A., Perrella M. A., Libermann T. A., Oettgen P. (2001) ESE-1 is a novel transcriptional mediator of inflammation that interacts with NF-κB to regulate the inducible nitric-oxide synthase gene. J. Biol. Chem. 276, 3302–3309 [DOI] [PubMed] [Google Scholar]
- 59. Grall F., Gu X., Tan L., Cho J. Y., Inan M. S., Pettit A. R., Thamrongsak U., Choy B. K., Manning C., Akbarali Y., Zerbini L., Rudders S., Goldring S. R., Gravallese E. M., Oettgen P., Goldring M. B., Libermann T. A. (2003) Responses to the proinflammatory cytokines interleukin-1 and tumor necrosis factor α in cells derived from rheumatoid synovium and other joint tissues involve nuclear factor κB-mediated induction of the Ets transcription factor ESE-1. Arthritis Rheum. 48, 1249–1260 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.