Background: WRN and DNA polymerase δ are involved in DNA replication and repair.
Results: WRN synergizes with Pol δ to degrade alternate DNA structures. WRN excises terminal mismatches to enable DNA chain extension by Pol δ.
Conclusion: WRN and Pol δ together minimize the accumulation of aberrant DNA structures and ensure unhindered DNA replication.
Significance: WRN contributes to the fidelity of DNA transactions, including replication.
Keywords: DNA Helicase, DNA Polymerase, DNA Repair, DNA Synthesis, DNA-Protein Interaction, Alternate DNA Structures, DNA Exonuclease, DNA Polymerase δ, Werner Syndrome Exonuclease
Abstract
DNA Polymerase δ (Pol δ) and the Werner syndrome protein, WRN, are involved in maintaining cellular genomic stability. Pol δ synthesizes the lagging strand during replication of genomic DNA and also functions in the synthesis steps of DNA repair and recombination. WRN is a member of the RecQ helicase family, loss of which results in the premature aging and cancer-prone disorder, Werner syndrome. Both Pol δ and WRN encode 3′ → 5′ DNA exonuclease activities. Pol δ exonuclease removes 3′-terminal mismatched nucleotides incorporated during replication to ensure high fidelity DNA synthesis. WRN exonuclease degrades DNA containing alternate secondary structures to prevent formation and enable resolution of stalled replication forks. We now observe that similarly to WRN, Pol δ degrades alternate DNA structures including bubbles, four-way junctions, and D-loops. Moreover, WRN and Pol δ form a complex with enhanced ability to hydrolyze these structures. We also present evidence that WRN can proofread for Pol δ; WRN excises 3′-terminal mismatches to enable primer extension by Pol δ. Consistent with our in vitro observations, we show that WRN contributes to the maintenance of DNA synthesis fidelity in vivo. Cells expressing limiting amounts (∼10% of normal) of WRN have elevated mutation frequencies compared with wild-type cells. Together, our data highlight the importance of WRN exonuclease activity and its cooperativity with Pol δ in preserving genome stability, which is compromised by the loss of WRN in Werner syndrome.
Introduction
Werner syndrome is an autosomal recessive disorder characterized by accelerated aging and genomic instability (1). At the molecular level, genomic instability is manifested by the accumulation of large DNA deletions (2) and at the organismal level by increased prevalence of mesenchymal cell-derived cancers, particularly sarcomas (3). WRN,2 the gene mutated in Werner syndrome, is a member of the RecQ family of DNA helicases. Like all members of this family, WRN encodes a helicase that unwinds DNA with 3′ → 5′ directionality (4). In addition to B-form DNA, WRN unwinds DNA structures such as triplex and quadruplex DNA, DNA containing nicks, gaps, hairpins, and bubbles, splayed arm structures, model DNA replication forks, and recombination intermediates such as 3- and 4-way junctions and D-loops (5–8). Unique among the RecQ family, WRN encodes a 3′ → 5′ DNA exonuclease with sequence similarity to Escherichia coli RNase D and to the proofreading exonuclease of E. coli DNA polymerase I (9, 10). Although WRN proficiently removes 3′-terminal mismatched nucleotides, it does not digest single-stranded DNA, a classical hallmark of proofreading exonucleases (11), or blunt DNA duplexes. However, it hydrolyzes non-canonical DNA substrates (12). These structures are recognized and bound with exquisite specificity by WRN. WRN binds at the junction of a four-way junction DNA structure and at junctions of single strand-double strand in replication forks (13). Cumulative evidence suggests that by virtue of its ability to unwind and degrade DNAs with alternate secondary structures, WRN helps to prevent replication forks from stalling and to resolve forks once stalled (14).
WRN has been shown to physically and functionally interact with multiple proteins, including DNA polymerases δ (Pol δ). Pol δ is one of the major replicative DNA polymerases in eukaryotic cells (15, 16). Extensive evidence indicates that Pol δ is responsible for lagging-strand DNA synthesis (17). In combination with proliferating cell nuclear antigen, Pol δ is highly processive and accurate. High fidelity DNA synthesis by Pol δ is ensured through selectivity at the level of nucleotide incorporation and by hydrolysis of mismatched nucleotides partitioned from the polymerase active site to the proofreading exonuclease active site (18). Pol δ is also a key enzyme in DNA repair and recombination, processes that can also require high fidelity DNA synthesis (18).
WRN and Pol δ interact both physically and functionally. Although Szekely et al. (16) demonstrated a physical interaction between WRN and Pol δ, we showed that WRN preferentially increases the rate of polymerization by Pol δ (15). The increase was observed only in the absence of proliferating cell nuclear antigen, suggesting that the WRN-Pol δ complex is most likely not involved in processive DNA replication but rather in proliferating cell nuclear antigen-independent processes. In addition, by virtue of its helicase activity, we showed that WRN resolves DNA structural roadblocks such as hairpins and quadruplexes to alleviate stalling and enable synthesis by Pol δ (19). Shah et al. (20) recently confirmed these results and showed in addition that progression of Pol δ-mediated DNA synthesis is hindered at hairpins and microsatellite sequences within the fragile FRA16D region. Inclusion of WRN increased the processivity of human Pol δ and alleviated its stalling. Both reports highlight the role of WRN helicase activity in assisting Pol δ-mediated DNA synthesis.
In previous studies on the functional interaction of WRN with Pol δ, we utilized polymerase holoenzyme from Saccharomyces cerevisiae, a heterotrimeric complex comprising of Pol 3p, Pol 31p, and Pol32p subunits (21). It lacks the fourth smallest subunit, homologous to the p12 subunit that is present in human Pol δ, and that modulates both the rate and fidelity of DNA synthesis by human Pol δ holoenzyme (22). The presence of this fourth subunit is likely to differently affect the functional interaction of human Pol δ with WRN. We have thus undertaken to explore in this study interactions between nearly homogeneous four-subunit human Pol δ holoenzyme and WRN. We report the following novel observations. (i) Pol δ can exonucleolytically digest blunt duplexes containing non-canonical DNA structures, and that WRN co-operates with Pol δ to degrade these structures. (ii) WRN exonuclease can substitute for Pol δ exonuclease to remove single 3′-terminal mismatched nucleotides. (iii) The spontaneous random point mutation frequency is higher in human cells depleted of WRN and, therefore, deficient in WRN helicase and exonuclease. We hypothesize that WRN and Pol δ co-operate to maintain the fidelity of DNA transactions and, thus, genomic stability.
EXPERIMENTAL PROCEDURES
Enzymes
His-tagged full-length wild-type, helicase-minus, and exonuclease-minus WRN were purified from baculovirus-infected insect cells as described previously (23). Recombinant four-subunit wild-type and exonuclease-defective human Pol δ holoenzymes were expressed and purified from E. coli using the protocols published by Matsumoto and co-workers (24) and implemented in our laboratory (25). Human and S. cerevisiae Pol δ lacking the p12 subunit were kindly provided by the Fanning (Vanderbilt University) and Burgers (Washington University) laboratories, respectively. His-tagged human Pol β was expressed and purified from E. coli. Pol η was purchased from Enzymax (Lexington, KY). Human Trex1 was a generous gift from Fred Perrino (Wake Forest University). All enzymes were >90% homogeneous. Molar amounts of WRN and Pol δ were calculated on the basis of monomeric molecular weights.
DNA Oligonucleotides
PAGE-purified oligonucleotides were purchased from Integrated DNA Technologies or Invitrogen (DL1-3). DNA oligonucleotides were 5′-end-labeled using [γ-32P]ATP and T4 polynucleotide kinase (26). DNA substrates were generated by annealing the indicated combinations of oligonucleotides (asterisks indicate the 5′-end labeled DNA oligonucleotide: 4-way/X junction DNA: *HJ1, HJ2, HJ3, and HJ4; bubble DNA: *B1 and B2; blunt duplex: *B1 and B3; D-loop: DL1, DL2, and *DL3; matched primer-template (P/T): *Match 1 or *Match 2 and B1; mismatched P/T: *Mismatch and B1. Sequences are HJ1, 5′-CGCGCCGAATTCCCGCTAGCAATATTCTGCAGCCAAGCTTCCGCGC; HJ2, 5′-GCGCGGAAGCTTGGCTGCAGAATATATAAGGATCCAATCATGAGGC; HJ3, 5′-GCCTCATGATTGGATCCTTATGTGTAAGTGCGAATGGCAGTTCGCC; HJ4, 5′-GGCGAACTGCCATTCGCACTTACACTGCTAGCGGGAATTCGGCGCG; B1, 5′-GCGCGGAAGCTTGGCTGCAGAATATTGCTAGCGGGAATTCGGCGCG; B2, 5′-CGCGCCGAATTCCCGCTAGTGGCGCCTTGCAGCCAAGCTTCCGCGC; B3, 5′-CGCGCCGAATTCCCGCTAGCAATATTCTGCAGCCAAGCTTCCGCGC; DL1, 5′-GCCAGGGACGGGGTGAACCTGCAGGTGGGCGGCTGCTCATCGTAGGTTAGTATCGACCTATTGGTAGAATTCGGCAGCGTCATGCGACGGC; DL2, 5′-GCCGTCGCATGACGCTGCCGAATTCTACCACGCTACTAGGGTGCCTTGCTAGGACATCTTTGCCCACCTGCAGGTTCACCCCGTCCCTGGC; DL3, 5′-AAGATGGGTCCTAGCAAGGCACCCTAGTAGC; Match1, 5′-CGCGCCGAATTCCCGCTAGCAAT; Match2, 5′CGCGCCGAATTCCCGCTAGCAATA; Mismatch, 5′-CGCGCCGAATTCCCGCTAGCAATG.
Enzyme Assays
Exonuclease
Reactions were carried out in mixtures that contained in a final volume of 10 μl 40 mm Tris-HCl, pH 7.5, 4 mm MgCl2, 5 mm DTT, 1 mm ATP, and 0.1 mg/ml BSA (23). The indicated amounts of WRN and Pol δ were incubated with 10 nm concentrations of a specified DNA substrate at 37 °C for 10–20 min. Reactions were terminated by the addition of formamide-EDTA stop solution. The mixtures were boiled, and aliquots were electrophoresed through urea, 14% polyacrylamide gels. Radioactivity was visualized on a PhosphorImager (GE Healthcare) and quantified by using ImageJ software. Percent hydrolysis was quantified by determining the ratio of degradation products to the sum of substrate plus products.
Polymerase
Primer extension reactions with matched or mismatched P/T DNA were carried out under identical conditions as the exonuclease reactions, except that ATP was substituted with 100 μm concentrations of each of the four dNTPs. WRN and Pol δ were used at concentrations indicated in figure legends.
Electrophoretic Mobility Shift Assay
Binding of WRN or Pol δ or both to HJ DNA was carried out in exonuclease reaction buffer containing the non-hydrolyzable analog ATPγS instead of ATP. WRN and Pol δ were preincubated on ice for 5 min before the addition of reaction components, including DNA (100 fmol), and reaction mixtures were subsequently incubated at room temperature for 15 min. For antibody supershift assays, anti-WRN antibody (195C, Sigma; 1 μg) was added, and the reactions were incubated for an additional 15 min at room temperature. Reactions were transferred to 0 °C for 10 min, and glycerol was added to a final concentration of 15%. Reaction aliquots were electrophoresed through 3% native acrylamide gels at a constant voltage of 290 V at 4 °C. Gels were dried and imaged on a PhosphorImager to visualize free DNA and protein-bound DNA complexes.
Depletion of WRN in Mammalian Cells
shRNA-mediated depletion of WRN was carried out in a SV40 immortalized human fibroblast cell line, GM639, by retroviral transduction. Briefly, pBABEpuro retrovirus expressing a short hairpin construct with homology to WRN cDNA nt 160–184, was obtained by transient transfection of 293T cells (27). The resulting virus was used to infect GM639 cells. After incubation with the virus for 24 h, puromycin (1 μg/ml) was added to select infected cells expressing the siRNA (28). Depletion was confirmed to be ∼90%.
Oligonucleotide Probe Retrieval Assay
In vivo mutation frequencies were determined by a sensitive quantitative PCR assay that measures the fidelity of DNA transactions on substrates introduced into cells.3 Briefly, control and WRN-depleted cells were transiently transfected with a primer-template DNA probe harboring a single TaqI (TCGA) restriction site and a 5′ biotin moiety. The in vivo extended DNA probe was retrieved 24 h after transfection by using streptavidin-coated magnetic beads (Invitrogen), and mutations at the TaqI site were quantified by quantitative PCR amplification (Refs. 29 and 30; see the supplemental Methods for a detailed description of the assay).
RESULTS
Human Pol δ Degrades DNA-containing Alternate Secondary Structures
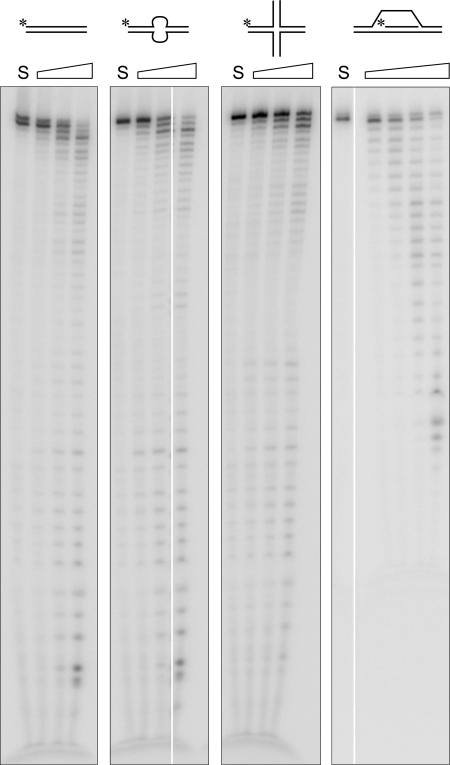
DNA Pol δ encodes 3′ → 5′ proofreading exonuclease activity that ensures high fidelity DNA synthesis during cellular DNA replication; it does so by removing 3′-terminal misincorporated nucleotides that impair the addition of subsequent nucleotides. In vitro, Pol δ displays robust exonuclease activity in the absence of dNTPs on primer-template (3′-recessed ends) DNA substrates and on single-stranded DNA (ssDNA). We asked whether blunt DNA duplexes or duplexes with internal secondary structures positioned at least 20 nt upstream of the 3′ terminus are substrates of Pol δ exonuclease. We observed that Pol δ can degrade in vitro a blunt duplex, a blunt duplex with an internal 8-nt bubble, 4-way junction DNA, and the 3′ invading strand in a D-loop mimic (Fig. 1). The efficiency of degradation under our assay conditions was D-loop ≫ bubble DNA > blunt duplex ≥ 4-way junction, with D-loop DNA being degraded 15–20-fold more efficiently than the other examined DNA substrates (65% of D-loop DNA was hydrolyzed by 18 fmol of Pol δ in 10 min versus 30–45% hydrolysis of the remaining substrates by 100 fmol in 20 min). The fact that the observed activity was not due to a contaminating exonuclease in our preparation of Pol δ was established by using a variant, D402A Pol δ, in which the active site Asp residue is replaced with Ala. D402A Pol δ retains wild-type DNA polymerase activity but is unable to hydrolyze ssDNA (25). We noted that D402A is also unable to hydrolyze DNA substrates with alternate secondary structures (see Fig. 3A).
FIGURE 1.
DNA Pol δ can degrade DNA with alternate structures. Increasing amounts of human 4-subunit Pol δ holoenzyme were incubated with 10 nm concentrations of either blunt duplex DNA, a blunt duplex with an internal 8-nt bubble, a model X-junction structure, or a D-loop mimic as described under “Experimental Procedures” (30, 100, and 300 fmol of Pol δ with all substrates, except D-loop that was incubated with 18, 60, and 180 fmol of Pol δ). The reactions were incubated at 37 °C for 20 min (10 min with D-loop DNA), terminated by the addition of formamide stop solution, boiled, and aliquots were electrophoresed through 14% urea polyacrylamide gels. The gels were dried, imaged, and quantified on a PhosphorImager (GE Healthcare).
FIGURE 3.
Synergistic degradation of bubble DNA requires exonuclease-proficient WRN and exonuclease-proficient Pol δ. Wild-type WRN (50 fmol) was mixed with three different amounts (30, 100, and 300 fmol; indicated as 1, 2, and 3, respectively) of exonuclease-minus Pol δ (D402A) (A), or wild-type Pol δ (60 fmol) was combined with increasing amounts (3–25 fmol; represented as 1–4) of exonuclease-minus WRN (B). Reactions were otherwise carried out as described in the legend to Fig. 2.
WRN Synergizes with DNA Pol δ to Degrade Alternate DNA Substrates
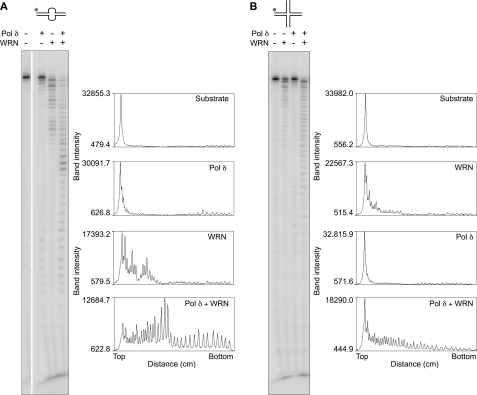
A hallmark of WRN exonuclease is its ability to hydrolyze DNA containing alternate secondary structures (12). Because we observed that Pol δ also degrades such DNA substrates, we looked at the extent of degradation of bubble and X-junction DNA in reaction mixtures that contained both WRN and Pol δ. Surprisingly, we observed more than an additive effect on DNA degradation by combining the two enzymes. Both the amount of substrate depleted (>95%) and the profile of digestion products differed markedly from those generated by each enzyme separately (Fig. 2, A and B). A ladder of degradation products corresponding to cleavage at each nucleotide position and spanning the length of the template was observed with WRN + Pol δ on both DNA substrates. Four-way junction DNA that was formed with a completely different sequence set of oligonucleotides also showed the same response (not shown), indicating that degradation is structure- but not sequence- dependent. Furthermore, four-way junction DNA with a recessed 3′-end was an equally effective substrate for revealing synergistic interactions between WRN and Pol δ exonucleases. Cooperativity was thus not limited to blunt-ended DNA substrates in which removal of the 3′-terminal paired nucleotide could be rate-limiting.
FIGURE 2.
WRN and DNA Pol δ co-operate to degrade bubble and X-junction DNA. WRN (50 fmol) and DNA Pol δ (60 fmol) were preincubated on ice for ∼2 min either separately or together as indicated. Reactions were initiated by the addition of DNA and incubated and processed as described above. The band intensities of exonucleolytic DNA products, obtained using ImageJ software, are plotted to better illustrate synergism between WRN and Pol δ exonucleases. A, bubble DNA. B, four-way junction DNA.
Requirements for Synergistic Hydrolysis
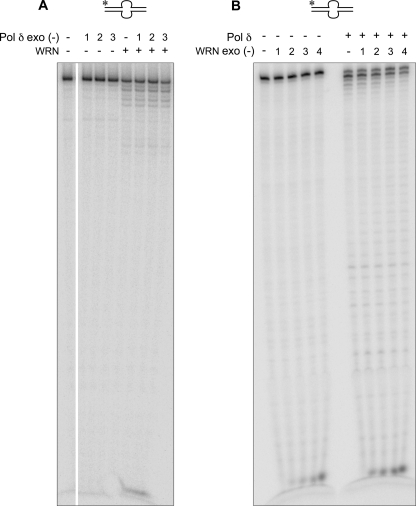
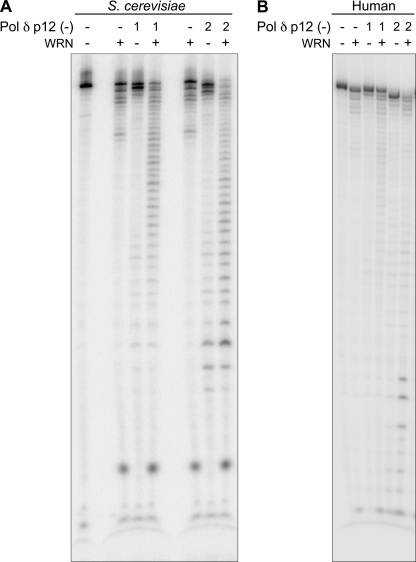
In characterizing requisite features of the WRN-Pol δ interaction that result in synergism of DNA hydrolysis, we noted that synergism requires the active co-operation of both exonucleases. Synergism was not observed when exonuclease-deficient D402A Pol δ was used in combination with wild-type WRN or significantly reduced when exonuclease-proficient Pol δ was used with exonuclease-deficient WRN (Fig. 3, A and B, respectively). Second, both WRN and Pol δ have to be present together to observe maximal degradation. If either Pol δ or WRN was preincubated with the DNA substrate and then heat-inactivated at 63 °C before the addition of the second enzyme, degradation was not synergistic (supplemental Fig. 1). Control experiments indicated that the heat treatment rendered the enzymes inactive without altering the DNA structure. Third, an N-terminal truncated variant of WRN encoding a minimal exonuclease (amino acids 1–240) failed to elicit the same response as full-length WRN (supplemental Fig. 2). Thus, co-operative interactions between WRN and Pol δ exonuclease require more than the exonuclease domain of WRN, consistent with the report that the Pol δ interaction domain on WRN maps to the C terminus (16). However, we determined that the helicase activity of WRN is not required for exonuclease synergism. Neither a point mutation in the helicase domain (K577M) that abolishes unwinding nor the omission of ATP that is absolutely essential for DNA unwinding eliminates cooperativity between Pol δ and WRN exonuclease (Fig. 4). Last, we examined the requirement of the p12 subunit that is reported to modulate the fidelity of Pol δ (22). We observed that both S. cerevisiae and human Pol δ lacking the p12 subunit are proficient at hydrolyzing alternate DNA and, importantly, retain the ability to synergize with WRN, indicating that this subunit is dispensable for functional interactions between Pol δ exonuclease and WRN exonuclease (Fig. 5, A and B).
FIGURE 4.
Synergism between WRN and Pol δ exonucleases is independent of DNA unwinding. WRN (20 fmol) and Pol δ (45 fmol) were incubated either individually or together with bubble DNA in reactions lacking ATP or containing the helicase minus WRN variant, K577M, as indicated. Exonucleolytic digestion of DNA was compared with control reactions with ATP.
FIGURE 5.
The p12 subunit of Pol δ is dispensable for exonuclease synergism with WRN. Reactions with bubble DNA were carried out as described, except that the 4-subunit Pol δ holoenzyme was replaced with either the S. cerevisiae (A) or human (B) 3-subunit Pol δ complex that lacks the p12 subunit. Lanes 1 and 2 refer to 0.5 and 1.0 fmol of S. cerevisiae Pol δ or 10 and 40 fmol of human Pol δ, respectively; WRN was used at 6 fmol per reaction.
Specificity of WRN-Pol δ Exonuclease Interaction
The synergism exhibited by WRN and Pol δ was limited to specific DNA substrates and was not observed when WRN was combined with another exonuclease.
DNA Substrates
Not all substrates revealed cooperative interactions between WRN and Pol δ. A blunt duplex lacking the internal 8-nt bubble, but otherwise of identical sequence as the bubble DNA substrate, is digested by Pol δ but not by WRN. Likewise, ssDNA is an efficient target of Pol δ exonuclease but is not digested by WRN. Exonucleolytic digestion of both these substrates by Pol δ was not altered in the presence of WRN (data not shown). Interestingly, the D-loop mimic is a substrate for both WRN and Pol δ exonuclease (Fig. 1), but unlike X-junction and bubble DNA, no cooperativity was observed in reactions containing both these exonucleases (supplemental Fig. 3A).
Other Exonucleases
Trex1 is a human 3′ → 5′ exonuclease that belongs to the same family of proofreading exonucleases as Pol δ and WRN (31). We observed that Trex1 also degrades four-way junction and bubble DNA. However, unlike the cooperativity between WRN and Pol δ, WRN and Trex1 (at amounts that yield comparable exonuclease activities) do not synergize to hydrolyze these DNA substrates (supplemental Fig. 3B and data not shown).
WRN and Pol δ Form a Complex with X-junction DNA
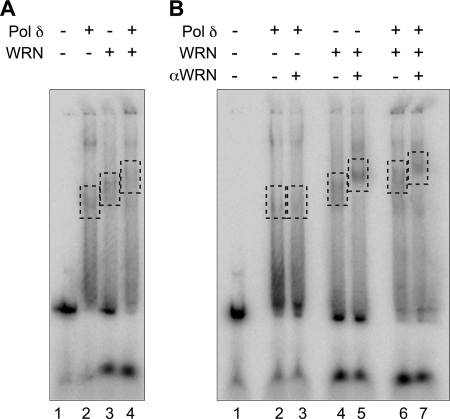
We performed electrophoretic mobility shift assays to determine whether Pol δ forms stable complexes with DNA containing alternate secondary structures. Our data indicate that similarly to WRN, Pol δ binds X-junction DNA to generate discrete slower migrating complexes on a non-denaturing gel (Fig. 6A, lane 2). Interestingly, when WRN and Pol δ were incubated together with DNA, we observed the emergence of a complex, larger than the major complexes formed by either enzyme alone (Fig. 6A, lane 4). An antibody against WRN that supershifted the WRN·X-junction DNA complex (Fig. 6B, lane 5) also slowed the mobility of the complex observed with Pol δ + WRN, indicating the presence of WRN in this complex (Fig. 6B, lane 7). These results are consistent with formation of a tripartite DNA-WRN-Pol δ complex.
FIGURE 6.
WRN and Pol δ form a complex on X-junction DNA. WRN (∼50 fmol) and Pol δ (∼1000 fmol) were preincubated either separately or together on ice for 5 min. After the addition of DNA (100 fmol) and ATPγS, reaction mixtures were incubated at room temperature for 15 min. For supershift assays, anti-WRN antibody was added, and the reactions were incubated for an additional 15 min at room temperature. The reactions were transferred to ice for 15 min, and after the addition of glycerol to a final concentration of 15%, aliquots were electrophoresed through 3% non-denaturing gels at 290 V at 4 °C until the bromphenol blue front was 7 cm from the point of application. Gels were dried and developed on a PhosphorImager to visualize free and protein-bound DNA complexes. Complexes formed by Pol δ and WRN in the absence (A) or presence (B) of anti-WRN antibody are highlighted by dashed rectangles.
WRN Enables Pol δ to Extend Past 3′-terminal Mismatches When Copying DNA Templates
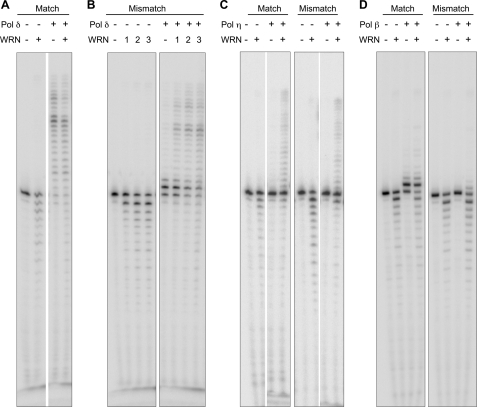
WRN degrades P/T DNA substrates containing a single 3′-terminal mismatched nucleotide at a greater efficiency than substrates without a mismatch (11). Therefore, we asked if by virtue of this ability, WRN enables Pol δ to extend DNA containing a 3′-terminal G:T mismatched nucleotide. To reveal maximal effects of this interaction, we chose to use exonuclease-deficient Pol δ. Surprisingly, exo (−) Pol δ is unable to extend the mismatched primer beyond a single nucleotide even in the presence of all four dNTPs (Fig. 7B). Under the assay conditions ∼50% of the primer remained unextended and another 40% was extended by just a single nucleotide. However, in the presence of WRN, >90% of the primer was extended to span the full-length of the template (Fig. 7B). Similar results were obtained with other mismatched nucleotide combinations (not shown). At concentrations equivalent to those of mismatched P/T, a matched primer-template was efficiently extended by exo (−) Pol δ, and the effect of WRN was less apparent (Fig. 7A). The effect of WRN was also not evident if the mismatch was positioned either 2 or 6 nt upstream of the 3′ terminus or if the primer-template contained two 3′-terminal mismatched nucleotides (not shown). Thus, the ability of WRN to enable primer extension by Pol δ is restricted to P/T DNA containing a single 3′-terminal mismatched nucleotide.
FIGURE 7.
WRN enables DNA Pol δ to preferentially extend a primer bearing a single 3′- terminal G:T mismatched nucleotide. Wild-type WRN and/or DNA Pol δ exo (−) were incubated with 10 nm primer-template DNA containing either a matched (A) or a single 3′-terminal mismatched nucleotide (B). DNA Pol δ exo (−) was used at 5 fmol per reaction, whereas WRN was used at 30 fmol (A) or at 6, 15, and 30 fmol (1, 2, and 3, respectively, in B). As controls, WRN (15 fmol) was also incubated with either Pol η (∼0.3 fmol; C) or Pol β (2 fmol; D). After 10 min at 37 °C in the presence of 100 μm concentrations of each of the four dNTPs, reactions were terminated, and aliquots were electrophoresed through urea-14% polyacrylamide gels. Gels were dried, and reaction products were visualized on a PhosphorImager.
We inquired whether the cooperation between WRN and Pol δ is reproduced when Pol δ is substituted with other human DNA polymerases. We used translesion DNA polymerase, Pol η, and the BER DNA polymerase, Pol β, both of which have been reported to functionally interact with WRN and that lack intrinsic proofreading exonuclease activity. Unlike Pol δ, the extension activity of Pol η was stimulated to the same extent by WRN on both matched and mismatched P/T; there was no preferential stimulation on the mismatched P/T (Fig. 7C). On the other hand, extension by Pol β on either of the two DNA substrates was not responsive to WRN. Pol β generated a predominant +1 extension product on the matched P/T but was unable to significantly extend this product in the presence of WRN. On the mismatched P/T, Pol β was unable to extend the primer by even a single nucleotide, and the addition of WRN had only a minimal effect despite having demonstrable exonuclease activity (Fig. 7D).
WRN Excises the Mismatch before Extension by Pol δ
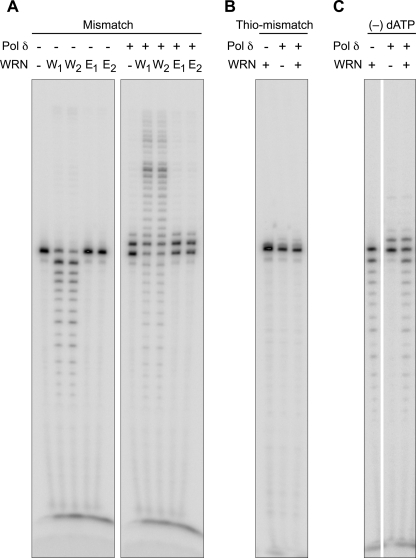
We used several approaches to demonstrate that WRN excises the mismatch before extension of the primer by Pol δ. First, we used exonuclease-deficient WRN; in stark contrast to the wild-type protein, exo (−) WRN is unable to assist Pol δ to extend the primer (Fig. 8A). Second, we employed a primer in which the terminal and penultimate nucleotide are bonded through a thio linkage. In the absence of excision of the thio linkage by WRN, Pol δ is again unable to extend the mismatched primer (Fig. 8B). Finally, we omitted dATP, complementary to template dTMP of the mismatch. Although WRN is able to excise the mismatch (revealed by the presence of a robust −1 product), Pol δ was unable to extend the excised product in the absence of the first complementary nucleotide (Fig. 8C). In aggregate, these lines of data indicate that excision of the mismatched terminal nucleotide by WRN is necessary for subsequent extension of the primer stem by Pol δ.
FIGURE 8.
Extension of a mismatched primer terminus by DNA Pol δ requires removal of the mismatch by WRN. Two equivalent amounts (30 and 60 fmol) of either wild-type (W1 and W2) or exonuclease-minus WRN (E1 and E2) were incubated with DNA Pol δ exo (−) (5 fmol) and G:T mismatched P/T DNA (A). Alternatively, fixed concentrations of WRN (15 fmol) and Pol δ (3 fmol) were incubated with P/T DNA in which the terminal two nucleotides were linked with a thio bond (Thio-mismatch, B) or in reaction mixtures lacking dATP (−) dATP, C). Reactions were carried out and processed as described.
WRN Contributes to DNA Replication Fidelity in Vivo
The fact that WRN can uniquely assist Pol δ to remove terminal mismatches and enable DNA synthesis led us to explore its contribution to the overall fidelity of DNA synthesis in vivo. We used an oligonucleotide probe retrieval assay that allows us to transfect human cells with short fragments of primer-template DNA and to quantify the frequency of errors generated during in vivo extension of the primer DNA across a TaqI restriction enzyme cleavage site by real time quantitative PCR (supplemental Methods).3
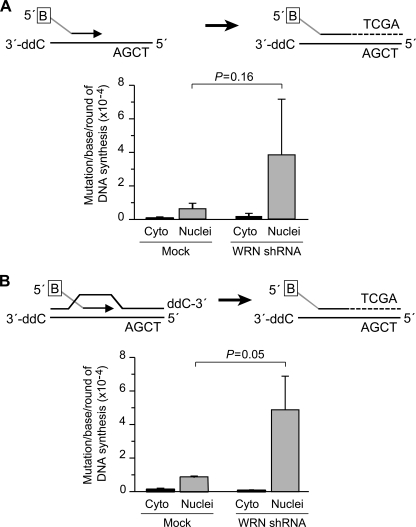
Immortalized human fibroblasts were infected with retrovirus expressing WRN-specific shRNA to selectively deplete WRN. Western blot analysis revealed that the efficiency of WRN depletion was ≥90% (27). By contrast, no depletion was observed in mock-transfected cells. We noted that primer extension on both DNA substrates was more error-prone in WRN-depleted cells than in control WRN-expressing cells (Fig. 9, A and B). A reproducible 5-fold increase in mutation frequency, measured by the ability to amplify full-length PCR products, was observed in the absence of WRN. Thus, depletion of WRN decreases the fidelity of chain elongation of transfected DNA substrates in vivo.
FIGURE 9.
WRN-depleted cells exhibit increased mutagenesis. SV40-immortalized fibroblasts were infected with pBabe retrovirus expressing WRN-specific shRNA sequences. Seven days post-infection, short oligonucleotides bearing TaqI restriction enzyme cleavage sites and resembling primer-template (A) or D-loop (B) DNA (as described in supplemental Methods) were introduced in to WRN-depleted or control cells. The in vivo extended oligonucleotide probes were retrieved with streptavidin-coated magnetic beads from whole cell extracts (cyto) or nuclei, digested with TaqI restriction enzyme to eliminate wild-type sequences, and amplified by quantitative PCR. Mutagenesis was scored by the ability to amplify full-length PCR products due to absence of cleavage of the extended oligonucleotide by TaqI restriction enzyme. B in the box, 5′-biotin; 3′-ddC, dideoxycytidine.
DISCUSSION
We present evidence in this report for functional interactions between WRN exonuclease and human DNA Pol δ holoenzyme. These interactions enable synergistic degradation of alternate DNA structures and facilitate extension of DNA containing a single mismatched nucleotide at the 3′-primer terminus. The cooperative actions of WRN and Pol δ could also play a role in the observed contribution of WRN to the maintenance of replication fidelity in vivo.
We have previously shown that WRN binds to and degrades DNA bearing alternate secondary structures (12). We now observe that Pol δ also binds stably to structured DNA substrates (Fig. 6). Moreover, similarly to WRN, Pol δ exonuclease degrades blunt-ended DNA with internal secondary DNA structures that resemble intermediates of DNA replication and recombination (Fig. 1). These previously unreported DNA substrates of Pol δ exonuclease implicate it in additional transactions besides replication of genomic DNA. In fact, genetic studies in S. cerevisiae have shown that Pol δ functions in both meiotic and mitotic DNA recombination (32, 33).
A noteworthy finding of our study is the synergism between WRN exonuclease and Pol δ exonuclease. This is perhaps the first report of a synergistic interaction between two 3′ → 5′ DNA exonucleases. We show that WRN and Pol δ form a complex on structured DNA substrates (Fig. 6) and that this complex of exonuclease-competent WRN and Pol δ enables the enzymes to cooperate in digesting bubble and X-junction DNA to completion (Fig. 2, A and B). This effect is not manifested when WRN is combined with Trex1 (supplemental Fig. 3B) and is also not observed with all DNA substrates; among those examined in this study, only bubble DNA and X-junction DNA were synergistically degraded (Fig. 2, A and B). Recent studies from Orren and co-workers (34, 35) have implicated WRN in regression of stalled replication forks to form four-stranded “chicken-foot” structures and in the reformation of functional replication forks from regressed structures. Synergism between WRN and Pol δ exonucleases at the fork may be required to facilitate these processes.
It has been reported that p12, the smallest subunit of human Pol δ, modulates the exonuclease activity of the complex (22). Pol δ lacking the p12 subunit exhibits decreased polymerase activity but increased exonuclease activity compared with the four-subunit holoenzyme complex (36, 37). We observed that Pol δ lacking the p12 subunit exhibits proficient exonuclease activity on alternate DNA structures. Moreover, it retains the ability to synergize with WRN in hydrolyzing bubble DNA (Fig. 5, A and B). Thus, from a mechanistic standpoint, the p12 subunit does not contribute to functional exonuclease interactions between Pol δ and WRN. Furthermore, the fact that we observe synergism between WRN exonuclease and S. cerevisiae 3-subunit Pol δ (Fig. 5B) is remarkable; it shows that this function of Pol δ is conserved through evolution. Depending on in vivo cellular conditions, human Pol δ could exist in different forms, i.e. with different subunit compositions, each serving a specific role. For instance, it has been shown that upon exposure to DNA-damaging agents, there is rapid loss of p12 and a concomitant increase in the level of three-subunit Pol δ (38). Although the four-subunit human Pol δ is optimal for processive polymerase activity, the three-subunit enzyme lacking p12 could be a potent exonuclease that is required for repair processes after DNA damage.
Cooperativity between WRN exonuclease and other exonucleases has not been previously reported. However, stimulation of WRN exonuclease by the Ku70/80 proteins that lack intrinsic exonuclease activity has been demonstrated (39, 40). Ku70/80 stimulates a minimal WRN exonuclease domain to the same extent as full-length WRN. In contrast, our data reveal that synergism with Pol δ necessitates the use of more than the WRN exonuclease domain (supplemental Fig. 2), but it does not require WRN helicase activity (Fig. 4). Mechanistically, this suggests that DNA unwinding and/or ATP hydrolysis by WRN is not necessary for synergistic interactions between the two exonucleases.
Our studies also revealed another facet of WRN exonuclease; that is, that it can substitute for Pol δ exonuclease when the latter is either limiting or absent (Fig. 7B). This was evident on P/T DNA containing a single mismatched nucleotide at the 3′ primer terminus. Whereas Pol δ exo (−) is unable to extend the 3′ G:T mismatched primer by more than a single nucleotide, WRN allows it to fully extend the primer (Fig. 7B). WRN does so by removing the terminal mismatch before extension by Pol δ (Fig. 8, A–C). This effect is preferential to the WRN-Pol δ holoenzyme combination. Neither WRN-Pol η nor WRN-Pol β was able to preferentially extend a mismatched primer terminus (Fig. 7, C and D). Harrigan et al. (41) have reported the contrary: that WRN can proofread for Pol β. Under steady state conditions with excess DNA substrate and Pol β amounts comparable with those used with Pol δ, we were unable to demonstrate that WRN facilitates extension of mismatched P/T DNA by Pol β. Differences in reaction conditions could likely be responsible for the discrepancy.
There is precedence for the involvement of autonomous 3′ → 5′ DNA exonucleases in cooperation with proofreading DNA polymerases in mutation avoidance mechanisms (42). It is hypothesized that this is especially important under conditions of genotoxic stress when proofreading by DNA polymerases becomes rate-limiting. Does an autonomous exonuclease such as WRN, therefore, contribute to maintenance of fidelity during DNA synthesis in vivo? We show that, unlike wild-type cells, cells that lack WRN exhibit a 5-fold elevated mutation frequency during extension of synthetic DNA substrates in vivo. Using a completely different reporter assay, Bacolla et al. (43) also recently noted that WRN deficiency increases the frequency of base substitution errors 2-fold in human cells. This spontaneous increase in mutation frequency may be low, but it is observed in unstressed cells that are proficient for Pol δ and ϵ proofreading. The dependence on WRN could be greater in cells that are exposed to DNA-damaging agents or that are deficient/limited in proofreading activity. Although the identity of the DNA polymerase whose fidelity is modulated by WRN in vivo is unknown, based on our cumulative in vitro findings (Figs. 7 and 8), we hypothesize that it could be Pol δ. The fact that we do not observe increased fidelity in the absence of WRN argues against the involvement of error-prone translesion DNA polymerases, whose activities we have shown are stimulated by WRN (44).
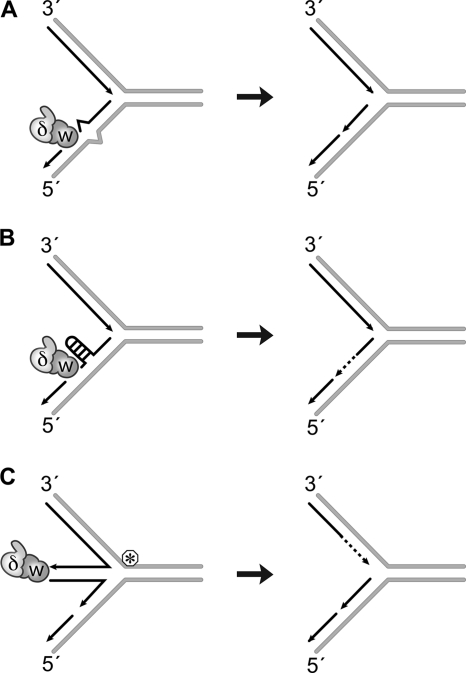
Are these seemingly disparate observations, WRN-Pol δ exonuclease synergism and proofreading capability of WRN- related? We believe that they are. We hypothesize that WRN and Pol δ cooperate with each other to prevent replication forks from stalling when it encounters roadblocks and to enable resumption of fork progression after collapse. The roadblocks could include DNA sequences that assume non-canonical secondary structures or DNA lesions/adducts and nucleotide mismatches that perturb the local environment and may be potentially perceived as alternate structures (Fig. 10). We have previously shown that the unwinding activity of WRN resolves DNA hairpins and tetraplex structures ahead of the fork to enable chain elongation by Pol δ (19). Our new observations now suggest that the exonuclease activity of WRN can likewise prevent Pol δ from stalling by hydrolyzing blocking structures in newly synthesized DNA. The multimeric nature of WRN could account for unwinding of roadblocks ahead of the fork and hydrolysis of daughter strands. If forks do stall, four-stranded DNA structures that could form to facilitate lesion repair can be hydrolyzed by the action of WRN and Pol δ exonucleases. Exonucleolytic processing may be necessary to ensure accurate synthesis during replication fork restart.
FIGURE 10.
Model for WRN and Pol δ exonuclease function at replication forks. Hydrolysis of a single mismatched nucleotide at the 3′-primer terminus (A) or that of alternate DNA structures assumed by the newly synthesized strand (B) could facilitate uninterrupted DNA synthesis by DNA Pol δ on the lagging strand. Alternately, replication forks stalled by a lesion/roadblock are believed to form four-stranded structures. Hydrolysis of DNA in such structures could ensure accurate synthesis after lesion repair (C). W, WRN; δ, Pol δ; circled asterisk, lesion/replication roadblock.
The importance of WRN exonuclease and Pol δ exonuclease in maintaining the fidelity of DNA transactions and preventing the accumulation of mutations and ensuing genomic instability is evident in the following reports. First, in sequencing the WRN exonuclease domain from individuals with sarcomas, one of the most frequently observed cancers in Werner syndrome patients, we identified a variant that results in a Pro-Leu substitution at amino acid 204 in WRN. The P204L variant protein is expressed at 10-fold lower levels and has a further 10-fold lower exonuclease activity than wild-type WRN. In effect, this individual with the sarcoma has a net 100-fold reduction of WRN exonuclease activity (45). Second, mice homozygous for the D400A mutation that inactivates Pol δ exonuclease activity exhibit elevated spontaneous mutation rates and develop cancers at an early age (46). These observations together with data presented in this report highlight the importance of WRN and Pol δ exonucleases in preserving genomic integrity.
Supplementary Material
Acknowledgments
We thank members of the Loeb laboratory and of the University of Washington Werner Syndrome Program Project for helpful comments and discussions and Dr. Michael Fry for critically reading the manuscript and providing insightful input throughout the course of this work. We are extremely grateful to Fred Perrino for the generous gift of human Trex 1 exonuclease and to the Monnat laboratory for providing bacterial strains with pBabe plasmids required to generate WRN-depleting retrovirus.
This work was supported, in whole or in part, by National Institutes of Health Grants P01-CA77852, R01-CA102029, and P01-AG0751 (to L. A. L.).

This article contains supplemental Methods, Table 1, and Figs. 1–3.
J. C. Shen and L. A. Loeb, submitted for publication.
- WRN
- Werner syndrome protein
- Pol δ
- DNA polymerase δ
- ssDNA
- single-stranded DNA
- ATPγS
- adenosine 5′-O-(thiotriphosphate)
- n
- nucleotide(s)
- P/T
- primer-template.
REFERENCES
- 1. Epstein C. J., Martin G. M., Schultz A. L., Motulsky A. G. (1966) Werner's syndrome. A review of its symptomatology, natural history, pathologic features, genetics, and relationship to the natural aging process. Medicine 45, 177–221 [DOI] [PubMed] [Google Scholar]
- 2. Fukuchi K., Martin G. M., Monnat R. J., Jr. (1989) Mutator phenotype of Werner syndrome is characterized by extensive deletions. Proc. Natl. Acad. Sci. U.S.A. 86, 5893–5897 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Goto M., Miller R. W., Ishikawa Y., Sugano H. (1996) Excess of rare cancers in Werner syndrome (adult progeria). Cancer Epidemiol. Biomarkers Prev. 5, 239–246 [PubMed] [Google Scholar]
- 4. Gray M. D., Shen J. C., Kamath-Loeb A. S., Blank A., Sopher B. L., Martin G. M., Oshima J., Loeb L. A. (1997) The Werner syndrome protein is a DNA helicase. Nat. Genet. 17, 100–103 [DOI] [PubMed] [Google Scholar]
- 5. Brosh R. M., Jr., Majumdar A., Desai S., Hickson I. D., Bohr V. A., Seidman M. M. (2001) Unwinding of a DNA triple helix by the Werner and Bloom syndrome helicases. J. Biol. Chem. 276, 3024–3030 [DOI] [PubMed] [Google Scholar]
- 6. Fry M., Loeb L. A. (1999) Human Werner syndrome DNA helicase unwinds tetrahelical structures of the fragile X syndrome repeat sequence d(CGG)n. J. Biol. Chem. 274, 12797–12802 [DOI] [PubMed] [Google Scholar]
- 7. Mohaghegh P., Karow J. K., Brosh R. M., Jr., Bohr V. A., Hickson I. D. (2001) The Bloom's and Werner's syndrome proteins are DNA structure-specific helicases. Nucleic Acids Res. 29, 2843–2849 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Orren D. K., Theodore S., Machwe A. (2002) The Werner syndrome helicase/exonuclease (WRN) disrupts and degrades D-loops in vitro. Biochemistry 41, 13483–13488 [DOI] [PubMed] [Google Scholar]
- 9. Moser M. J., Holley W. R., Chatterjee A., Mian I. S. (1997) The proofreading domain of Escherichia coli DNA polymerase I and other DNA and/or RNA exonuclease domains. Nucleic Acids Res. 25, 5110–5118 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Mushegian A. R., Bassett D. E., Jr., Boguski M. S., Bork P., Koonin E. V. (1997) Positionally cloned human disease genes. Patterns of evolutionary conservation and functional motifs. Proc. Natl. Acad. Sci. U.S.A. 94, 5831–5836 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Kamath-Loeb A. S., Shen J. C., Loeb L. A., Fry M. (1998) Werner syndrome protein. II. Characterization of the integral 3' → 5' DNA exonuclease. J. Biol. Chem. 273, 34145–34150 [DOI] [PubMed] [Google Scholar]
- 12. Shen J. C., Loeb L. A. (2000) Werner syndrome exonuclease catalyzes structure-dependent degradation of DNA. Nucleic Acids Res. 28, 3260–3268 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Compton S. A., Tolun G., Kamath-Loeb A. S., Loeb L. A., Griffith J. D. (2008) The Werner syndrome protein binds replication fork and holliday junction DNAs as an oligomer. J. Biol. Chem. 283, 24478–24483 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Rossi M. L., Ghosh A. K., Bohr V. A. (2010) Roles of Werner syndrome protein in protection of genome integrity. DNA Repair 9, 331–344 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Kamath-Loeb A. S., Johansson E., Burgers P. M., Loeb L. A. (2000) Functional interaction between the Werner syndrome protein and DNA polymerase δ. Proc. Natl. Acad. Sci. U.S.A. 97, 4603–4608 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Szekely A. M., Chen Y. H., Zhang C., Oshima J., Weissman S. M. (2000) Werner protein recruits DNA polymerase δ to the nucleolus. Proc. Natl. Acad. Sci. U.S.A. 97, 11365–11370 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Nick McElhinny S. A., Gordenin D. A., Stith C. M., Burgers P. M., Kunkel T. A. (2008) Division of labor at the eukaryotic replication fork. Mol. Cell 30, 137–144 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Hubscher U., Spadari S., Villani G., Maga G. (2010) DNA Polymerases: Discovery, Characterization and Functions in Cellular DNA Transactions, World Scientific Publishing Co., Singapore [Google Scholar]
- 19. Kamath-Loeb A. S., Loeb L. A., Johansson E., Burgers P. M., Fry M. (2001) Interactions between the Werner syndrome helicase and DNA polymerase δ specifically facilitate copying of tetraplex and hairpin structures of the d(CGG)n trinucleotide repeat sequence. J. Biol. Chem. 276, 16439–16446 [DOI] [PubMed] [Google Scholar]
- 20. Shah S. N., Opresko P. L., Meng X., Lee M. Y., Eckert K. A. (2010) DNA structure and the Werner protein modulate human DNA polymerase δ-dependent replication dynamics within the common fragile site FRA16D. Nucleic Acids Res. 38, 1149–1162 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Gerik K. J., Li X., Pautz A., Burgers P. M. (1998) Characterization of the two small subunits of Saccharomyces cerevisiae DNA polymerase δ. J. Biol. Chem. 273, 19747–19755 [DOI] [PubMed] [Google Scholar]
- 22. Meng X., Zhou Y., Lee E. Y., Lee M. Y., Frick D. N. (2010) The p12 subunit of human polymerase δ modulates the rate and fidelity of DNA synthesis. Biochemistry 49, 3545–3554 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Shen J. C., Gray M. D., Oshima J., Kamath-Loeb A. S., Fry M., Loeb L. A. (1998) Werner syndrome protein. I. DNA helicase and DNA exonuclease reside on the same polypeptide. J. Biol. Chem. 273, 34139–34144 [DOI] [PubMed] [Google Scholar]
- 24. Fazlieva R., Spittle C. S., Morrissey D., Hayashi H., Yan H., Matsumoto Y. (2009) Proofreading exonuclease activity of human DNA polymerase δ and its effects on lesion-bypass DNA synthesis. Nucleic Acids Res. 37, 2854–2866 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Schmitt M. W., Matsumoto Y., Loeb L. A. (2009) High fidelity and lesion bypass capability of human DNA polymerase delta. Biochimie 91, 1163–1172 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Sambrook J., Russell D. W. (2001) Molecular Cloning: A Laboratory Manual, 3rd Ed., pp. 10.17–10.19, Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY [Google Scholar]
- 27. Grandori C., Wu K. J., Fernandez P., Ngouenet C., Grim J., Clurman B. E., Moser M. J., Oshima J., Russell D. W., Swisshelm K., Frank S., Amati B., Dalla-Favera R., Monnat R. J., Jr. (2003) Werner syndrome protein limits MYC-induced cellular senescence. Genes Dev. 17, 1569–1574 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Sidorova J. M., Li N., Folch A., Monnat R. J., Jr. (2008) The RecQ helicase WRN is required for normal replication fork progression after DNA damage or replication fork arrest. Cell Cycle 7, 796–807 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Bielas J. H., Loeb L. A. (2005) Quantification of random genomic mutations. Nat. Methods 2, 285–290 [DOI] [PubMed] [Google Scholar]
- 30. Wright J. H., Modjeski K. L., Bielas J. H., Preston B. D., Fausto N., Loeb L. A., Campbell J. S. (2011) A random mutation capture assay to detect genomic point mutations in mouse tissue. Nucleic Acids Res. 39, e73. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Mazur D. J., Perrino F. W. (1999) Identification and expression of the TREX1 and TREX2 cDNA sequences encoding mammalian 3' → 5' exonucleases. J. Biol. Chem. 274, 19655–19660 [DOI] [PubMed] [Google Scholar]
- 32. Maloisel L., Bhargava J., Roeder G. S. (2004) A role for DNA polymerase δ in gene conversion and crossing over during meiosis in Saccharomyces cerevisiae. Genetics 167, 1133–1142 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Maloisel L., Fabre F., Gangloff S. (2008) DNA polymerase δ is preferentially recruited during homologous recombination to promote heteroduplex DNA extension. Mol. Cell Biol. 28, 1373–1382 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Machwe A., Karale R., Xu X., Liu Y., Orren D. K. (2011) The Werner and Bloom syndrome proteins help resolve replication blockage by converting (regressed) holliday junctions to functional replication forks. Biochemistry 50, 6774–6788 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Machwe A., Xiao L., Lloyd R. G., Bolt E., Orren D. K. (2007) Replication fork regression in vitro by the Werner syndrome protein (WRN). Holliday junction formation, the effect of leading arm structure, and a potential role for WRN exonuclease activity. Nucleic Acids Res. 35, 5729–5747 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Meng X., Zhou Y., Zhang S., Lee E. Y., Frick D. N., Lee M. Y. (2009) DNA damage alters DNA polymerase δ to a form that exhibits increased discrimination against modified template bases and mismatched primers. Nucleic Acids Res. 37, 647–657 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Podust V. N., Chang L. S., Ott R., Dianov G. L., Fanning E. (2002) Reconstitution of human DNA polymerase δ using recombinant baculoviruses. The p12 subunit potentiates DNA polymerizing activity of the four-subunit enzyme. J. Biol. Chem. 277, 3894–3901 [DOI] [PubMed] [Google Scholar]
- 38. Zhang S., Zhou Y., Trusa S., Meng X., Lee E. Y., Lee M. Y. (2007) A novel DNA damage response. Rapid degradation of the p12 subunit of DNA polymerase δ. J. Biol. Chem. 282, 15330–15340 [DOI] [PubMed] [Google Scholar]
- 39. Li B., Comai L. (2000) Functional interaction between Ku and the Werner syndrome protein in DNA end processing. J. Biol. Chem. 275, 28349–28352 [DOI] [PubMed] [Google Scholar]
- 40. Li B., Comai L. (2001) Requirements for the nucleolytic processing of DNA ends by the Werner syndrome protein-Ku70/80 complex. J. Biol. Chem. 276, 9896–9902 [DOI] [PubMed] [Google Scholar]
- 41. Harrigan J. A., Wilson D. M., 3rd, Prasad R., Opresko P. L., Beck G., May A., Wilson S. H., Bohr V. A. (2006) The Werner syndrome protein operates in base excision repair and cooperates with DNA polymerase β. Nucleic Acids Res. 34, 745–754 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Shevelev I. V., Hübscher U. (2002) The 3' → 5' exonucleases. Nat. Rev. Mol. Cell Biol. 3, 364–376 [DOI] [PubMed] [Google Scholar]
- 43. Bacolla A., Wang G., Jain A., Chuzhanova N. A., Cer R. Z., Collins J. R., Cooper D. N., Bohr V. A., Vasquez K. M. (2011) Non-B DNA-forming sequences and WRN deficiency independently increase the frequency of base substitution in human cells. J. Biol. Chem. 286, 10017–10026 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Kamath-Loeb A. S., Lan L., Nakajima S., Yasui A., Loeb L. A. (2007) Werner syndrome protein interacts functionally with translesion DNA polymerases. Proc. Natl. Acad. Sci. U.S.A. 104, 10394–10399 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Hsu J. J., Kamath-Loeb A. S., Glick E., Wallden B., Swisshelm K., Rubin B. P., Loeb L. A. (2010) Werner syndrome gene variants in human sarcomas. Mol. Carcinog. 49, 166–174 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Goldsby R. E., Lawrence N. A., Hays L. E., Olmsted E. A., Chen X., Singh M., Preston B. D. (2001) Defective DNA polymerase-δ proofreading causes cancer susceptibility in mice. Nat. Med. 7, 638–639 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.