Background: Human SAMHD1 protein restricts HIV/SIV infection of myeloid cells and is targeted for proteasomal degradation by HIV-2 Vpx protein.
Results: Vpx binds the divergent C terminus of human SAMHD1 and loads it onto DCAF1 substrate receptor of CRL4 E3 ubiquitin ligase.
Conclusion: Vpx programs SAMHD1 degradation by loading it onto CRL4DCAF1.
Significance: Learning how viruses overcome innate anti-viral mechanisms is critical for the conception of new antiviral therapeutics.
Keywords: E3 Ubiquitin Ligase, HIV, Protein Complexes, Protein Degradation, Virus, AIDS, SIV, Innate Immunity, Restriction Factor
Abstract
The sterile alpha motif and HD domain-containing protein-1 (SAMHD1) inhibits infection of myeloid cells by human and related primate immunodeficiency viruses (HIV and SIV). This potent inhibition is counteracted by the Vpx accessory virulence factor of HIV-2/SIVsm viruses, which targets SAMHD1 for proteasome-dependent degradation, by reprogramming cellular CRL4DCAF1 E3 ubiquitin ligase. However, the precise mechanism of Vpx-dependent recruitment of human SAMHD1 onto the ligase, and the molecular interfaces on the respective molecules have not been defined. Here, we show that human SAMHD1 is recruited to the CRL4DCAF1-Vpx E3 ubiquitin ligase complex by interacting with the DCAF1 substrate receptor subunit in a Vpx-dependent manner. No stable association is detectable with DCAF1 alone. The SAMHD1 determinant for the interaction is a short peptide located distal to the SAMHD1 catalytic domain and requires the presence of Vpx for stable engagement. This peptide is sufficient to confer Vpx-dependent recruitment to CRL4DCAF1 and ubiquitination when fused to heterologous proteins. The precise amino acid sequence of the peptide diverges among SAMHD1 proteins from different vertebrate species, explaining selective down-regulation of human SAMHD1 levels by Vpx. Critical amino acid residues of SAMHD1 and Vpx involved in the DCAF1-Vpx-SAMDH1 interaction were identified by mutagenesis. Our findings show that the N terminus of Vpx, bound to DCAF1, recruits SAMHD1 via its C terminus to CRL4, in a species-specific manner for proteasomal degradation.
Introduction
The innate immune system is the first line of defense against infection by pathogens. Myeloid cells such as dendritic cells and macrophages play important roles in immune response to viruses by linking innate detection of an invading virus to anti-viral adaptive immune response (1, 2). HIV-1 differs from HIV-2 and related simian viruses in its inability to transduce myeloid cells. HIV-1 is unable to transduce dendritic cells and infects macrophages very inefficiently, because of a cellular mechanism that curtails infection at an early post-entry step by interfering with efficient synthesis of viral cDNA (3, 4). In contrast, HIV-2 and a subset of SIV strains, including SIV from sootymangabey (SIVsm), transduce myeloid cells more efficiently, because of the presence of the virion-associated Vpx accessory protein that counteracts the restriction (3, 5–7). Interestingly, HIV-2 infection is not pandemic, in contrast to HIV-1, suggesting that the ability of HIV-2 to establish infection of myeloid cells may contribute to a better adaptive immune response and hence more effective control of the invading virus by the immune system of the host.
The restriction factor inhibiting HIV/SIV infection of human primary myeloid cells was recently identified as the cellular sterile alpha motif (SAM)2 domain HD domain protein 1 (SAMHD1) (8–10). SAMHD1 possesses deoxynucleotide triphosphohydrolase activity conferred by its extended HD domain, and probably inhibits HIV/SIV infection by maintaining deoxynucleotide triphosphate concentrations in non-dividing myeloid cells below the threshold required for robust viral reverse transcription (3, 11–15).
SAMHD1-mediated restriction plays an important role in primate lentivirus infection. Depletion of SAMHD1 levels, by RNA interference or by transduction with SIVmac or HIV-2 Vpx protein, relieves restriction of HIV-1, and Vpx-deficient HIV-2/SIVmac infection of DC, monocyte-derived macrophages (MDM), and terminally differentiated myeloid THP-1 cells (8–10). Furthermore, lack of endogenous SAMHD1 expression in monocytes from an individual with Aicardi-Goutières Syndrome, because of a premature stop codon mutation, renders these cells permissive for HIV-1 replication, thus corroborating earlier evidence that SAMHD1 protects myeloid cells from infection by primate lentiviruses and providing persuasive evidence for the importance of SAMHD1 in HIV-1 pathogenesis (10). Finally, the fact that HIV-2 and SIV viruses evolved virion-associated Vpx proteins that target SAMHD1 for degradation, and thereby counteract the restrictive mechanism, clearly illustrates the key role of SAMHD1 in defending the monocyte-derived cell lineages from HIV-2/SIV infection (8–10).
Vpx relieves the restriction of HIV/SIV infection of myeloid cells by directing SAMHD1 for degradation by the proteasome (8–10). Such a strategy is commonly used by HIV and other viruses, whose accessory virulence factors often usurp cellular E3 ubiquitin ligases to program degradation of cellular proteins with anti-viral activities (16–20, reviewed in Ref. 21). In particular, Vpx tightly associates with DDB1- and the CUL4-associated factor 1 (DCAF1) substrate receptor subunit of the CRL4 E3 ubiquitin ligase complex, and this interaction is linked to Vpx ability to relieve the inhibition of HIV/SIV infection of myeloid cells (11, 13, 22). Moreover, a recent proteomic study demonstrated that SAMHD1 associates with the DDB1-DCAF1 module of the CRL4 complex, but only in the presence of Vpx, suggesting that Vpx uses this E3 to program SAMHD1 degradation (8). Here we provide extensive genetic and biochemical evidence supporting this model. Specifically, we show that Vpx down-regulates human SAMHD1 levels via its C-terminal region and that this stretch of peptide is sufficient for Vpx-mediated loading of heterologous proteins onto CRL4DCAF1 and ubiquitination in vitro. Together, our studies validate a mechanistic model for the relief of SAMHD1-mediated restriction of primate lentivirus infection of myeloid cells by Vpx.
EXPERIMENTAL PROCEDURES
Mammalian Expression Plasmids
Human SAMHD1 deletion and point mutants and SIVmac 239 vpx point mutants were constructed using standard techniques and subcloned into pCG (23) and pCDNA3.1 (Invitrogen) plasmids encoding N-terminal myc-, HA-, FLAG-, or V5- epitope tags, and/or into MSCV(puro) retroviral vector. pCDNA3.1 plasmids expressing DCAF1 and Vpx were previously described (20).
Mammalian Cell Lines and Transient Transfection
Human embryonic kidney cells (HEK 293T) were cultured in DMEM medium supplemented with 10% fetal bovine serum, and antibiotics. HEK 293T cells were transfected with mixtures of pCDNA3.1 plasmids encoding expressing DCAF1, SAMHD1, and/or Vpx using LipofectAMINES 2000 (Invitrogen), or calcium phosphate co-precipitation method, and detergent extracts were prepared 24 to 48 h post-transfection, as described previously (20, 24). U937 cells were cultured in RPMI 1640 medium supplemented with 10% fetal bovine serum.
Immunoprecipitations and Western Blotting
Protein complexes were immunoprecipitated via FLAG or HA epitope tag as described previously (20, 24). Cell extracts and immunoprecipitates were separated by SDS-PAGE and transferred to PVDF membrane for immunoblotting. Proteins were detected with monoclonal antibodies specific for epitope tags: anti-HA (Covance), anti-Myc (Covance), anti-V5, and anti-FLAG M2 (Sigma), or reacting directly with human DDB1 (Invitrogen), DCAF1 (11), and Vpx (25). Immune complexes were revealed with HRP-conjugated antibodies specific for the Fc fragment of mouse or rabbit immunoglobulin G. (1:5,000; Jackson ImmunoResearch Laboratories) and enhanced chemiluminescence (Amersham Biosciences), or with fluorescent antibodies to mouse or rabbit immunoglobulin G (KPL) and Odyssey Infrared Imager (Licor).
Retroviral Vectors, SIV VLP, and Infections
VSV-G pseudotyped MSCV(puro) viral particles were produced from transiently transfected HEK 293T cells. 48 h after infection cells were selected with and then cultured in the continuous presence of puromycin (2 μg/ml). SIV VLP loaded with Vpx (VLP(Vpx)) were produced as described previously (8).
Escherichia coli Expression Plasmids
The cDNAs encoding residues 1–626, 115–585, and 115–626 of human SAMHD1 were cloned into the pET21 vector (EMD4Biosciences) with His6 tag at the C terminus. The thioredoxin (Trx) fusion constructs comprising SAMHD1 residues 575–600, 575–605, 575–610, 575–615, 575–620, 575–626, 585–626, 595–626, and internal deletion constructs (Δ596–600, Δ601–605, Δ606–610, Δ611–615) of residues 575–626 were cloned into the pET32 vector (EMD4Biosciences) with the His6 tag connecting Trx and SAMHD1 moieties. The SIVmac Vpx residues 1–112, 1–102, 1–89, 13–90, and 13–112 were cloned into the pET43 vector (EMD4Bioscienes) as NusA fusions.
Protein Expression and Purification
Proteins including His6-tagged SAMHD1, Trx fusion proteins comprising the C-terminal SAMHD1 peptides, and NusA-Vpx fusion proteins were expressed in E. coli Rosetta 2 (DE3) cultured in Luria-Bertani medium using 0.4 mm IPTG for induction at 18 °C for 16 h. Proteins were first purified using a 5-ml Ni-NTA column (GE Healthcare), then followed by gel-filtration column chromatography (Hi-Load Superdex200 16/60, GE Healthcare) equilibrated with a buffer containing 25 mm sodium phosphate, pH 7.5, 150 mm NaCl, 1 mm DTT, 10% glycerol, and 0.02% sodium azide. The proteins were further purified over a 5 ml Hi-Trap QP column (GE Healthcare) at pH 7.5 using a 0–1 m NaCl gradient. The C-terminal DCAF1 fragment (DCAF1c, residues 1045–1396) and DDB1-DCAF1c complex were expressed and purified from SF21 insect cells infected with recombinant baculoviruses as described previously (20).
In Vitro Ubiquitination Assays
Typically, E1 (UBA1, 0.2 μm), E2 (UbcH5b, 2.5 μm), and E3 complexes (mixtures of DDB1-DCAF1c-Vpx and CUL4A-RBX1 at 0.3 μm) were incubated with 0.6 μm of SAMHD1 and 2.5 μm of His6-FLAG-tagged ubiquitin in a buffer containing 10 mm Tris-HCl, pH 7.5, 150 mm NaCl, 5% glycerol, 20 units/ml pyrophosphatase, 2 mm DTT, and 5 mm ATP at 37 °C for indicated time. The extent of ubiquitination was assessed by immunoblotting with anti-FLAG antibody after separation of reaction mixture on 10–20% gradient SDS-PAGE, and transfer to PVDF membrane.
Analytical Size Exclusion Column Chromatography of Protein Complexes
Purified proteins or protein mixtures (100 μl) at concentration of 10 or 20 μm in 25 mm sodium phosphate buffer, pH 7.5 containing 150 mm NaCl, 5% glycerol and 0.02% sodium azide were injected into a 24-ml analytical superdex200 or superdex75 column and separated at a flow rate of 0.8 ml/min. Peak fractions were collected at 0.5-ml intervals and concentrated 10-fold with Amicon concentrators (Millipore). Proteins in each fraction were separated by 10–20% gradient SDS-PAGE, and stained with Coomassie Brilliant Blue.
RESULTS
The Divergent C Terminus of Human SAMHD1 Mediates Down-regulation by Vpx
The SAMHD1 molecule comprises a N-terminal sterile alpha motif (SAM) domain, followed by a HD/COG1078 phosphohydrolase domain and a C-terminal region (Fig. 1A). To determine which of these domains mediates Vpx-induced SAMHD1 degradation, we constructed variants lacking the N-terminal or C-terminal region and expressed them in human promonocytic U937 cells by transduction with MSCV(puro) retroviral vector. Two days after infection, positively transduced cells were selected with puromycin, and cultured for 2 days in the presence of phorbol 12-myristate 13-acetate (PMA) to initiate their withdrawal from the cell cycle and differentiation toward macrophages (26). Differentiated U937 cells were then infected with VSV-G pseudotyped SIV virus like particles loaded with SIVmac 239 Vpx (VLP(Vpx)) (8, 27) and SAMHD1 levels were assessed 2 days later by Western blotting.
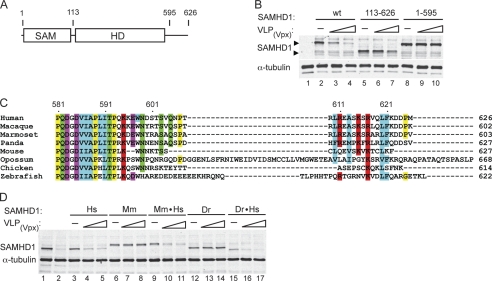
FIGURE 1.
C terminus of human SAMHD1 mediates Vpx-directed down-regulation of SAMHD1 levels. A, schematic representation of SAMHD1. The location of the SAM and HD/COG1078 phosphohydrolase domains in the 626 amino acid residues long human SAMHD1 protein is shown. B, U937 cells stably expressing wild type or truncated FLAG-tagged SAMHD1 proteins were either not infected (−), or infected with increasing amounts of VLP(Vpx). Detergent extracts prepared after 2 days from the infected cells were separated by SDS-PAGE and SAMHD1 levels were determined by immunoblotting with anti-FLAG antibody. C, alignment of the amino acid sequences of SAMHD1 proteins from different species was generated with ClustalW and Jalview (43). Amino acid residues are assigned Clustal color code. D, FLAG-tagged human (Hs), mouse (Mm), zebrafish (Dr) SAMHD1 proteins, or chimeric proteins comprising the C-terminal region (residue 585–626) of human SAMHD1 (Mm·Hs, Dr·Hs) were expressed stably in U937 cells, and then tested for Vpx sensitivity, as described for B, above.
As shown in Fig. 1B, infection with VLP(Vpx) reduced the levels of the full-length human SAMHD1 protein in a dose-dependent manner. Similarly, the N-terminally truncated SAMHD1 variant (residues 113–626, SAMHD1(113–626)) was also readily down-regulated following VLP-mediated delivery of Vpx, indicating that the SAM domain is not the target of Vpx (compare lanes 2–4 with lanes 5–7). In contrast, the levels of human SAMHD1 variant lacking the C-terminal 31 amino acid residues, SAMHD1(1–595) was not down-regulated by Vpx (compare lanes 2–4 with lanes 8–10). We therefore conclude that the C-terminal region is required for Vpx-induced human SAMHD1 degradation.
Alignment of amino acid sequences of SAMHD1 proteins from various vertebrate species revealed sequence divergence in the C-terminal regions (Fig. 1C, supplemental Fig. S1). This observation raised the possibility that the vpx function has evolved to selectively target primate SAMHD1 via its C terminus. To explore this possibility, we expressed in U937 cells mouse and zebrafish SAMHD1 proteins, whose C-terminal regions share only limited amino acid sequence identity with that of human SAMHD1 protein (Fig. 1C), and tested whether they are down-regulated by Vpx. As shown in Fig. 1D, the levels of both mouse and zebrafish SAMHD1 proteins were not affected following infection with VLP(Vpx), in contrast to human SAMHD1 (compare lanes 3–5 with lanes 6–8 and lanes 12–14), consistent with the notion that Vpx targets human SAMHD1 C-terminal region.
To further confirm the involvement of human SAMHD1 C terminus we constructed chimeric mouse and zebrafish SAMHD1 proteins, in which their C-terminal domains were exchanged for that of the human SAMHD1 (human SAMHD1 residues 585–626). As shown in Fig. 1D, the chimeric proteins were readily depleted in the presence of Vpx similar to what was seen with human SAMHD1 (compare lanes 3–5 with lanes 9–11 and lanes 15–17). Therefore, we conclude that the divergent C-terminal region of human SAMHD1 mediates SAMHD1 recognition by Vpx, and degradation, in a species-specific manner.
Vpx Recruits SAMHD1 to DCAF1-DDB1 Complex via the SAMHD1 C-terminal Region
Vpx was shown to mediate recruitment of human SAMHD1 to the CRL4DCAF1 E3 ubiquitin ligase complex (8). CRL4DCAF1 is a modular protein complex comprising a RING finger H2 protein (RBX1/ROC1), Cullin 4 (CUL4), DDB1, and DCAF1 subunits (28, 29). DCAF1 is a substrate receptor subunit that recruits proteins with defined structural elements (30–33). Some viral accessory proteins bind to substrate receptors and modify their structure for recruitment of anti-viral host cell factors for proteasomal degradation (21). Here we tested whether the C-terminal region of human SAMHD1 is required for Vpx-mediated SAMHD1 recruitment to DCAF1, or rather for another step in SAMHD1 degradation.
We began by asking whether Vpx can recruit mouse and zebrafish SAMHD1 proteins to DCAF1 and, if not, whether this can be achieved by replacing their C-terminal region with that of human SAMHD1. Human (Hs), mouse (Mm), and zebrafish (Dr) SAMHD1 proteins and their chimeras that were shown to be sensitive to Vpx-mediated down-regulation (see Fig. 1D) were transiently expressed in HEK 293T cells in the absence or presence of Vpx. SAMHD1 and associated proteins were then immunoprecipitated from detergent extracts, and immune complexes were analyzed for the presence of the DCAF1 and DDB1 subunits by SDS-PAGE and immunoblotting.
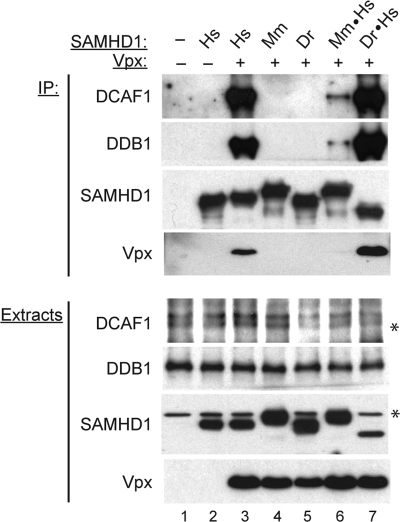
As shown in Fig. 2, human SAMHD1 co-precipitated DCAF1 and DDB1 efficiently, only in the presence of Vpx, as we reported previously (compare lane 2 with lane 3, (8)). In contrast, this was not observed for the mouse and zebrafish SAMHD1 proteins, even though they were expressed at levels similar to that seen for human SAMHD1 (compare lane 3 with lanes 4–5, in Fig. 2, panels A and B). Significantly, the C-terminal region of human SAMHD1 permitted the Vpx-directed assembly of CRL4DCAF1 protein complexes containing chimeric Mm·Hs, or Dr·Hs SAMHD1 proteins (compare lanes 4–5 with lanes 6–7). Interestingly, the steady state level of the Dr·Hs chimera complex that comprises the zebrafish SAMHD1 SAM and HD1 domains followed by the C-terminal region of human SAMHD1, was significantly higher than that of the Mm·Hs chimera complex, and similar to the level of the human SAMHD1 complex. This confirms that the human SAMHD1 C-terminal domain is specifically recognized by Vpx and mediates SAMHD1 recruitment to the CRL4DCAF1 E3 complex. The observation that both the Mm·Hs and Dr·Hs proteins were readily down-regulated by Vpx, despite the apparent differences in steady state levels of their complexes with DCAF1-DDB1, suggests that degradation of these chimeric proteins programmed by Vpx via CRL4DCAF1 may be influenced by other region(s) of the SAMHD1 polypeptide and/or other factors.
FIGURE 2.
Vpx recruits SAMHD1 to the DCAF1-DDB1 complex via the SAMHD1 C-terminal region. Human (Hs), mouse (Mm), and zebrafish (Dr) FLAG-tagged SAMHD1 proteins, or chimeric SAMHD1 proteins in which mouse (Mm·Hs) and zebrafish (Dr·Hs) C-terminal domains were exchanged for that of human SAMHD1 were transiently co-expressed with Vpx in HEK 293T cells. SAMHD1, and its associated proteins, were precipitated from detergent extracts via SAMHD1 FLAG-tag. Extracts and immune complexes were separated by SDS-PAGE and immunoblotted for DCAF1, DDB1, SAMHD1, and Vpx. (*) indicates endogenous proteins that cross-react with SAMHD1 and DCAF1 antibodies.
DCAF1-Vpx Complex Binds Directly to the C Terminus of SAMHD1
In general, recruitment of substrate proteins to E3 ubiquitin ligases is mediated by substrate receptor subunits, such as DCAF1 (34, 35). Vpx was thought to bind DCAF1 and thereby modify its surface to permit selective loading of target proteins (11, 13, 36). Hence, we reasoned that Vpx recruits SAMHD1, via the latter's C terminus to CRL4DCAF1 and nucleates a hetero-trimeric complex comprising Vpx, DCAF1, and human SAMHD1. We therefore set out to test this hypothesis in vitro using recombinant proteins.
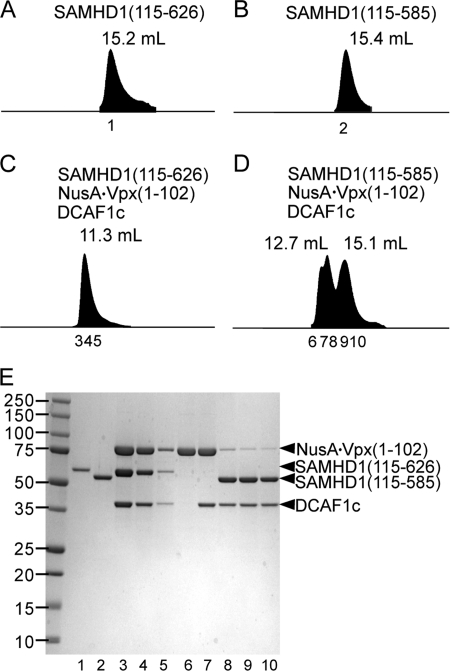
For these experiments, previously delineated interaction domains (data not shown) were employed, in particular SIVmac 239 Vpx (residues 1 to 102; Vpx(1–102)) as a NusA fusion to improve its solubility (NusA·Vpx(1–102)) and the C-terminal DCAF1 fragment (residue 1045–1396; DCAF1c) that contains both the DDB1 and Vpx interaction motifs. These proteins and human SAMHD1 variants with (SAMHD1(115–626)), or without (SAMHD1(115–585)) C-terminal residues 586–626 were used for complex assembly. DCAF1c-NusA·Vpx(1–102)-SAMHD1 complex formation was assessed by analytical gel filtration column chromatography followed by SDS-PAGE analyses of column fractions containing the eluted proteins. As shown in Fig. 3, panels C and E, NusA·Vpx(1–102), DCAF1c, and SAMHD1(115–626) co-eluted in fractions 3–5, with the peak at 11.3 ml elution volume, while SAMHD1(115–626) alone eluted at 15.2 ml (Fig. 3A). Significantly, the relative ratios of the three proteins appeared approximately constant across the elution volume (Fig. 3E, lanes 3–5), indicating that they formed a stoichiometric complex. In contrast, no evidence for formation of a stable ternary complex was observed with SAMHD1(115–585) that lacked the C-terminal residues (Fig. 3, D and E, lanes 6–10). This confirms that the C-terminal region of human SAMHD1 directly binds to the DCAF1-Vpx complex.
FIGURE 3.
The DCAF1-Vpx complex binds the C terminus of SAMHD1. A and B, SAM-domain deleted SAMHD1(115–626) (A) and SAM-domain and the C terminus-deleted SAMHD1(115–585) (B) were separated on an analytical size exclusion column at a flow rate of 0.8 ml/min. C and D, mixtures of NusA·Vpx(1–102), DCAF1c, and SAMHD1(115–626) (C) or SAMHD1(115–585) (D) were analyzed as described for panel A. Peak elution volumes are indicated above the peaks. Numbers and positions of peak fractions are indicated below each peak. E, proteins in peak fractions were concentrated 10-fold, resolved by SDS-PAGE, and stained with Coomassie Brilliant Blue. The SDS-PAGE gel lane numbers correspond to the peak fractions numbers indicated in panels A–D.
The Interaction with Vpx Reveals a Degradation Signal in the C Terminus of SAMHD1
The SAMHD1 C-terminal region contains conserved amino acid sequence stretches interspersed with more divergent ones (Fig. 1C). To address the molecular basis for the specific recruitment of human SAMHD1 to DCAF1 by Vpx, we determined which amino acid residues confer this effect. Initially, we performed a systematic alanine scan of the SAMHD1 C-terminal region distal to proline residue P593 and evaluated the mutant proteins, expressed in U937 cells, for Vpx-mediated down-regulation. As shown in Fig. 4A, several variants including SAMHD1(TR608AA), SAMHD1(LRE610AAA), SAMHD1(SKS614AAA), and SAMHD1(KDD622AAA) were partially refractory to down-regulation by Vpx. In particular, SAMHD1(L617A) and SAMHD1(LF620AA) variants were refractory to Vpx-dependent down-regulation, tentatively mapping the effect to a SAMHD1 region bordered by residues Leu-608 and Lys-625.
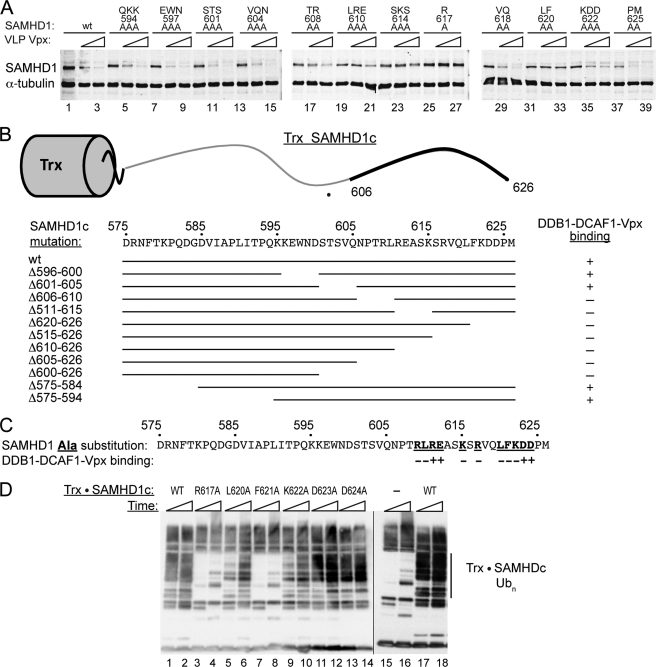
FIGURE 4.
Mutational analyses of the human SAMHD1 C terminus identify a Vpx-dependent degradation signal. A, U937 cells stably expressing wild type or mutant FLAG-SAMHD1 proteins with the alanine substitutions at the indicated positions in their C-terminal domains were either not infected (−), or infected (triangle) with increasing amounts of VLP(Vpx). SAMHD1 levels were determined 2 days later by immunoblotting with anti-FLAG antibody. B, preassembled DDB1-DCAF1c-Vpx(1–102) complex was incubated in vitro with recombinant fusion proteins comprising wild type or mutated SAMHD1 C-terminal peptide residues 575–626 fused to thioredoxin (Trx·SAMHD1c). The ability of mutant SAMHD1c peptides to bind DDB1-DCAF1-Vpx(1–102) was analyzed by analytical gel filtration chromatography as described in the legend to Fig. 3, and results are summarized with + (binding) or − (no binding). A schematic representation of the Trx·SAMHD1c fusion protein is shown at the top of the panel and the SAMHD1c region mediating binding to DDB1-DCAF1c-Vpx(1–102) complex is indicated by a bold line. Mutant SAMHD1c moieties are aligned with the wild type SAMHD1c amino acid sequence to visualize the location of the deleted regions. C, panel of Trx·SAMHD1c fusion proteins with single alanine substitution at the indicated positions within the SAMHD1c peptide (residues 575–626) was tested for binding to preassembled DDB1-DCAF1-Vpx(1–102), as described in the Fig. 3 legend, and results are summarized with + (binding) or − (no binding). The substituted residues are underlined and listed in bold. D, in vitro ubiquitination assays of wild type (lanes 1, 2, 17, 18) and single alanine-substituted Trx·SAMHD1c fusion proteins (lanes 3–14). Trx·SAMHD1c proteins were incubated with DDB1-DCAF1c-Vpx(1–102) in the presence of E1 (UBA1), E2 (UbcH5b), CUL4A-RBX1, and FLAG-tagged ubiquitin at 37 °C. A reaction assembled and incubated without Trx·SAMHD1c substrate (−) (lanes 15 and 16) provided a negative control to identify bands corresponding to ubiquitinated Trx·SAMHD1c, indicated on the right side of the panel (Trx·SAMHD1c Ubn). The ubiquitinated species seen in the negative control reaction probably reflect ubiquitination of the CRL4 subunits and other reaction components. The reactions were terminated after 10 min (odd numbered lanes) and 30 min (even numbered lanes) by the addition of Laemmli buffer and incubation at 95 °C for 5 min. The reaction mixtures were separated by 10–20% SDS-PAGE, transferred to PVDF, and ubiquitin was revealed with anti-FLAG antibody.
Next, in vitro binding assays were performed to corroborate our findings from the in vivo studies. We constructed and purified fusion proteins comprising a thioredoxin (Trx) moiety and SAMHD1 C-terminal peptides (residues D575-M626; SAMHD1c), containing N-terminal, internal and C-terminal deletions, and tested their ability to bind to recombinant DDB1-DCAF1c-Vpx(1–102) protein complex. The Trx·SAMHD1c fusions were incubated with pre-assembled DDB1-DCAF1c-Vpx(1–102) and complex formation was assessed by analytical gel filtration chromatography as described in Fig. 3. As summarized in Fig. 4B, no binding was detected for Trx·SAMHD1c proteins with deletions distal to SAMHD1 glutamine Gln-606. These results are consistent with the alanine-scanning data, presented in Fig. 4A.
For final identification of the pivotal SAMHD1 amino acids important for DDB1-DCAF1c-Vpx(1–102) binding, a panel of recombinant Trx·SAMHD1c fusions with single alanine substitutions was tested in binding assays. As illustrated in Fig. 4C, we found that SAMHD1 residues Arg-617, Leu-620, and Phe-621 have key role in binding to the DDB1-DCAF1c-Vpx(1–102) complex. These findings are in good agreement with the results of in vivo assays performed with full length human SAMHD1 protein, shown in Fig. 4A.
Finally, to evaluate whether the in vitro binding of SAMHD1 to the DDB1-DCAF1-Vpx(1–102) complex positions SAMHD1 correctly for ubiquitination by the catalytic core of the CRL4DCAF1 E3 complex in vitro ubiquitination experiments were performed. Wild type and selected single alanine substituted Trx·SAMHD1c fusion proteins were incubated with preassembled DDB1-DCAF1c-Vpx(1–102) complex in the presence of recombinant E1 (UBA1), E2 (UbcH5b), CUL4A-RBX1, and FLAG-tagged ubiquitin, in the ubiquitination buffer. The reaction mixtures were then separated by SDS-PAGE and FLAG·Ubiquitin revealed by immunoblotting for the FLAG epitope tag. As shown in Fig. 4D, the wild type SAMHD1 C-terminal region directed robust poly-ubiquitination of Trx·SAMHD1c fusion protein (compare WT, lanes 17 and 18 with (−), lanes 15 and 16). Significantly, Trx·SAMHD1c ubiquitination in vitro was disrupted by alanine substitutions for SAMHD1 Arg-617, Leu-620, and Phe-621. These data are in agreement with our observation that these mutations compromised Trx·SAMHD1c binding to the DDB1-DCAF1c-Vpx(1–102) complex in vitro (Fig. 4C) and SAMHD1 down-regulation by Vpx in vivo (Fig. 4A, lanes 25–27 and 31–33)). Alanine substitutions at other positions that disrupted binding to DDB1-DCAF1c-Vpx(1–102), such as Lys-622, also negatively affected Trx·SAMHD1c ubiquitination, albeit to a lesser extent (compare lane 1 and 9 in Fig. 4D). Thus, taken together, all three assays (in vivo depletion by VLP(Vpx), in vitro binding and ubiquitination assays) yielded consistent results. Importantly, the pertinent residues are not strictly conserved in mouse and zebrafish SAMHD1 proteins, explaining the selective targeting of the human SAMHD1 by Vpx (Fig. 1C).
The N Terminus of Vpx Mediates SAMHD1 Loading onto DCAF1 and Down-regulation
To determine which region of Vpx is critical for SAMHD1 loading onto DCAF1 we created N- and C-terminal deletions of Vpx. Previous reports showed that substitutions of amino acid residues in the central part of Vpx disrupted binding to DCAF1 (11, 13). In contrast, deletions and mutations in the N-terminal region of Vpx abolished the relief of restriction without interfering with DCAF1 binding (37), suggesting that the Vpx N terminus binds to and recruits the restriction factor via DCAF1 to CRL4. We therefore characterized the effects of N- and C-terminal Vpx deletions on its ability to recruit SAMHD1 to the DDB1-DCAF1 complex in vitro (see supplemental Fig. S2, panels A–G), and noted that N-terminal deletions disrupted SAMHD1 incorporation into the DDB1-DCAF1-Vpx complex. By contrast, C-terminal deletions distal to the cysteine Cys-89 had no effect on complex formation.
The role of the Vpx N terminus was then investigated in an in vitro ubiquitination assay (see supplemental Fig. S2, panel H). Pre-assembled complexes comprising DDB1-DCAF1c and Vpx(1–102), full-length Vpx(1–112), or N terminus-deleted Vpx(13–112) were incubated with SAMHD1 in the presence of E1, E2, CUL4A-RBX1, and N-terminally FLAG-tagged ubiquitin, to allow Vpx-dependent SAMHD1 ubiquitination. SDS-PAGE and immunoblot analyses of the reaction products revealed that an intact Vpx N terminus is required for efficient SAMHD1 ubiquitination, consistent with the inability of the Vpx(13–112) fragment to recruit SAMHD1 to DDB1-DCAF1 complex (supplemental Fig. S2, panel E and lanes 12–14 of panel G).
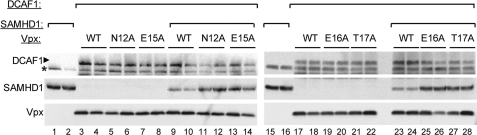
Next, we characterized the effect of amino acid substitutions in the Vpx N terminus that were previously shown to interfere with restriction relief of HIV-1 infection in dendritic cells (37). In vitro binding analyses combined with analytical gel filtration revealed that Asn-12, Glu-15, Glu-16, Thr-17 are essential for efficient recruitment of SAMHD1 to the DDB1-DCAF1-Vpx complex (data not shown), and substitutions at these positions disrupted Vpx ability to down-regulate SAMHD1 levels in HEK 293T cells, when tested either individually (Fig. 5), or in combination (supplemental Fig. S2I). These results unequivocally demonstrate that the N terminus of Vpx binds SAMHD1 and loads it onto CRL4DCAF1 E3 ubiquitin ligase complex, thus programming SAMHD1 for degradation.
FIGURE 5.
The N terminus of Vpx mediates DCAF1-dependent down-regulation of SAMHD1 levels. HEK 293T cells were transiently co-transfected with plasmids expressing DCAF1, wild type, or mutant Vpx (N12A, E15A, E16A, and T17A), and/or human SAMHD1, as indicated. Each transfection was performed in duplicate. Extracts prepared from the transfected cells were separated on 10–20% SDS-PAGE and Vpx, SAMHD1, and DCAF1 were revealed by immunoblotting. A nonspecific protein band (*) cross-reacting with the DCAF1 antibody (*) is shown as a loading control.
DISCUSSION
The SAMHD1 dNTPase protects a subset of hematopoietic cell lineages from infection by primate lentiviruses. Efficient transduction of these cells is mediated by Vpx, which directs SAMHD1 for proteasome-dependent degradation. Previous findings, in particular the presence of SAMHD1, DCAF1, and DDB1 in Vpx immunoprecipitates (8–10), implicated CRL4DCAF1 as the cellular E3 ubiquitin ligase enzyme usurped by Vpx to direct SAMHD1 degradation. We now provide extensive biochemical evidence in support of this model.
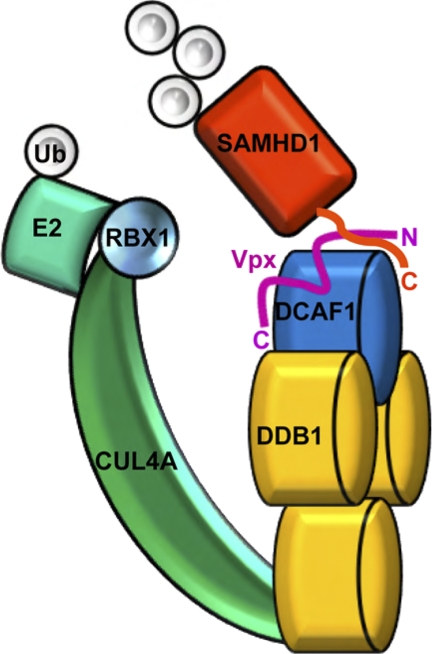
Our findings suggest a molecular mechanism for Vpx-mediated recruitment of SAMHD1 to CRL4DCAF1. Vpx readily assembles a complex with DCAF1, which binds SAMHD1 (Fig. 3). Vpx binding with DCAF1 involves a central region in the Vpx molecule, whereas the N-terminal region of Vpx is critical for association with SAMHD1, without influencing Vpx binding to DCAF1. Indeed, in complex with DDB1-DCAF1, the N-terminal region of Vpx interacts directly with the C terminus of SAMHD1 (Fig. 6). DCAF1 also participates in the binding to SAMHD1 directly, or indirectly, since a stable Vpx-SAMHD1 binary complex is not observed in the absence of DCAF1 (data not shown). Therefore, by interacting with DCAF1, the structure of Vpx is modulated and a unique interaction surface for SAMHD1 is created that allows for recruitment of SAMHD1 to CRL4 for ubiquitination. Future structural studies will provide more detailed atomic understanding of the DCAF1-Vpx-SAMHD1 interaction.
FIGURE 6.
Model for SAMHD1 recruitment to CRL4DCAF1-Vpx. The N terminus of Vpx interacts with the C-terminal region of SAMHD1 and recruits SAMHD1 to CRL4DCAF1-Vpx E3 ubiquitin ligase for ubiquitination. The relative positioning of the individual subunits in the CRL4DCAF-Vpx complex is based on the known architecture of CRL4 E3 ubiquitin ligases (44).
Recruitment of SAMHD1 involves a short sequence stretch located at the C terminus of the human SAMHD1 molecule (Fig. 4). This sequence is sufficient when added to heterologous proteins, such as thioredoxin, to mediate Vpx-dependent recruitment to the CRL4 E3 for ubiquitination. Therefore, this C-terminal peptide can be considered as a Vpx-dependent degron (38, 39), an autonomous sequence element that controls protein levels via interacting with the N terminus of Vpx (N12-T17, see Fig. 5). It should be pointed out that this amino acid sequence is distinct from that in the Vpr accessory protein of HIV/SIV, and the degron-binding interface of Vpr is probably located in its C-terminal region (36, 40). Thus, Vpr and Vpx have evolved unique degron-binding interfaces to recruit different substrates to CRL4DCAF1 and future work is expected to shed more light on their precise molecular interfaces for substrate binding.
The species-specific targeting of human SAMHD1 by HIV/SIV Vpx can be explained by the sequence variability of the C-terminal regions of SAMHD1 proteins from different species (Fig. 1C). It is tempting to speculate that SAMHD1 sequence divergence reflects evolution under positive selection pressure exerted by interactions with invading pathogens, while maintaining its normal cellular function. This possibility is consistent with the findings from a recent study of Vpx interactions with primate SAMHD1 proteins (41, 42). The precise function of the SAMHD1 C-terminal domain is not yet known, but it does not appear to be required for SAMHD1 dNTPase activity in vitro (data not shown). Interestingly, in the recent crystal structure of SAMHD1, the C terminus was found to be disordered (14), suggesting that it only assumes a stably folded conformation in the presence of a yet to be identified, possible regulatory ligand.
Significantly, genetic evidence from previous studies linked Vpx ability to counteract SAMHD1 restriction to the interaction with CRL4DCAF1 in a range of myeloid cell types, including monocyte-derived macrophages, dendritic cells, as well as established myeloid cell lines (8, 9, 22, data not shown) and both Vpx mutations that abolish DCAF1 binding and recruitment of SAMHD1 to DCAF1 severely diminish Vpx ability to rescue HIV-1 transduction of dendritic cells from the interferon-indued antiviral state (22, 37). Therefore, the main mechanism employed by Vpx to counteract restriction of primate lentivirus infection imposed by SAMHD1 appears to be associated with reprogramming of CRL4DCAF1 for inducing SAMHD1 degradation.
Supplementary Material
Acknowledgments
We thank Teresa Brosenitsch and Linda Van Aelst for critical reading of the manuscript and editorial help.
This work was supported, in whole or in part, by National Institutes of Health Grants P50GM082251 (to A. M. G.), R01 AI077459 (to J. S.), and Case CFAR funds (to J. S.).

This article contains supplemental Figs. S1 and S2.
- SAM
- sterile alpha motif
- SAMHD1
- sterile alpha motif domain- and HD domain-containing protein 1
- DDB1
- damage-specific DNA-binding protein 1
- DCAF1
- DDB1- CUL4-associated factor 1
- Ub
- ubiquitin
- E1
- ubiquitin-activating enzyme
- E2
- ubiquitin-conjugating enzyme
- E3
- ubiquitin ligase
- CRL
- cullin-RING ubiquitin ligase
- AGS
- Aicardi-Goutieres Syndrome
- HIV
- human immunodeficiency virus
- SIV
- simian immunodeficiency virus
- VSV-G
- vesicular stomatitis virus glycoprotein
- CTD
- C-terminal domain
- VLP
- virus-like particles
- PMA
- phorbol 12-myristate 13-acetate
- Trx
- thioredoxin
- HRP
- horseradish peroxidase
- IgG
- immunoglobulin G.
REFERENCES
- 1. Steinman R. M., Hemmi H. (2006) Dendritic cells: translating innate to adaptive immunity. Curr. Top Microbiol. Immunol. 311, 17–58 [DOI] [PubMed] [Google Scholar]
- 2. Takeuchi O., Akira S. (2009) Innate immunity to virus infection. Immunol. Rev. 227, 75–86 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Goujon C., Rivière L., Jarrosson-Wuilleme L., Bernaud J., Rigal D., Darlix J. L., Cimarelli A. (2007) SIVSM/HIV-2 Vpx proteins promote retroviral escape from a proteasome-dependent restriction pathway present in human dendritic cells. Retrovirology 4, 2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Kaushik R., Zhu X., Stranska R., Wu Y., Stevenson M. (2009) A cellular restriction dictates the permissivity of nondividing monocytes/macrophages to lentivirus and gammaretrovirus infection. Cell Host Microbe 6, 68–80 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Yu X. F., Yu Q. C., Essex M., Lee T. H. (1991) The vpx gene of simian immunodeficiency virus facilitates efficient viral replication in fresh lymphocytes and macrophage. J. Virol. 65, 5088–5091 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Gibbs J. S., Regier D. A., Desrosiers R. C. (1994) Construction and in vitro properties of SIVmac mutants with deletions in “nonessential” genes. AIDS Res. Hum. Retroviruses 10, 607–616 [DOI] [PubMed] [Google Scholar]
- 7. Fletcher T. M., 3rd, Brichacek B., Sharova N., Newman M. A., Stivahtis G., Sharp P. M., Emerman M., Hahn B. H., Stevenson M. (1996) Nuclear import and cell cycle arrest functions of the HIV-1 Vpr protein are encoded by two separate genes in HIV-2/SIV(SM). EMBO J. 15, 6155–6165 [PMC free article] [PubMed] [Google Scholar]
- 8. Hrecka K., Hao C., Gierszewska M., Swanson S. K., Kesik-Brodacka M., Srivastava S., Florens L., Washburn M. P., Skowronski J. (2011) Vpx relieves inhibition of HIV-1 infection of macrophages mediated by the SAMHD1 protein. Nature 474, 658–661 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Laguette N., Sobhian B., Casartelli N., Ringeard M., Chable-Bessia C., Ségéral E., Yatim A., Emiliani S., Schwartz O., Benkirane M. (2011) SAMHD1 is the dendritic- and myeloid cell-specific HIV-1 restriction factor counteracted by Vpx. Nature 474, 654–657 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Berger A., Sommer A. F., Zwarg J., Hamdorf M., Welzel K., Esly N., Panitz S., Reuter A., Ramos I., Jatiani A., Mulder L. C., Fernandez-Sesma A., Rutsch F., Simon V., König R., Flory E. (2011) SAMHD1-deficient CD14+ cells from individuals with Aicardi-Goutières syndrome are highly susceptible to HIV-1 infection. PLoS Pathog. 7, e1002425. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Srivastava S., Swanson S. K., Manel N., Florens L., Washburn M. P., Skowronski J. (2008) Lentiviral Vpx accessory factor targets VprBP/DCAF1 substrate adaptor for cullin 4 E3 ubiquitin ligase to enable macrophage infection. PLoS Pathog. 4, e1000059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Fujita M., Otsuka M., Miyoshi M., Khamsri B., Nomaguchi M., Adachi A. (2008) Vpx is critical for reverse transcription of the human immunodeficiency virus type 2 genome in macrophages. J. Virol. 82, 7752–7756 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Bergamaschi A., Ayinde D., David A., Le Rouzic E., Morel M., Collin G., Descamps D., Damond F., Brun-Vezinet F., Nisole S., Margottin-Goguet F., Pancino G., Transy C. (2009) The human immunodeficiency virus type 2 Vpx protein usurps the CUL4A-DDB1 DCAF1 ubiquitin ligase to overcome a postentry block in macrophage infection. J. Virol. 83, 4854–4860 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Goldstone D. C., Ennis-Adeniran V., Hedden J. J., Groom H. C., Rice G. I., Christodoulou E., Walker P. A., Kelly G., Haire L. F., Yap M. W., de Carvalho L. P., Stoye J. P., Crow Y. J., Taylor I. A., Webb M. (2011) HIV-1 restriction factor SAMHD1 is a deoxynucleoside triphosphate triphosphohydrolase. Nature 480, 379–382 [DOI] [PubMed] [Google Scholar]
- 15. Powell R. D., Holland P. J., Hollis T., Perrino F. W. (2011) Aicardi-Goutieres syndrome gene and HIV-1 restriction factor SAMHD1 is a dGTP-regulated deoxynucleotide triphosphohydrolase. J. Biol. Chem. 286, 43596–43600 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Margottin F., Bour S. P., Durand H., Selig L., Benichou S., Richard V., Thomas D., Strebel K., Benarous R. (1998) A novel human WD protein, h-beta TrCp, that interacts with HIV-1 Vpu connects CD4 to the ER degradation pathway through an F-box motif. Mol. Cell 1, 565–574 [DOI] [PubMed] [Google Scholar]
- 17. Yu X., Yu Y., Liu B., Luo K., Kong W., Mao P., Yu X. F. (2003) Induction of APOBEC3G ubiquitination and degradation by an HIV-1 Vif-Cul5-SCF complex. Science 302, 1056–1060 [DOI] [PubMed] [Google Scholar]
- 18. Douglas J. L., Viswanathan K., McCarroll M. N., Gustin J. K., Früh K., Moses A. V. (2009) Vpu directs the degradation of the human immunodeficiency virus restriction factor BST-2/Tetherin via a {β}TrCP-dependent mechanism. J. Virol. 83, 7931–7947 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Mangeat B., Gers-Huber G., Lehmann M., Zufferey M., Luban J., Piguet V. (2009) HIV-1 Vpu neutralizes the antiviral factor Tetherin/BST-2 by binding it and directing its beta-TrCP2-dependent degradation. Plos Pathog 5, e1000574. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Ahn J., Vu T., Novince Z., Guerrero-Santoro J., Rapic-Otrin V., Gronenborn A. M. (2010) HIV-1 Vpr loads uracil DNA glycosylase-2 onto DCAF1, a substrate recognition subunit of a cullin 4A-ring E3 ubiquitin ligase for proteasome-dependent degradation. J. Biol. Chem. 285, 37333–37341 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Barry M., Früh K. (2006) Viral modulators of cullin RING ubiquitin ligases: culling the host defense. Sci. STKE 335, pe21. [DOI] [PubMed] [Google Scholar]
- 22. Pertel T., Reinhard C., Luban J. (2011) Vpx rescues HIV-1 transduction of dendritic cells from the antiviral state established by type 1 interferon. Retrovirology 8, 49. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Tanaka M., Herr W. (1990) Differential transcriptional activation by Oct-1 and Oct-2: interdependent activation domains induce Oct-2 phosphorylation. Cell 60, 375–386 [DOI] [PubMed] [Google Scholar]
- 24. Janardhan A., Swigut T., Hill B., Myers M. P., Skowronski J. (2004) HIV-1 Nef binds the DOCK2-ELMO1 complex to activate rac and inhibit lymphocyte chemotaxis. PLoS Biol. 2, E6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Hrecka K., Gierszewska M., Srivastava S., Kozaczkiewicz L., Swanson S. K., Florens L., Washburn M. P., Skowronski J. (2007) Lentiviral Vpr usurps Cul4-DDB1[VprBP] E3 ubiquitin ligase to modulate cell cycle. Proc. Natl. Acad. Sci. U.S.A. 104, 11778–11783 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Harris P., Ralph P. (1985) Human leukemic models of myelomonocytic development: a review of the HL-60 and U937 cell lines. J. Leukoc Biol. 37, 407–422 [DOI] [PubMed] [Google Scholar]
- 27. Goujon C., Jarrosson-Wuillème L., Bernaud J., Rigal D., Darlix J. L., Cimarelli A. (2006) With a little help from a friend: increasing HIV transduction of monocyte-derived dendritic cells with virion-like particles of SIV(MAC). Gene Ther. 13, 991–994 [DOI] [PubMed] [Google Scholar]
- 28. Higa L. A., Zhang H. (2007) Stealing the spotlight: CUL4-DDB1 ubiquitin ligase docks WD40-repeat proteins to destroy. Cell Div. 2, 5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Lee J., Zhou P. (2007) DCAFs, the missing link of the CUL4-DDB1 ubiquitin ligase. Mol. Cell 26, 775–780 [DOI] [PubMed] [Google Scholar]
- 30. Angers S., Li T., Yi X., MacCoss M. J., Moon R. T., Zheng N. (2006) Molecular architecture and assembly of the DDB1-CUL4A ubiquitin ligase machinery. Nature 443, 590–593 [DOI] [PubMed] [Google Scholar]
- 31. He Y. J., McCall C. M., Hu J., Zeng Y., Xiong Y. (2006) DDB1 functions as a linker to recruit receptor WD40 proteins to CUL4-ROC1 ubiquitin ligases. Genes Dev. 20, 2949–2954 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Higa L. A., Wu M., Ye T., Kobayashi R., Sun H., Zhang H. (2006) CUL4-DDB1 ubiquitin ligase interacts with multiple WD40-repeat proteins and regulates histone methylation. Nat. Cell Biol. 8, 1277–1283 [DOI] [PubMed] [Google Scholar]
- 33. Jin J., Arias E. E., Chen J., Harper J. W., Walter J. C. (2006) A family of diverse Cul4-Ddb1-interacting proteins includes Cdt2, which is required for S phase destruction of the replication factor Cdt1. Mol. Cell 23, 709–721 [DOI] [PubMed] [Google Scholar]
- 34. Petroski M. D., Deshaies R. J. (2005) Function and regulation of cullin-RING ubiquitin ligases. Nat. Rev. Mol. Cell Biol. 6, 9–20 [DOI] [PubMed] [Google Scholar]
- 35. Zimmerman E. S., Schulman B. A., Zheng N. (2010) Structural assembly of cullin-RING ubiquitin ligase complexes. Curr. Opin. Struct. Biol. 20, 714–721 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Le Rouzic E., Belaïdouni N., Estrabaud E., Morel M., Rain J. C., Transy C., Margottin-Goguet F. (2007) HIV1 Vpr arrests the cell cycle by recruiting DCAF1/VprBP, a receptor of the Cul4-DDB1 ubiquitin ligase. Cell Cycle 6, 182–188 [DOI] [PubMed] [Google Scholar]
- 37. Gramberg T., Sunseri N., Landau N. R. (2010) Evidence for an activation domain at the amino terminus of simian immunodeficiency virus Vpx. J. Virol. 84, 1387–1396 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Varshavsky A. (1991) Naming a targeting signal. Cell 64, 13–15 [DOI] [PubMed] [Google Scholar]
- 39. Ravid T., Hochstrasser M. (2008) Diversity of degradation signals in the ubiquitin-proteasome system. Nat. Rev. Mol. Cell Biol. 9, 679–690 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. DeHart J. L., Zimmerman E. S., Ardon O., Monteiro-Filho C. M., Argañaraz E. R., Planelles V. (2007) HIV-1 Vpr activates the G2 checkpoint through manipulation of the ubiquitin proteasome system. Virol. J. 4, 57. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Laguette N., Rahm N., Sobhian B., Chable-Bessia C., Münch J., Snoeck J., Sauter D., Switzer W. M., Heneine W., Kirchhoff F., Delsuc F., Telenti A., Benkirane M. (2012) Evolutionary and functional analyses of the interaction between the myeloid restriction factor SAMHD1 and the lentiviral Vpx protein. Cell Host Microbe 11, 205–217 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Lim E. S., Fregoso O. I., McCoy C. O., Matsen F. A., Malik H. S., Emerman M. (2012) The ability of primate lentiviruses to degrade the monocyte restriction factor SAMHD1 preceded the birth of the viral accessory protein Vpx. Cell Host Microbe 11, 194–204 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Waterhouse A. M., Procter J. B., Martin D. M., Clamp M., Barton G. J. (2009) Jalview Version 2-a multiple sequence alignment editor and analysis workbench. Bioinformatics 25, 1189–1191 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Fischer E. S., Scrima A., Böhm K., Matsumoto S., Lingaraju G. M., Faty M., Yasuda T., Cavadini S., Wakasugi M., Hanaoka F., Iwai S., Gut H., Sugasawa K., Thomä N. H. (2011) The molecular basis of CRL4DDB2/CSA ubiquitin ligase architecture, targeting, and activation. Cell 147, 1024–1039 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.