Abstract
The basic unit of skeletal muscle in all metazoans is the multinucleate myofiber, within which individual nuclei are regularly positioned1. The molecular machinery responsible for myonuclear positioning is not known. Improperly positioned nuclei are a hallmark of numerous muscles diseases2, including centronuclear myopathies3, but it is unclear whether correct nuclear positioning is necessary for muscle function. Here we identify the microtubule-associated protein Ensconsin(Ens)/MAP7 and Kinesin Heavy Chain (Khc)/Kif5b as essential, evolutionary conserved regulators of myonuclear positioning in Drosophila and cultured mammalian myotubes. We find that these proteins physically interact and that expression of the Kif5b motor domain fused to the MAP7 microtubule binding domain rescues nuclear positioning defects in MAP7 depleted cells. This suggests that MAP7 links Kif5b to the microtubule cytoskeleton to promote nuclear positioning. Finally we demonstrate that myonuclear positioning is physiologically important. Drosophila ens mutant larvae display decreased locomotion and incorrect myonuclear positioning, and these phenotypes are rescued by muscle specific expression of Ens. We conclude that improper nuclear positioning contributes to muscle dysfunction in a cell autonomous fashion.
Keywords: Muscle, nuclear positioning, MAP, Kinesin, Drosophila, C2C12, primary myotubes, nuclear movement
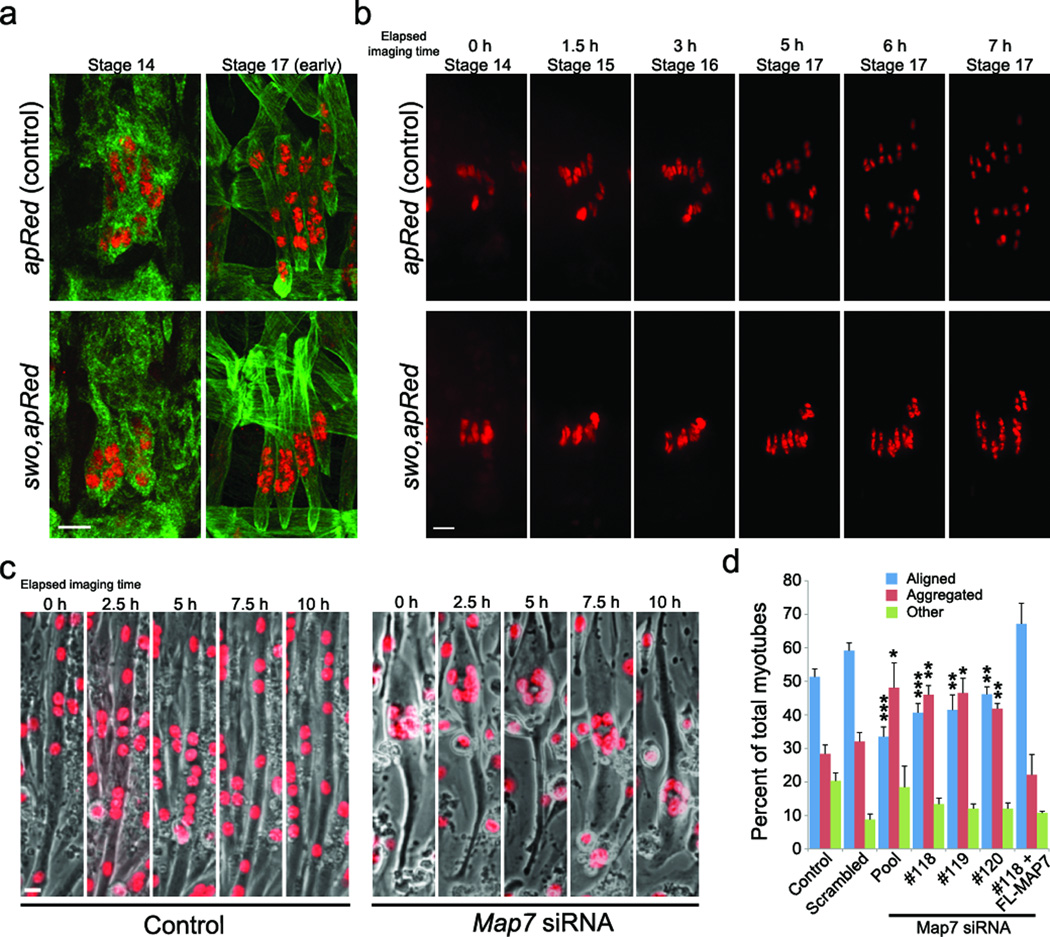
To identify mechanisms governing nuclear positioning in muscle, we performed an F3 recessive EMS mutagenesis screen using the apterousME-NLS::dsRed transgenic line (apRed)4, which expresses dsRed in the nuclei of the 4 lateral transverse (LT) muscles of the Drosophila embryo. At the end of embryogenesis, the myonuclei are distributed throughout the muscles (Fig. 1a, b). One mutation resulted in clustered nuclei near the ventral end of each LT muscle. Within each hemi-segment these clusters resembled the Nike trademark so we named the mutant swoosh (swo) (Fig. 1a).
Figure 1. Myonuclear positioning requires Ensconsin/MAP7.
a. Hemi-segments from control and ensswo embryos at beginning (stage 14) and end (stage 17) of nuclear migration. Muscles, tropomyosin (green), Nuclei, dsRed (red). Bar, 10 µm.
b. Timelapse of nuclear migration in control and ensswo hemisegment. Bar, 10 µm.
c. Timelapse of nuclear migration in control and Map7 depleted C2C12-H1B-GFP myotubes. Nuclei, red; Bar, 15 um.
d. Nuclear distribution in C2C12 myotubes that were untreated (control) or treated with indicated siRNA and Map7 depleted cells expressing full length MAP7 (#118 + FL-MAP7). Error bars, s.e.m. ***p < 0.001, **p < 0.01, *p < 0.05 (scrambled vs. experimental condition).
Further analysis of swo mutant embryos indicated that myoblast specification5, fusion5, muscle elongation6,7, and attachment7 occur normally. The only notable difference in swo mutant muscles is a ventral bulge in the cell body that correlates with the cluster of nuclei (Supplementary Figs. 1,2). These data suggest that the clustering of nuclei in swo mutants is a bona fide nuclear positioning defect.
To examine nuclear movement, we performed in vivo time-lapse imaging on apRed control and swo,apRed mutant embryos (Fig. 1b). In stage 14 apRed control embryos (10.5 h after egg laying [AEL]), nuclei in the 4 LT muscles group near the ventral end of each growing myotube in the hemi-segment. During stage 15 (11.5 h AEL) after the completion of myoblast fusion and muscle attachment, the nuclei within each myotube begin to separate into distinct dorsal and ventral clusters. These clusters finish their migration during stage 16 (15 h AEL) and each cluster resides at an opposite end of the myotube. Finally, during stage 17 (18 h AEL), the 6–8 nuclei in each LT muscle move from the clusters and evenly distribute throughout each LT muscle. The entire process of nuclear movement spans 7 hours (Supplemental Movie 1). Time lapse imaging of swo,apRed mutant embryos demonstrated that the nuclei fail to undergo the initial separation into dorsal and ventral clusters during stage 15 and remain clustered at stage 17 indicating that swo does not simply cause a delay in myonuclear positioning (Supplementary Movie 2). Additionally, the nuclei in every other somatic muscle examined in swo mutant embryos also failed to separate (Supplementary Fig. 1e).
We determined that the swo flies carry a nonsense mutation in ensconsin (ens), a gene encoding a microtubule associated protein (MAP). Expression of HA-tagged Ens specifically in the developing mesoderm and muscle rescued the nuclear positioning phenotype in swo embryos, confirming that the mutation in ens is responsible for the nuclear positioning defect. RNAi mediated depletion of ens solely in the developing mesoderm and muscle recapitulated the phenotype we observed in the swo mutant (renamed ensswo) and known ens mutant alleles failed to complement ensswo (Supplementary Fig. 1c, 2a, data not shown). These data indicate that ens is required autonomously within muscle for proper myonuclear positioning. We also found that nuclear position was disrupted in ensswo oocytes (Supplementary Fig. 3a,b and 8), but not in ensswo photoreceptor cells (Supplementary Fig. 3c, data not shown)9, indicating that Ens is necessary for some, but not all, nuclear positioning processes in Drosophila.
Four different genes in the mouse genome encode Ens orthologs: Map7 (E-MAP-115, Ensconsin), Map7D1, Map7D2 and Map7D3 (Supplementary Fig. 2c). Using cultures of C2C12 myoblasts and primary mouse myoblasts, we examined the effect on nuclear positioning in myotubes depleted of each Ens ortholog by siRNA (Fig. 1, Supplementary Fig. 4). MAP7 depletion caused a significant increase in the aggregation of nuclei within a myotube but did not affect myoblast fusion or myotube differentiation (Fig. 1c,d, Supplementary Fig. 4d,e, Supplementary Movies 3–6). Depletion of MAP7D1, MAP7D2 or MAP7D3 did not affect nuclear positioning (Supplementary Fig. 4b). Expression of full length MAP7 in MAP7 depleted myotubes restored nuclear alignment (Fig. 1d, Supplementary Fig 6h). Thus, MAP7 is required for nuclear positioning in both Drosophila muscles and cultured mammalian myotubes.
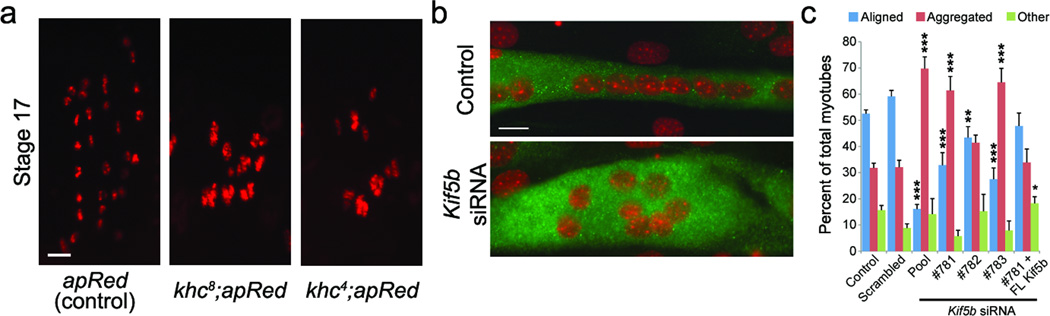
No major defects on the microtubule network were observed in ensswo mutant embryos or MAP7 depleted myotubes (Supplementary Fig. 2g-h, 6g). Therefore, to gain mechanistic insight, we conducted a yeast-2-hybrid screen to find Ens binding proteins and identified Kinesin heavy chain (Khc) (Supplementary Fig. 5). Both a kinesin null mutation (khc8) and a motor dead mutation (khc4)10 disrupted myonuclear positioning without affecting muscle elongation and attachment. (Fig. 2a, Supplementary Fig. 5a). Likewise, siRNA mediated depletion of the mammalian Khc ortholog Kif5b11 in primary and C2C12 myotubes disrupted nuclear alignment similarly to MAP7 depletion without affecting myotube formation or differentiation (Fig. 2b, c, Supplementary Fig. 4d-e, 6, Supplementary Movies 7,8). Expression of full length Kif5b in Kif5b depleted myotubes restored nuclear alignment (Fig. 2c, Supplementary Fig.6h). These data indicate that Khc, with a functional motor domain, is required for proper nuclear positioning in both Drosophila muscles and cultured mammalian myotubes.
Figure 2. Kinesin is required for myonuclear positioning.
a. Single hemi-segments from stage 17 (22h AEL) embryos of indicated genotypes; Bar, 10 µm.
b. Representative immunofluorescence images of control and Kif5b depleted C2C12 myotubes differentiated for 4 days and immunostained for MHC (green) and DAPI (red). Bars, 15 µm.
c. Histogram of nuclear distribution in C2C12 myotubes that were untreated (control) or treated with the indicated siRNA or Kif5b depleted cells expressing full length Kif5b (#781 + FL-Kif5b). Error bars, s.e.m. ***p < 0.001, **p < 0.01, *p < 0.05 (scrambled vs. experimental condition).
We next examined whether Ens and Khc functionally interact to position myonuclei. Embryos heterozygous for ens or khc position their myonuclei normally. However, embryos that were doubly heterozygous for ensswo and khc displayed defective nuclear positioning similar to the homozygous ens embryos (Supplementary Fig. 5). This demonstrates that ens and khc functionally interact and that the dosage of these genes is important for proper nuclear positioning.
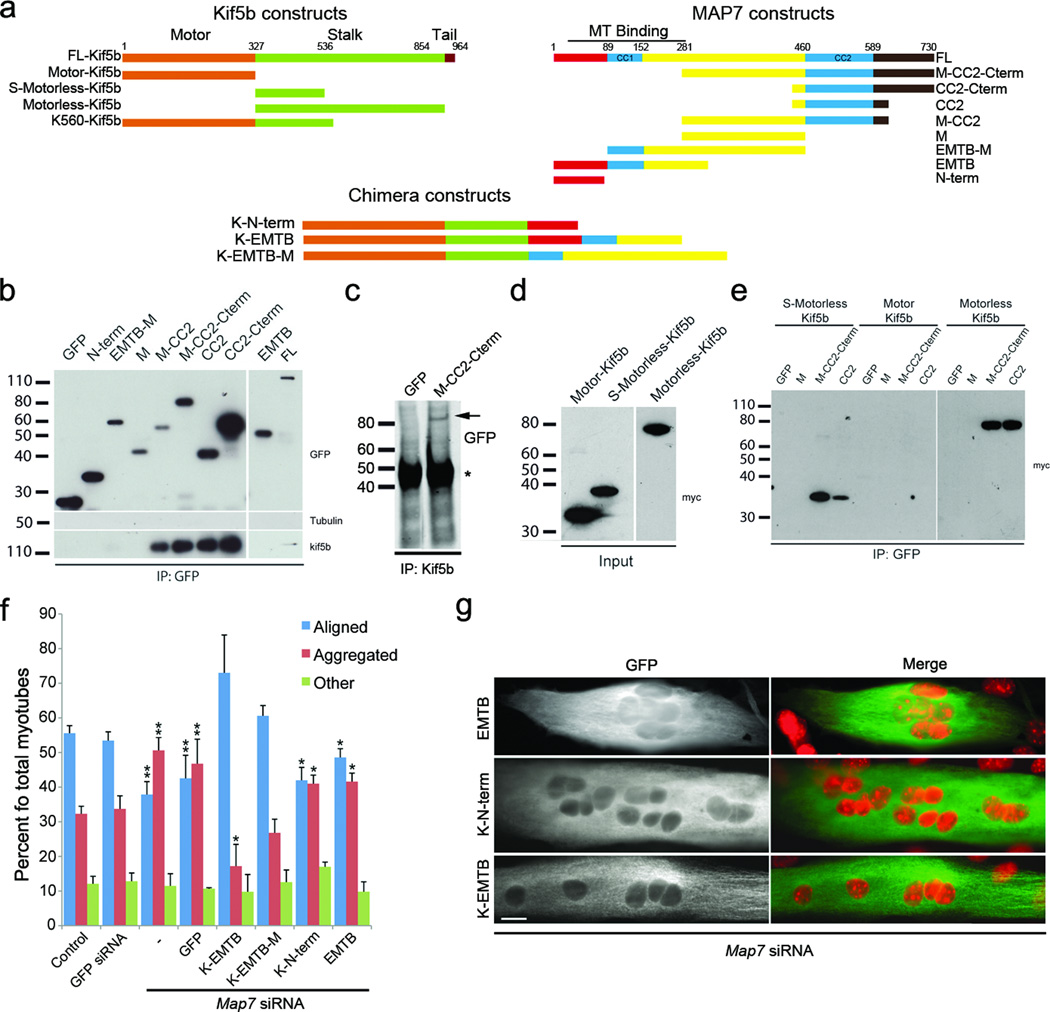
We next tested whether Ens/MAP7 and Khc/Kif5b physically interact. Endogenous Kif5b co-immunoprecipitated with expressed full length GFP-tagged MAP7, independent of microtubules (Fig. 3a,b). Using fragments of MAP7, we mapped the Kif5b interaction to the C-terminal coiled-coil domain of MAP7 (CC2), which was confirmed with the reciprocal co-immunoprecipitation (Fig. 3 a-c). Similarly, fragments of Kif5b were used to identify the C-terminal region (Kif5b-motorless) as the MAP7 interacting domain (Fig. 3a,d,e). Therefore, C-terminal coiled-coil domain of MAP7 interacts with the C-terminal region of Kif5b (Fig. 3).
Figure 3. Kinesin and Ensconsin/MAP7 interact to regulate nuclear position.
a. Kif5b, MAP7 and chimera constructs N-terminally tagged with GFP (MAP7/chimeras) or c-Myc (Kif5b). Numbers indicate amino acids.
b. Western blot with indicated antibodies (right) of GFP-MAP7 (top) immunoprecipitations.
c. Anti-GFP Western blot of Kif5b immunoprecipitations from C2C12s expressing indicated constructs (top). (*) anti-Kif5b IgG.
d. Anti-c-Myc Western blot from C2C12s expressing Kif5b constructs.
e. Anti-c-Myc Western blot from GFP immunoprecipitations using C2C12s expressing indicated constructs.
f. Nuclear distribution in Map7 depleted C2C12s expressing indicated chimeras. Error bars s.e.m. **p < 0.01, *p < 0.05
g. Map7 depleted C2C12s expressing indicated chimeras. Anti-GFP (green), Nuclei, DAPI (red). Bar, 15 µm.
To test whether the specific interaction identified above between Ens/MAP7 and Khc/Kif5b is required for nuclear positioning, we expressed chimeras consisting of the Kif5b motor domain12 fused to fragments of MAP7 that contained (K-EMTB and K-EMTB-M) or lacked (K-N-term) the microtubule binding domain (EMTB)13 in Map7 siRNA depleted myotubes (Fig. 3 a, f, g). Expression of chimeras that contained the MAP7 EMTB rescued the alignment of nuclei in MAP7 depleted myotubes, whereas expression of the EMTB domain alone, or a chimera that lacked the MAP7 EMTB (K-N-term) did not (Fig. 3f,g). These results indicate that the function of MAP7 in nuclear positioning is to link Kif5b to microtubules.
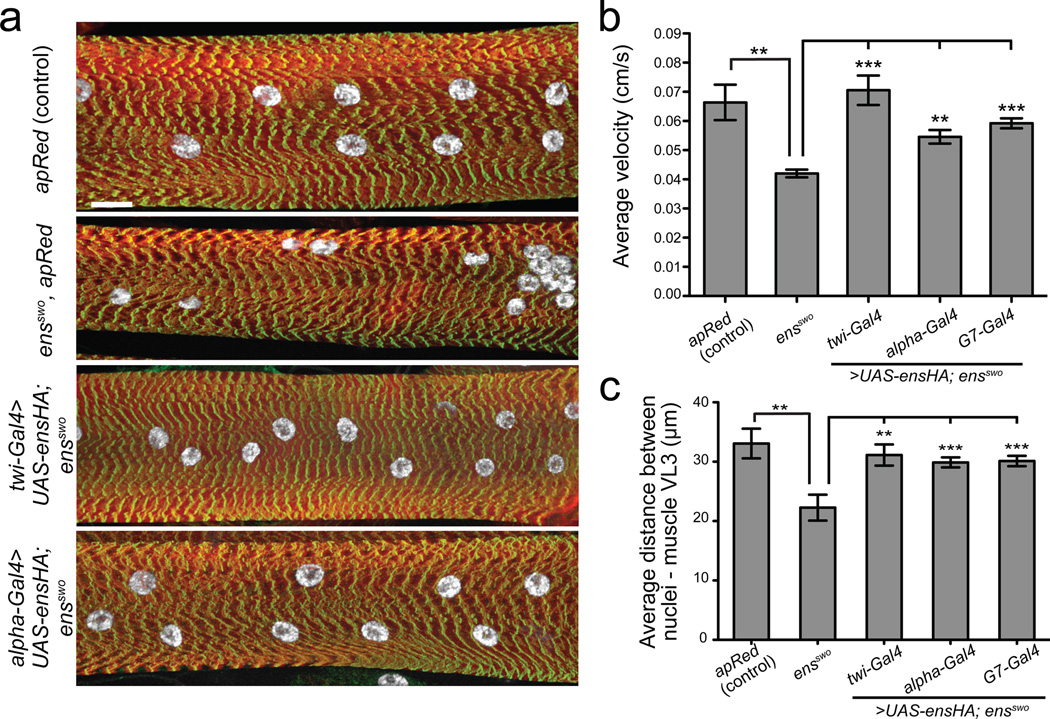
To investigate the physiological impact of mispositioned nuclei on muscle function in vivo, we examined larval motility14. ens mutant larvae move significantly slower than controls (Fig. 4b, Supplementary Fig. 7d). Moreover, analysis of the muscle structure in tracked larvae revealed that the myonuclei in ensswo mutants were 33% closer to one another than in controls (Fig. 4a, c, Supplementary Fig. 7a, e). Sarcomere structure15, t-tubules16, neuromuscular junctions17, nuclei number (Supplementary Fig. 7c, e, f, h, i) and mitochondrial localization (Supplementary Fig. 8) were unchanged in ens mutants compared to controls. Importantly, expression of Ens in the mesoderm and muscle during embryonic development or in muscle starting at the L2 larval stage in ens mutants rescued the nuclear positioning and motility defects in L3 larvae (Fig. 4a-c and Supplementary Fig. 7a,d,g) demonstrating a strong correlation between aberrant myonuclear position and reduced muscle function.
Figure 4. Ensconsin/MAP7 is required for intracellular muscle organization and efficient larval locomotion.
a. Maximum intensity XY projections of muscle VL3 from segment A3 of L3 larvae from the indicated genotypes stained for actin (red), nuclei (white), and Z-bands (green). Bar, 20 µm.
b. Average velocity of migration for L3 larvae of the indicated genotypes. Each of the indicated Gal4 drivers is expressing UAS-ensHA in a homozygous ensswo mutant background. Error bars, s.e.m. ***p < 0.001, **p < 0.01.
c. Nearest neighbor analysis of nuclei within muscle VL3 from segment A3 from L3 larvae of the indicated genotype. Error bars, s.e.m. ***p < 0.001, **p < 0.01.
These experiments define, for the first time, a subset of the nuclear behaviors that occur during muscle differentiation and demonstrate conservation across species at both the cellular and molecular levels. We find that Ens/MAP7 and Khc/Kif5b are critical for myonuclear positioning and are not required for fusion and myofiber formation. Moreover these proteins interact genetically and physically, and the physical interaction is necessary for proper nuclear positioning. Furthermore, our results argue that the correct spacing of myonuclei is required for proper muscle function.
Within a myotube, distinct microtubule networks emanate from each myonucleus producing regions of overlapping anti-parallel microtubules18–20. Furthermore, Ens/MAP7 functions to load Khc/Kif5b onto microtubules8. Therefore, the data presented here suggest a molecular mechanism for nuclear positioning. Adjacent nuclei could be positioned relative to each other by Ens/MAP7 and Khc/Kif5b interacting with and sliding anti-parallel microtubules that have their minus ends anchored to the nuclear envelope, similar to the mechanism by which the Eg5 Kinesin facilitates spindle elongation21 (Supplementary Fig 10). This mechanism is distinct from the Kinesin-1 dependent nuclear position in the C. elegans hypodermis where Khc is anchored to the nucleus by Kinesin light chain (Klc) and the KASH-domain protein Unc-83, which drive the nucleus unidirectionally on microtubules22. We find that klc mutants in Drosophila do not exhibit defects in the positioning of nuclei that we describe here. Additionally, over-expression of dn-KASH domain constructs in C2C12 myotubes that remove KASH proteins from the nuclear envelope and disrupt LINC complex (linker of nucleoskeleton and cytoskeleton) function and nuclear anchoring23,24 does not affect nuclear positioning (Supplementary Figure 9). Together these data suggest that Kinesin is not directly interacting with the nucleus, and therefore provide evidence for the sliding mechanism.
Patients suffering from various muscle diseases, including centronuclear myopathies, display muscle weakness with mislocalized myonuclei3. However, it remains unclear whether these mispositioned nuclei contribute to muscle weakness or are simply a result of impaired myofiber function3. Our results show that the ability to correctly position myonuclei correlates with better muscle function. Because nuclear misposition is the first observable defect in zygotic ens mutant embryos, we hypothesize that this is a cause of the observed muscle weakness. Mispositioned nuclei might lead to the observed muscle weakness due to a disruption in the size and spacing of myonuclear domains throughout the muscle25 or in the distribution of sub-cellular structures whose positioning depends upon correct nuclear spacing. We rescue nuclear position, muscle function, and viability not only by supplying Ens during embryogenesis but also by expressing Ens solely in mature muscles later in larval development. Thus, we propose that correcting nuclear positioning defects in patients with muscle diseases might benefit muscle strength and improve muscle function. The model systems that we present here provide a novel platform to identify additional nuclear positioning genes and assess their function and to screen for potential drugs to alleviate muscle weakness due to disease.
Methods Summary
Drosophila stocks used included apME-NLS::dsRed4, Df(3L)GN3426, ensHP36480 (Bloomington), ensf07121 (Harvard), Df-ensΔ3277, ensΔN, ensΔC (ref. 8), khc8, khc4, khc23 (ref. 10), klc8ex94 (ref. 27), twi-Gal44, alpha-Gal4, G7-Gal428 (from K. Broadie), and UAS-ens-IR lines 106207 and 18491 (VDRC). UAS-ensHA transgenic flies were generated by BestGene Inc. Mouse cells were transfected with siRNA using lipofectamine RNAiMAX (Invitrogen) or DNA using lipofectamine 2000 (Invitrogen). Primary antibodies used were: rabbit anti-dsRed (Clontech), rat anti-tropomyosin (Abcam), mouse anti-myosin heavy chain (from S. Abmayr), rabbit anti-Zasp (from F. Schöck), chicken anti-βgal (Abcam), mouse anti-α-Tubulin (Sigma), rat anti-Ensconsin (from P. Rørth), guinea pig anti-Krüppel (from J. Reinitz), rabbit anti-Eve (from M. Frasch), mouse anti-βPS Integrin (DSHB), rabbit anti-Vestigial (from S. Carroll), FITC conjugated anti-HRP (Jackson ImmunoResearch), mouse anti-Discs large (DSHB), rabbit anti-ATP synthase29 (from H. Duan), rat anti-DE-Cadherin (DSHB), mouse anti-Chaoptin 24B10 (DSHB), rat anti-Elav (DSHB), Alexa Fluor 488 conjugated wheat germ agglutinin (Invitrogen), MF20 (DSHB), rabbit anti-KHC (Santa Cruz), mouse anti-c-Myc (Roche), rabbit anti-GFP (Invitrogen). Secondary antibodies were either biotinylated (Vector Laboratories and Jackson ImmunoResearch) or conjugated to Alexa Fluor 488, 555, or 647. The fusion index30, sarcomere length15, bouton number17, and larval velocity14 were quantified as described with minor modifications (see methods). The yeast 2-hybrid was performed with full length Ens by Hybrigenics S.A Services using a 0–24 hour Drosophila cDNA library. Standard protocols were used for immunoprecipitation, western blot, and qPCR experiments and are described in Methods.
METHODS
Flies and husbandry
Stocks were grown and maintained under standard conditions on standard cornmeal medium. Crosses were performed at 25°C. Drosophila stocks: apME-NLS::dsRed4, Df(3L)GN3426, ensHP36480 (Bloomington), ensf07121 (Harvard), ensΔN, ensΔC, Df-ensΔ3277 (ref. 8), UAS-ensHA (this study), khc8, khc4, khc23 (ref. 10), klc8ex94 (ref. 27), and UAS-ens-IR lines 106207 and 18491 (VDRC). Mutant alleles were balanced and identified using CyO P[w+en11lacZ] or CTG [CyO, twi-GAL4, UAS-2xeGFP]26, TM3 Sb1Dfd-lacZ or TTG [TM3, twi-GAL4, UAS-2xeGFP]31. Gal4 lines used in this work were twi-Gal44 that is specifically expressed in the mesoderm and developing muscle (2.5 h AEL to 13 h AEL), and alpha-Gal4, G7-Gal428 (gift of K. Broadie) that are specifically expressed in the larval muscles beginning at the late L1 & early L2 larval stages and perdures until pupation.
EMS mutagenesis and screening
A detailed description of the screen and its findings will be described elsewhere. Briefly, males from an isogenized stock carrying the apME-NLS::dsRed transgene on the third chromosome were mutagenized with 35mM EMS dissolved in 1% sucrose according to standard protocol32. Individual mutant lines were established over the TTG balancer. Live F3 progeny were screened under fluorescence, and embryos homozygous for the mutant chromosome were identified by an absence of the balancer GFP. Deficiency mapping, complementation, and sequencing were used to identify the genetic lesion.
Phylogenetic analysis
Drosophila, human and mouse MAP7 orthologs were aligned using LALIGN (http://www.ch.embnet.org/software/LALIGN_form.html) and the phylogenetic tree was created using Vector NTI (Invitrogen).
Cell culture
C2C12 myoblasts were grown and differentiated for 4 days as described33. Primary myoblasts were isolated and differentiated for 3 days as described30,34.
Constructs
Map7 fragments were cloned from a C2C12 cDNA library into pEGFP-C1 vector (Invitrogen) using the following primers:
N-term (1–2 44): forward 5’-ATGGCGGAGCAGGGAGC-3’, reverse 5’-CTAGCTGTTTTTCTCGTTCCTC-3’;
EMTB-M (261–1351): forward 5’-ATGTGGCTAGAGAGAGAAGAACGAG-3’, reverse 5’-TAGGAACTACACCGACAGTCACAG-3’;
M-CC2-Cterm (859–2187): forward 5’-ATGACCATTCATGGACTAGCGAG-3’, reverse 5’-AACTTCTGCGGTCTGTTGT-3’;
M-CC2 (859–1806): forward 5’-ATGACCATTCATGGACTAGCGAG-3’, reverse 5’-TTTCTTATCAGCGGTCTCTGTCCTC-3’;
CC2-Cterm (1337–2187):forward 5’-ATGGGCGGTGTAGTTCCTAAGACTTCTG-3’, reverse 5’-AACTTCTGCGGTCTGTTGTG-3’;
CC2 (1337–1806): forward 5’-ATGGGCGGTGTAGTTCCTAAGACTTCTG-3’, reverse 5’-TTTCTTATCAGCGGTCTCTGTCCTC-3’;
M (859–1351): forward 5’-ATGACCATTCATGGACTAGCGAG-3’, reverse 5’-TAGGAACTACACCGACAGTCACAG-3’.
Full length Map7 (NM_008635) was made from the Map7 fragments and was cloned into pEGFP-C1. EMTB-Map7 was made by cloning the 1–947 bp fragment from full length Map7 into the pEGFP-C1 vector. K-N-term, K-EMTB and K-EMTB-M chimera constructs were made by cloning the K560 fragment from pET17b-K560-GFP (gift from Ron Vale) into N-Term, EMTB and EMTB-M Map7 pEGFP constructs, respectively. Motor-Kif5b, S-Motorless-Kif5b and Motorless-Kif5b c-Myc were a gift from Geri Kreitzer. FL-Kif5b was made by cloning the K560 fragment with a C-terminal region of full length Kif5b (gift from Casper C. Hoogenraad) into pEGFP C1 construct. EGFP-KASH2 and EGFP-KASH2ext were a gift from Didier Hodzic35.
siRNA sequences (AMBION):
Map7: #118: 5'-CAGAUUAGAUGUCACCAAUTT-3', #119: 5'-CCAUGAAUCUUUCGAAACATT-3',#120: 5'-ACUUACCUGUUGGAUCAAATT-3';
Map7D1: #710: 5'-GGAACAGAGGGAACGCGAATT-3', #711:5'-ACGUGGACUCUAUAAUCAATT-3'.#712: 5'-GCAAUCCAGCGGUCAGUGATT-3';
Map7D2: #436: 5'-CUUUCAACAUCAACCAUGATT-3', #437: 5'-CUUGAUGACUGUAACAAAATT-3',#438: 5'-GGUCUCCUGUGAAGUAUUATT-3';
Map7D3: #000: 5'-GGACGACAUCUCUAAAGUUTT-3', #001: 5'-GAACUAUCCUCCAUAGUGATT-3',#002: 5'-CUGAUGAGGUUGAUACCAATT-3',
Kif5b:, #781: 5’-GCAAGAAGUAGACCGGAUATT-3’, #782: 5’-GCUGUUAAUUAUGAUCAGATT-3’,#783:5’-GACAUGUCGCAGUUACAAATT-3’
Transfections
Myoblasts were transfected with siRNA using lipofectamine RNAiMAX (Invitrogen) or DNA using lipofectamine 2000 (Invitrogen). H1B-GFP stable cell line was made by transfecting cells with H1B-GFP (gift from Richard Vallee).
qPCR
mRNA from C2C12 cells was isolated using the RNeasy Micro Kit (QIAGEN) and cDNA was prepared using the Transcriptor 1st strand cDNA synthesis kit (Roche). Quantitative analyses (qPCR) was performed using CyberGreen kit in a LightCycler 480 II system (Roche), using the following primers:
Kif5b-F, 5’-GGAGGCAAGCAGTCGTAAAC-3;
Kif5b-R, 5’-TCTAGTGTTGGGAAGCAGCA-3’;
Map7-F, 5’-TGAAACCAATTTTGGCCTTT-3’;
Map7-R, 5’-AAAACCAGAGGGAGGAGCTG-3’;
Hprt1-F, 5’-GTTAAGCAGTACAGCCCCAAA-3’;
Hprt1-R, 5’-AGGGCATATCCAACAACAAACTT-3’.
Immunoprecipitations
C2C12 cells were transfected with GFP or c-Myc tagged constructs. After 24 hours, cell lysates were prepared using lysis buffer (10 mM Tris, 150 mM NaCl, 0.5mM EDTA, 0.5% NP40, 1mM PMSF, pH 7.4) containing protease inhibitor cocktail (Roche) and 0.6 µM nocodazole (Sigma). Immunoprecipitation of GFP-fusion proteins or endogenous Kif5b were performed at 4°C using the magnetic GFP-Trap kit (Chromotek) or protein A Magnetic Beads (Millipore) coated with Khc antibody, respectively.
Western blot
Extracts were loaded on a 4–12% gradient gel (Invitrogen) and transferred using iBlot (Invitrogen).
Immunohistochemistry, immunofluorescence, microscopy, and image analysis
Embryos were collected at 25°C on apple juice agar plates and were fixed as described4 except for tubulin staining where the embryos were fixed in equal volumes of 10% Formalin (Sigma) and heptane (Fisher Scientific). Larvae were dissected and fixed in 4% paraformaldedhyde (EMS) in relaxing buffer as previously described15,36,37. Eye discs were dissected from wandering third-instar larvae and fixed in 4% paraformaldedhyde. Ovaries were dissected from. Ovaries from females that were fed fresh yeast for two days were dissected in PBS and fixed in 4% paraformaldedhyde. All fluorescent stainings were mounted in Prolong Gold (Molecular Probes), otherwise embryos were mounted in Araldite.
Mouse cells were fixed using paraformaldehyde 3.7% (Sigma) and permeabilized with 0.5% Triton. All fluorescent stainings were mounted in Fluoromount G (Southern Biotech).
Antibodies were preabsorbed (PA) where noted and used at the specified final dilutions: rabbit anti-dsRed (1:400, Clontech), rat anti-tropomyosin (1:1000, Abcam), mouse anti-myosin heavy chain (1:400, gift from S. Abmayr), rabbit anti-Zasp (1:400, gift from F. Schöck), chicken anti-βgal (1:1000, Abcam), mouse anti-α-Tubulin (1:500, Sigma), rat anti-Ensconsin (1:100, gift from P. Rørth), guinea pig anti-Krüppel (PA, 1:2000, gift from J. Reinitz), rabbit anti-Eve (PA, 1:3000, gift from M. Frasch), mouse anti-βPS Integrin (1:50, DSHB), rabbit anti-Vestigial (PA, 1:50, gift from S. Carroll), FITC conjugated anti-HRP (1:500, Jackson ImmunoResearch), mouse anti-Discs large (1:200, DSHB), rabbit anti-ATP synthase (1:100, gift from H. Duan)(REF- Pena & Garesse), rat anti-DE-Cadherin (1:100, DSHB), mouse anti-Chaoptin 24B10 (1:200, DSHB), rat anti-Elav (1:50, DSHB), mouse anti-Myosin Heavy Chain MF20 (1:500, DSHB), rabbit anti-KHC (1:2500, Santa Cruz), mouse anti-c-Myc (1:5000, Roche), rabbit anti-GFP (1:2500, Invitrogen). Biotinylated secondary antibodies (Vector Laboratories and Jackson ImmunoResearch) and the Vectastain ABC kit (Vector Laboratories) were applied for non-fluorescent stainings. Alexa Fluor 488-, Alexa Fluor 555-, and Alexa Fluor 647-conjugated secondary antibodies, Alexa Fluor 546-, and Alexa Fluor 647-conjugated phalloidin, Alexa Fluor 488 conjugated wheat germ agglutinin (Invitrogen), and Hoechst 33342 or DAPI were used for fluorescent staining (Invitrogen).
Fluorescent images of Drosophila were acquired on a Leica SP5 laser scanning confocal microscope running the LAS AF 2.2 software (objectives used: 20x 0.70 NA HC PL APO multi-immersion, 40x 1.25 NA, 63x 1.4 NA, or 100x 1.43 NA HCX PL APO oil). Non-fluorescent images were acquired on a Zeiss Axiophot microscope. Image stacks were analyzed and processed using Volocity (Improvision), ImageJ, and Photoshop CS4 (Adobe).
Fluorescent images of mouse cells were acquired using a Nikon Ti microscope equipped with a CoolSNAP HQ2 camera (Roper Scientific), an XY motorized stage (Nikon), and a 40x 1.0 NA PL APO oil objective using Metamorph Software (Molecular Devices).
Quantification methods
Nuclear positioning was quantified in mammalian myotubes containing ≥3 nuclei, and myotubes were classified as follows: “aligned”, > 70% of the nuclei in a myotube align along the same axis; “aggregated”, >70% of the nuclei do not align along the same axis; “other”, nuclei in a myotube do not fall into either category. Fusion index was quantified as described30.
A nearest neighbor analysis in Drosophila was conducted to determine the average distance between a nucleus and its single closest neighbor within a muscle. To identify the closest neighbor for each nucleus in a muscle, the distance between the center of a nucleus and the center of each of the closely surrounding nuclei (usually 4 – 8) was measured. The shortest distance identified the closest nucleus or “nearest neighbor” and this distance was recorded. A nearest neighbor was identified for each nucleus in every muscle analyzed and the nearest neighbor distances were averaged to give an average “nearest neighbor” distance for the average space between one nucleus and the next closest nucleus in a muscle.
Sarcomere length was defined as the distance between the Z-lines labeled with the Anti-Zasp antibody15. Sarcomere length was measured at 5 separate regions per muscle and in the same 5 regions for each muscle examined.
Neuromuscular junctions were visualized by HRP or Dlg staining and quantified as previously described17.
The position of the oocyte nucleus was determined by measuring the distance from the anterior border of the oocyte to the center of the nucleus.
Time lapse imaging
Drosophila embryos were prepared as described4,38. Time-lapse sequences were acquired using a Zeiss Axio Imager.Z1 with a 20X NA 0.75 Plan-Apochromat dry objective. All time-lapse series were taken as a set of z-stacks over time (4D imaging) with optical sections every 3 µm. Only a single z-section with the greatest number of nuclei in focus was selected for each time point. Images were processed using ImageJ and compiled into movie using Apple Quicktime. Single images of live embryos were acquired on a Zeiss Axiophot microscope.
Time lapse imaging of mouse cells was done as reported30.
Yeast 2-hybrid
The yeast two-hybrid screening was based on a full length Ens as bait and performed by Hybrigenics S.A Services using a 0–24 hour embryonic Drosophila cDNA library. 99 million interactions were analyzed. 10% of the high confidence clones that were recovered from the screen were Khc.
Larval behavior
Larval behavior was assessed as previously described with minor modifications14,39. Briefly, embryos (stage 16 and 17) were selected for the presence of clustered apRed nuclei and/or the absence of the fluorescent balancer. Selected embryos were placed on a yeast-coated apple juice plated overnight at 25°C, and L1 larvae were selected the following day and placed into vials of standard food containing bromophenol blue. Larvae were picked from the vial 3 days later and tracked. Larvae were tracked individually as they migrated towards a single odor source (0.25M ethyl butyrate, Sigma) and recorded with a CCD camera for 5 minutes, until they reached odor source, or until they contacted any of the walls of the apparatus. Images were processed by Ethovision software (Noldus).
Germline transformation and constructs
UAS-ensHA DNA was constructed by PCR amplifying the full length cDNA from the ens RA transcript with primers to add an HA tag to the C-terminal end of the protein followed by an EcoR1 restriction site. Primers used were:
5’: GCCGAATTCCACCATGGCGAGTCTTGGGGGC,
3’:GCCGAATTCTTATCAAGCGTAATCTGGAACATCGTATGGGTACAGCAGCGATATATCTTTATTTTCGTG. Amplified cDNA was introduced into the Uni-ZAP XR vector (a derivative of pBlueScript SK-, Stratagene) using EcoRI/XhoI. This DNA was used by BestGene Inc. to generate transgenic flies.
Statistics
Statistical analysis was performed using Prism (GraphPad Software Inc.). Pairwise comparisons were made using a Student’s t test, and group comparisons were made by ANOVA followed by Tukey’s posthoc analysis. In nuclear positioning analysis in C2C12 cells, student’s t tests were performed between scrambled siRNA and experimental condition for each category (aligned, aggregated and other).
Supplementary Material
Acknowledgements
We thank K. Anderson, K. Hadjantonakis, A. Hall, and D. Sassoon for comments on the manuscript. We thank the Baylies and Gomes labs for discussions and Rodrigo Fernandez-Gonzalez for his assistance in computational analysis. The initial screens in Drosophila were supported by NIH GM056989 and GM0781318 to MB; the Drosophila nuclear positioning analysis was supported by Muscular Dystrophy Association (MDA) to MB. TM was supported initially by the NIH Training Grant T32 BM008539. BC was supported initially by a Fondation pour la Recherche Médicale (FRM) fellowship. VG was supported initially by a Region Ile-de-France fellowship. Mammalian work was supported by Muscular Dystrophy Association (MDA), INSERM Avenir program and Agence Nationale de la Recherche (ANR) grants to ERG.
Footnotes
Author contributions: MB, MX, TM, EG, VG and BC conceived, designed, and analyzed the experiments. TM, MX, ES and BR conducted the Drosophila experimental work; VG and BC conducted the mouse primary cultures and C2C12 experimental work. The manuscript was written by TM, ES, VG, EG and MB with assistance from other authors.
Competing Financial Interests: The authors declare no competing financial interests.
References
- 1.Bruusgaard JC, Liestøl K, Ekmark M, Kollstad K, Gundersen K. Number and spatial distribution of nuclei in the muscle fibres of normal mice studied in vivo. J. Physiol. (Lond.) 2003;551:467–478. doi: 10.1113/jphysiol.2003.045328. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Cohn RD, Campbell KP. Molecular basis of muscular dystrophies. Muscle Nerve. 2000;23:1456–1471. doi: 10.1002/1097-4598(200010)23:10<1456::aid-mus2>3.0.co;2-t. [DOI] [PubMed] [Google Scholar]
- 3.Jungbluth H, Wallgren-Pettersson C, Laporte J. Centronuclear (myotubular) myopathy. Orphanet J Rare Dis. 2008;3:26. doi: 10.1186/1750-1172-3-26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Richardson BE, Beckett K, Nowak SJ, Baylies MK. SCAR/WAVE and Arp2/3 are crucial for cytoskeletal remodeling at the site of myoblast fusion. Development. 2007;134:4357–4367. doi: 10.1242/dev.010678. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Beckett K, Baylies MK. The development of the Drosophila larval body wall muscles. Int Rev Neurobiol. 2006;75:55–70. doi: 10.1016/S0074-7742(06)75003-6. [DOI] [PubMed] [Google Scholar]
- 6.Prokop A, Martín-Bermudo MD, Bate M, Brown NH. Absence of PS integrins or laminin A affects extracellular adhesion, but not intracellular assembly, of hemiadherens and neuromuscular junctions in Drosophila embryos. Dev Biol. 1998;196:58–76. doi: 10.1006/dbio.1997.8830. [DOI] [PubMed] [Google Scholar]
- 7.Volk T. Singling out Drosophila tendon cells: a dialogue between two distinct cell types. Trends Genet. 1999;15:448–453. doi: 10.1016/s0168-9525(99)01862-4. [DOI] [PubMed] [Google Scholar]
- 8.Sung HH, et al. Drosophila ensconsin promotes productive recruitment of Kinesin-1 to microtubules. Developmental Cell. 2008;15:866–876. doi: 10.1016/j.devcel.2008.10.006. [DOI] [PubMed] [Google Scholar]
- 9.Fischer-Vize JA, Mosley KL. Marbles mutants: uncoupling cell determination and nuclear migration in the developing Drosophila eye. Development. 1994;120:2609–2618. doi: 10.1242/dev.120.9.2609. [DOI] [PubMed] [Google Scholar]
- 10.Brendza KM, Rose DJ, Gilbert SP, Saxton WM. Lethal kinesin mutations reveal amino acids important for ATPase activation and structural coupling. J Biol Chem. 1999;274:31506–31514. doi: 10.1074/jbc.274.44.31506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Vale RD, Reese TS, Sheetz MP. Identification of a novel force-generating protein, kinesin, involved in microtubule-based motility. Cell. 1985;42:39–50. doi: 10.1016/s0092-8674(85)80099-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Vale RD, et al. Direct observation of single kinesin molecules moving along microtubules. Nature. 1996;380:451–453. doi: 10.1038/380451a0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Masson D, Kreis TE. Identification and molecular characterization of E-MAP-115, a novel microtubule-associated protein predominantly expressed in epithelial cells. The Journal of Cell Biology. 1993;123:357–371. doi: 10.1083/jcb.123.2.357. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Louis M, Huber T, Benton R, Sakmar TP, Vosshall LB. Bilateral olfactory sensory input enhances chemotaxis behavior. Nat Neurosci. 2008;11:187–199. doi: 10.1038/nn2031. [DOI] [PubMed] [Google Scholar]
- 15.Bai J, Hartwig JH, Perrimon N. SALS, a WH2-domain-containing protein, promotes sarcomeric actin filament elongation from pointed ends during Drosophila muscle growth. Developmental Cell. 2007;13:828–842. doi: 10.1016/j.devcel.2007.10.003. [DOI] [PubMed] [Google Scholar]
- 16.Razzaq A, et al. Amphiphysin is necessary for organization of the excitation-contraction coupling machinery of muscles, but not for synaptic vesicle endocytosis in Drosophila. Genes & Development. 2001;15:2967–2979. doi: 10.1101/gad.207801. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.McCabe BD, et al. The BMP homolog Gbb provides a retrograde signal that regulates synaptic growth at the Drosophila neuromuscular junction. Neuron. 2003;39:241–254. doi: 10.1016/s0896-6273(03)00426-4. [DOI] [PubMed] [Google Scholar]
- 18.Tassin AM, Maro B, Bornens M. Fate of microtubule-organizing centers during myogenesis in vitro. The Journal of Cell Biology. 1985;100:35–46. doi: 10.1083/jcb.100.1.35. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Guerin CM, Kramer SG. RacGAP50C directs perinuclear gamma-tubulin localization to organize the uniform microtubule array required for Drosophila myotube extension. Development. 2009;136:1411–1421. doi: 10.1242/dev.031823. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Bugnard E, Zaal KJM, Ralston E. Reorganization of microtubule nucleation during muscle differentiation. Cell Motil Cytoskeleton. 2005;60:1–13. doi: 10.1002/cm.20042. [DOI] [PubMed] [Google Scholar]
- 21.Glotzer M. The 3Ms of central spindle assembly: microtubules, motors and MAPs. Nat Rev Mol Cell Biol. 2009;10:9–20. doi: 10.1038/nrm2609. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Meyerzon M, Fridolfsson HN, Ly N, McNally FJ, Starr DA. UNC-83 is a nuclear-specific cargo adaptor for kinesin-1-mediated nuclear migration. Development. 2009;136:2725–2733. doi: 10.1242/dev.038596. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Grady RM, Starr DA, Ackerman GL, Sanes JR, Han M. Syne proteins anchor muscle nuclei at the neuromuscular junction. Proc Natl Acad Sci USA. 2005;102:4359–4364. doi: 10.1073/pnas.0500711102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Lei K, et al. SUN1 and SUN2 play critical but partially redundant roles in anchoring nuclei in skeletal muscle cells in mice. Proc Natl Acad Sci USA. 2009;106:10207–10212. doi: 10.1073/pnas.0812037106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Pavlath GK, Rich K, Webster SG, Blau HM. Localization of muscle gene products in nuclear domains. Nature. 1989;337:570–573. doi: 10.1038/337570a0. [DOI] [PubMed] [Google Scholar]
- 26.Feng G, Deák P, Kasbekar DP, Gil DW, Hall LM. Cytogenetic and molecular localization of tipE: a gene affecting sodium channels in Drosophila melanogaster. Genetics. 1995;139:1679–1688. doi: 10.1093/genetics/139.4.1679. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Gindhart JG, Desai CJ, Beushausen S, Zinn K, Goldstein LS. Kinesin light chains are essential for axonal transport in Drosophila. The Journal of Cell Biology. 1998;141:443–454. doi: 10.1083/jcb.141.2.443. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Zhang YQ, et al. Drosophila fragile X-related gene regulates the MAP1B homolog Futsch to control synaptic structure and function. Cell. 2001;107:591–603. doi: 10.1016/s0092-8674(01)00589-x. [DOI] [PubMed] [Google Scholar]
- 29.Peña P, Garesse R. The beta subunit of the Drosophila melanogaster ATP synthase: cDNA cloning, amino acid analysis and identification of the protein in adult flies. Biochem. Biophys. Res. Commun. 1993;195:785–791. doi: 10.1006/bbrc.1993.2114. [DOI] [PubMed] [Google Scholar]
- 30.Mitchell KJ, et al. Identification and characterization of a non-satellite cell muscle resident progenitor during postnatal development. Nat Cell Biol. 2010;12:257–266. doi: 10.1038/ncb2025. [DOI] [PubMed] [Google Scholar]
Additional References
- 31.Halfon MS, et al. New fluorescent protein reporters for use with thedrosophila gal4 expression system and for vital detection of balancer chromosomes. Genesis. 2002;34:135–138. doi: 10.1002/gene.10136. [DOI] [PubMed] [Google Scholar]
- 32.Lewis E. Method of feeding ethane methylsulfonate (EMS) to Drosophila males. (Dros. Inf. Serv: 1968) [Google Scholar]
- 33.Parlakian A, et al. Skeletal muscle phenotypically converts and selectively inhibits metastatic cells in mice. PLoS ONE. 2010;5:e9299. doi: 10.1371/journal.pone.0009299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.De Palma C, et al. Nitric oxide inhibition of Drp1-mediated mitochondrial fission is critical for myogenic differentiation. Cell Death Differ. 2010;17:1684–1696. doi: 10.1038/cdd.2010.48. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Khatau SB, et al. A perinuclear actin cap regulates nuclear shape. Proc Natl Acad Sci USA. 2009;106:19017–19022. doi: 10.1073/pnas.0908686106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Brent J, Werner K, Mccabe B. Drosophila Larval NMJ Dissection. Journal of Visualized Experiments. 2009 doi: 10.3791/1107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Brent J, Werner K, McCabe BD. Drosophila larval NMJ immunohistochemistry. Journal of visualized experiments : JoVE. 2009 doi: 10.3791/1108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Richardson BE, Beckett K, Baylies MK. Live imaging of Drosophila myoblast fusion. Methods Mol Biol. 2008;475:263–274. doi: 10.1007/978-1-59745-250-2_15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Louis M, Piccinotti S, Vosshall LB. High-resolution measurement of odor-driven behavior in Drosophila larvae. Journal of visualized experiments : JoVE. 2008 doi: 10.3791/638. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.