Abstract
More information is needed about genetic factors that initiate development of pancreatic intraepithelial neoplasms (PanINs)—the most common precursors of pancreatic ductal adenocarcinoma. We show that >99% of the earliest stage, lowest-grade, PanIN-1 lesions contain mutations in KRAS, p16/CDKN2A, GNAS, or BRAF. These findings could improve our understanding the development and progression of these pre-malignant lesions.
Keywords: Pancreatic cancer, tumorigenesis, transformation, neoplasm
Pancreatic cancer is the 4th leading cause of cancer death in the USA1. PanINs are the most common precursor to invasive pancreatic adenocarcinoma2. They are microscopic lesions (<5 mm diameter), and almost always too small to be identified by current imaging. Low-grade PanINs (PanIN-1) are common and their prevalence increases with age, while high-grade PanINs are uncommon and are usually found in pancreata with invasive pancreatic cancer. Multiple PanINs of all grades are frequently observed in individuals with inherited susceptibility to pancreatic cancer3. Over 90% of invasive invasive adenocarcinomas of the pancreas harbor oncogenic mutations in KRAS while BRAF mutations occur in a small subset of KRAS-wild-type pancreatic cancers1,4. Almost all invasive pancreatic cancers inactivate p16/CDKN2A. GNAS is mutated in ~60% of intraductal papillary mucinous neoplasms (IPMNs), and in some invasive pancreatic cancers arising in association with an IPMN5. Several genetic alterations identified in invasive pancreatic adenocarcinomas are also present in PanINs, with evidence of increasing prevalence of these alterations with PanIN grade2. However, the genes responsible for early PanIN development remain poorly understood. Thus, a meta-analysis evaluating studies of mutant KRAS prevalence in PanINs found that among patients with pancreatic ductal adenocarcinoma, KRAS mutations were detected by conventional methods in 36% of PanIN-1A, 44% of PanIN-1B, and 87% of high-grade PanIN lesions (PanIN-2 and -3)6. Data such as these indicate that KRAS mutations are more involved after PanIN initiation; genetic alterations that initiate tumorigenesis should have the same prevalence, independent of grade.
The goal of the current study was to employ more sensitive mutation detection methods to obtain a more detailed genetic understanding of early PanIN development. For this purpose, first we used laser capture to microdissect 169 PanINs (50 PanIN-1A, 52 PanIN-1B, 45 PanIN-2 and 22 PanIN-3 lesions) from 89 patients with benign and malignant pancreatic diseases (Supplementary Table 1), invasive pancreatic ductal adenocarcinomas from 12 patients, and normal pancreatic ducts from 20 patients. After DNA isolation and whole genome amplification, DNA was analyzed for somatic mutations in KRAS, BRAF, GNAS, and p16/CDKN2A using pyrosequencing and high-resolution melt-curve analysis. The limit of detection of these assays is ~5%, i.e. mutant alleles can be detected at concentrations of 5% or more (mutant: wild-type alleles: 1:20, cells: 1 in 10) (See Supplementary Methods).
Using pyrosequencing, KRAS codon 12 mutations were detected in 46 (92.0%) of 50 PanIN-1A, 48 (92.3%) of 52 PanIN-1B, 42 (93.3%) of 45 PanIN-2, and 21 (95.4%) of 22 PanIN-3 lesions (Supplementary Table 2). Occasional mutations of KRAS codon 13 and codon 61 were identified (Supplementary Table 2) and a 2nd non-dominant KRAS mutation was found in 6 of 169 PanINs. No evidence of KRAS amplification was found. Melt-curve analysis confirmed the presence of KRAS gene mutations in every sample that was positive by pyrosequencing (Supplementary Fig. 1). Overall, 163/169 (96.4%) of PanINs harbored KRAS mutations. No KRAS mutations were identified in normal pancreatic duct samples (Supplementary Table 2). Five of the 169 PanIN lesions tested by pyrosequencing had a 2nd minor KRAS codon 12 mutation. Many PanIN-1 lesions had low mutant KRAS concentrations (mean ~20% of alleles by pyrosequencing, Supplementary Table 3), perhaps explaining why prior studies reported a lower prevelance of KRAS mutations in low-grade PanINs. To check the purity of our laser capture microdissection, we repeated the microdissections from 16 of the PanINs, using another set of slides. The mutant KRAS concentrations in DNA from the second microdissection were not significantly different from those of the first microdissections (Supplemental methods). We also analyzed mutant KRAS concentrations in invasive pancreatic adenocarcinomas, and these samples had close to the concentrations of mutant KRAS one would expect if they consisted entirely of KRAS-mutant cancer cells without any contaminating wild-type (and presumably non-neoplastic) cells (mean 42.5% of KRAS alleles, not significantly different from the mean mutant KRAS allele concentration in PanIN-3 lesions). Indeed, we found that the average concentration of mutant KRAS alleles in PanINs increased significantly with increasing grade of PanIN (Supplementary Fig. 1C, Figure 2).
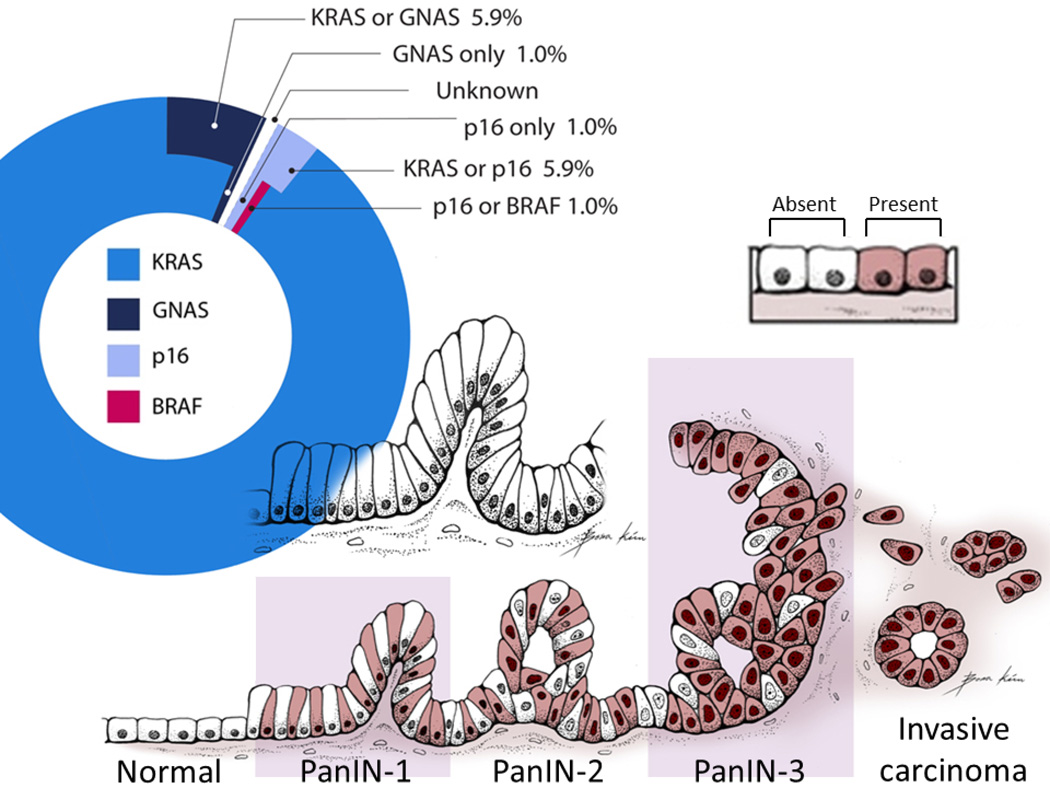
Figure 2. Initial mutations of PanIN-1 lesions.
The pie chart in the upper portion of the figure is indicates the percentage of mutations in each of the genes tested (KRAS, GNAS, p16, BRAF) identified in PanIN-1 lesions. For PanIN-1 lesions with more than one mutation, we could not determine which gene was mutated first. For example, a few PanIN-1 lesions had both a KRAS mutation and a GNAS mutation so the initial mutation in these PanINs was indicated as arising in either “KRAS or GNAS”.
The bottom portion of the figure is a schematic model illustrating the increasing percentage of mutant KRAS cells within PanIN lesions as they progress from a low-grade to a high-grade PanIN and to an invasive ductal adenocarcinoma, based on measurements of the average mutant KRAS concentrations per PanIN. Shaded cells (Present) represent PanIN cells with mutant KRAS, non-shaded cells (absent) represent PanIN cells without mutant KRAS (wild-type).
PanIN = pancreatic intraepithelial neoplasia.
These results indicate that virtually all PanINs harbor KRAS mutations. However, in the earliest PanIN lesions, these mutations are generally present in only a fraction of the cells comprising the lesion. The percentage of mutant KRAS cells in the PanIN progressively increases with the PanIN grade, consistent with a gradual expansion of the KRAS-mutant clone as the PanIN progresses.
We then sought to determine if mutations in other genes are present in the few KRAS-wild-type PanINs, particularly low-grade PanINs. As prior studies have found that TP53 and SMAD4 mutations do not appear until late in the neoplastic progression, we focused on BRAF, because it is sometimes mutant in KRAS wild-type cancers; p16/CDKN2A, because loss of p16/CDKN2A expression has been found in some early PanINs, and GNAS, because it is commonly mutated in another type of pre-malignant pancreatic lesion (IPMNs) 5.
Although p16/CDKN2A mutations were identified in only 17/147 of PanIN-1/2 lesions (11.5%), they were detected more often in KRAS-wild-type PanINs than in KRAS-mutant PanINs (P=0.0209). A similar trend was noted for GNAS mutations which were the only mutation identified in 2 PanINs (P=0.0886, Figure 2). Interestingly, similar to what was found for some IPMNs5, among PanIN-1/2 lesions with both GNAS and KRAS mutations, mutant GNAS concentrations were higher than mutant KRAS (P=0.0084, paired t-test)(Supplementary Table 2), suggesting that low mutant KRAS concentrations in PanIN samples was not simply the result of contamination with DNA from nearby stromal cells. It also suggests that in some PanINs, the KRAS mutation arose later than the GNAS mutation. There were no histological differences in cell morphology within PanIN-1 lesions with low vs. high concentrations of either mutant KRAS or GNAS (Supplementary Figure 1). GNAS mutations were detected more often in PanINs from patients with a diagnosis other than pancreatic adenocarcinoma (P=0.0398). Overall, we were able to identify at least one mutation in KRAS, GNAS p16/CDKN2A or BRAF, in all but 1 of 169 PanINs (Supplementary Tables 1 and 2). No mutant KRAS or GNAS was detected in this one wild-type PanIN (Patient 72) even with the sensitive techniques we used (detection limit <1%).
To confirm the prevalence of KRAS and GNAS mutations in PanINs, we also conducted an independent analysis of an additional 37 PanIN lesions (11 PanIN-1, 20 PanIN-2, and 7 PanIN-3 lesions) using two additional ultra-sensitive technologies - Digital ligation (limit of detection 1/200 alleles) and BEAMing (limit of detection 1/1000 alleles)(Supplementary Methods) and found KRAS mutations in 94.6% of PanINs and GNAS mutations in 11.4% (Supplementary Table 4), with complete concordance of the mutation results with both platforms. A 2nd non-dominant KRAS mutation was found more often using these methods than by pyrosequencing, consistent with the lower limit of detection of these assays.
These results indicate that somatic mutations are required for the early development of virtually all PanINs. Our results are consistent with observations in genetically engineered mouse models in which mouse PanINs can be initiated by oncogenic KRAS7. Although low-grade PanIN cells have some metaplastic features, our results do not support the hypothesis that PanINs begin as metaplasias and only subsequently acquire genetic alterations. If this were true, more low-grade (early) PanINs would lack oncogenic mutations. (We found only one of 102 PanIN-1 lesions lacked a mutation). In prior studies, we have examined the metaplastic lesion known as acinar-to-ductal metaplasia (ADM) for evidence of genetic alterations (mutant KRAS and telomere length analysis) and did not find evidence from this analysis that ADMs are precursors to PanINs. The findings that many low-grade PanINs contain mixtures of mutant and wild-type KRAS cells, that GNAS mutation concentrations can be higher than KRAS mutation concentrations in the same PanIN, and that the average proportion of mutant KRAS within PanINs increases with PanIN grade, suggests that mutant KRAS alone provides only a modest selective advantage over neighboring cells. This finding suggests that the KRAS-mutant clone is partially restrained within the PanIN, possibly by oncogene-induced senescence8,9 and this restraint is likely maintained until additional genetic and or epigenetic events (such as p16/CDKN2A inactivation) occur. The driving force behind the expansion of cells within PanINs that do not harbor mutant KRAS is not certain. One possibility is that PanIN-initiating event(s) precede oncogenic KRAS mutations. However, our genome4 and methylome10 studies indicate there are no other commonly mutated or epigenetically silenced10 genes in pancreatic cancers that stand out as candidate initiators of PanIN development. Telomere shortening is observed in almost all low-grade PanINs11 but this phenomenon could be a consequence of activation of oncogene stress-induced senescence programs12 rather than an initiator of PanINs.
One unifying hypothesis to explain all these observations is that KRAS, and occasionally p16/CDKN2A, GNAS, or BRAF, mutations can initiate PanIN development, and that these mutant cells induce surrounding cells to proliferate. Such proliferation could come from autocrine and paracrine influences from KRAS-mutant PanIN cells, such as expression of Shh, and other developmental genes,13,14 as well as stress-inducing signals that lead to senescence15, and induce metaplastic features in PanIN epithelial cells including adjacent cells lacking KRAS mutations.
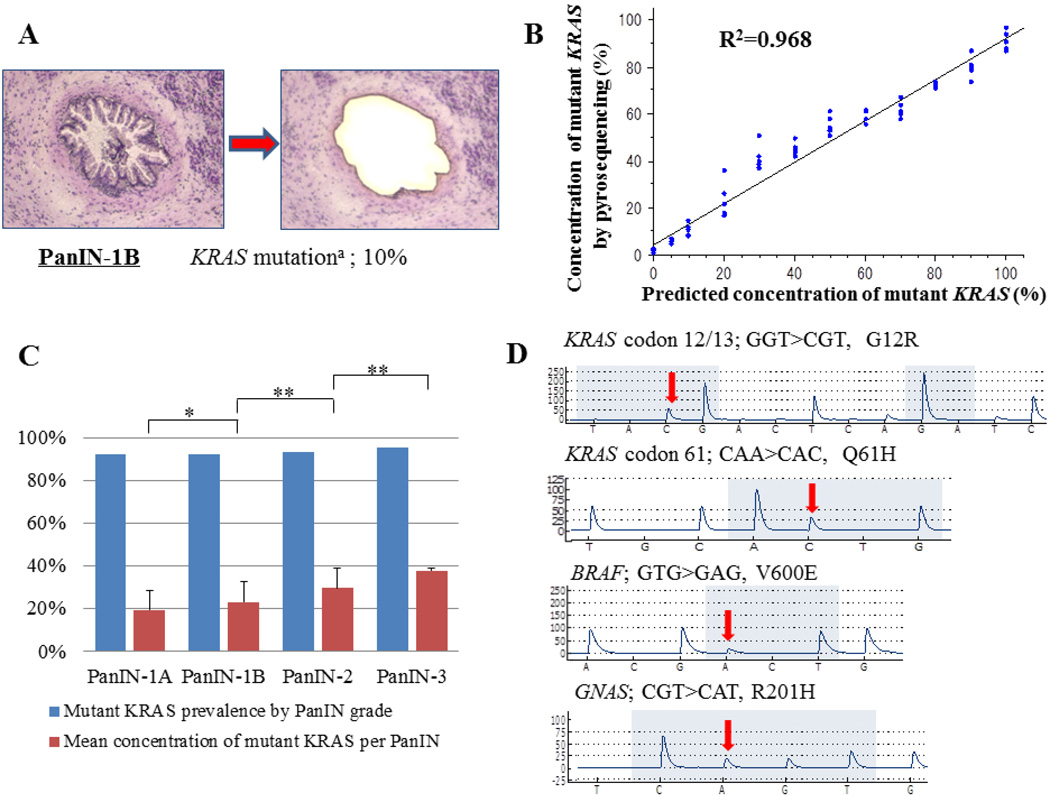
Figure 1.
(A) An example of a H + E stained PanIN before and after laser capture microdissection. Microdissection was performed at the duct epithelial cell borders to avoid contamination with stromal cells. (B) Scatter-plot graph of actual and predicted concentrations of mutant DNA by pyrosequencing. (C) Prevalence of KRAS codon 12 mutations and concentration of mutations per PanIN by grade of PanIN. KRAS codon 12 mutations were found in more than 92% of PanINs in every group. The average percentage of mutant KRAS alleles within a PanIN increased at each PanIN grade. (D) Representative pyrosequencing traces with mutant sequences highlighted by the arrows. * P<0.05, **P<0.001. a = Concentrations of mutant DNA by pyrosequencing.
Acknowledgements
We thank Ms. Bona Kim for providing the PanIN illustration (Figure 2).
Grant Support: This work was supported by NIH grants (CA62924, R01CA97075, R01CA120432, RC2CA148376, P01CA134292), the Stringer Foundation, the Michael Rolfe Foundation, the Lustgarten Foundation for pancreatic cancer research, and German Cancer Aid (Deutsche Krebshilfe e.V.).
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
The authors report no conflicts of Interest.
Author Contributions
Mitsuro Kanda; acquisition of data; analysis and interpretation of data; drafting of the manuscript
Hanno Matthaei: acquisition of data; analysis and interpretation of data; revision of the manuscript; obtained funding;
Jian Wu: acquisition of data; analysis and interpretation of data;
Seung-Mo Hong: acquisition of data;
Jun Yu: material support, acquisition of data;
Michael Borges: material support, acquisition of data;
Ralph Hruban: material support; acquisition and interpretation of data; revision of the manuscript
Anirban Maitra: material support; acquisition and interpretation of data; study supervision; revision of the manuscript
Ken Kinzler: study supervision; revision of the manuscript
Bert Vogelstein: study supervision; interpretation of data; revision of the manuscript: obtained funding;
Michael Goggins: study concept and design; study supervision; interpretation of data; drafting of the manuscript; obtained funding;
References
- 1.Vincent A, et al. Lancet. 2011;378:607–620. doi: 10.1016/S0140-6736(10)62307-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Hruban RH, et al. Clin Cancer Res. 2000;6:2969–2972. [PubMed] [Google Scholar]
- 3.Shi C, et al. Clin Cancer Res. 2009;15:7737–7743. doi: 10.1158/1078-0432.CCR-09-0004. Epub. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Jones S, et al. Science. 2008;321:1801–1806. doi: 10.1126/science.1164368. Epub 2008 Sep 1804. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Wu J, et al. Sci Transl Med. 2011;3:92ra66. doi: 10.1126/scitranslmed.3002543. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Lohr M, et al. Neoplasia. 2005;7:17–23. doi: 10.1593/neo.04445. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Hingorani SR, et al. Cancer Cell. 2003;4:437–450. doi: 10.1016/s1535-6108(03)00309-x. [DOI] [PubMed] [Google Scholar]
- 8.Caldwell ME, et al. Oncogene. 2011 epub aug 22. [Google Scholar]
- 9.Lee KE, et al. Cancer Cell. 2010;18:448–458. doi: 10.1016/j.ccr.2010.10.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Vincent A, et al. Clin Cancer Res. 2011;17:4341–4354. doi: 10.1158/1078-0432.CCR-10-3431. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.van Heek NT, et al. Am J Pathol. 2002;161:1541–1547. doi: 10.1016/S0002-9440(10)64432-X. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Ben-Porath I, et al. J Clin Invest. 2004;113:8–13. doi: 10.1172/JCI200420663. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Strobel O, et al. Gastroenterology. 2010;138:1166–1177. doi: 10.1053/j.gastro.2009.12.005. (epub dec1121 2009) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Prasad NB, et al. Cancer Res. 2005;65:1619–1626. doi: 10.1158/0008-5472.CAN-04-1413. [DOI] [PubMed] [Google Scholar]
- 15.Kuilman T, et al. Nat Rev Cancer. 2009;9:81–94. doi: 10.1038/nrc2560. [DOI] [PubMed] [Google Scholar]