Abstract
Chromosomal DNA replication requires one daughter strand—the lagging strand—to be synthesised as a series of discontinuous, RNA-primed Okazaki fragments, which must subsequently be matured into a single covalent DNA strand. Here, we describe the reconstitution of Okazaki fragment maturation in vitro using proteins derived from the archaeon Sulfolobus solfataricus. Six proteins are necessary and sufficient for coupled DNA synthesis, RNA primer removal and DNA ligation. PolB1, Fen1 and Lig1 provide the required catalytic activities, with coordination of their activities dependent upon the DNA sliding clamp, proliferating cell nuclear antigen (PCNA). S. solfataricus PCNA is a heterotrimer, with each subunit having a distinct specificity for binding PolB1, Fen1 or Lig1. Our data demonstrate that the most efficient coupling of activities occurs when a single PCNA ring organises PolB1, Fen1 and Lig1 into a complex.
Keywords: archaea, DNA replication, Okazaki fragment, PCNA
Introduction
All cellular organisms replicate the bulk of their DNA using the replication fork mechanism. The two strands of the DNA double helix are arranged in anti-parallel configuration and yet all known DNA polymerases can only synthesise DNA with 5′–3′ polarity. Consequently, while one daughter strand, the leading strand, can be synthesised continuously, the second daughter strand, the lagging strand, is synthesised discontinuously in short RNA-primed molecules; the Okazaki fragments. The RNA-containing primers of the Okazaki fragments must be removed during the maturation of the lagging strand and the fragments need to be ligated together to impart covalent integrity to the nascent DNA. An extensive series of biochemical reconstitution experiments have provided strong evidence for a number of distinct pathways operating in eukaryotic organisms to resolve discontinuous Okazaki fragments (for review see Burgers, 2009; Beattie and Bell, 2011a).
The crenarchaeon Sulfolobus solfataricus has been shown to possess a simplified complement of the eukaryotic DNA replication machinery, and thus serves as a model system for analysis of replication proteins (Dionne et al, 2003a). Consistent with a eukaryotic-like replication machinery, archaeal Okazaki fragments are found in a distribution of lengths centred around 100 nucleotides (nt), and possess a 5′ RNA primer of ∼10 nt, similar to eukaryotes (Matsunaga et al, 2003). Interestingly, S. solfataricus only appears to possess a subset of the eukaryotic Okazaki fragment maturation factors; most notably DNA polymerase, the flap endonuclease, Fen1, and an ATP-dependent DNA ligase, Lig1, as well as a DNA sliding clamp orthologous to eukaryal proliferating cell nuclear antigen (PCNA).
PCNA is a ring-shaped protein that encircles DNA and, via protein-binding sites located on the outer surface of the ring, functions as a molecular platform at the replication fork to recruit numerous replication-associated enzymes (Vivona and Kelman, 2003; Moldovan et al, 2007). The highly conserved mode of interaction between almost all PCNA-binding proteins and PCNA involves docking of hydrophobic and aromatic residues from a short PCNA-interacting protein (PIP) motif on the interacting protein into a hydrophobic pocket formed largely by the interdomain connector loop of each PCNA subunit.
Our previous work has revealed that S. solfataricus PCNA is unusually composed of three distinct subunits (Dionne et al, 2003b). The heterotrimeric PCNA has a precise order of assembly—PCNA1 and PCNA2 form a heterodimer that then recruits PCNA3—and thus has a defined stereochemistry. These initial biochemical studies have been confirmed by a number of X-ray crystallographic studies of heterotrimers and subassemblies thereof (Doré et al, 2006; Pascal et al, 2006; Williams et al, 2006; Hlinkova et al, 2008). Interestingly, the distinct S. solfataricus PCNA subunits have preferred interaction partners. Most intriguingly with relevance to Okazaki fragment maturation, PCNA1 specifically interacts with Fen1, PCNA2 is bound by the replicative DNA polymerase PolB1, and PCNA3 preferentially interacts with Lig1. Furthermore, our previous work revealed that the PCNA heterotrimer can act as a bridge between Fen1 and PolB1 and/or Lig1 (Dionne et al, 2003b). However, the stoichiometry and functional relevance of this potential coordination have not yet been addressed. As with other sliding clamps, S. solfataricus PCNA can also interact with a range of other proteins, and interactions between the lesion-bypass polymerase Dpo4 and PCNA1, between a type 4 uracil DNA glycosylase and PCNA3, and between the DNA repair factor Xpf and PCNA1 and 3 have been reported (Roberts et al, 2003; Dionne and Bell, 2005; Dionne et al, 2008).
In this study, we describe the in vitro reconstitution of DNA synthesis-dependent RNA primer removal and subsequent DNA ligation in a coupled system that is highly dependent on PCNA. A combination of mutational analyses and the use of an alternate DNA polymerase provide support for a model in which a single PCNA ring acts as the assembly platform for an Okazaki fragment maturation complex composed of PolB1, Fen1 and Lig1.
Results
All three subunits of S. solfataricus heterotrimeric PCNA are expressed throughout S-phase
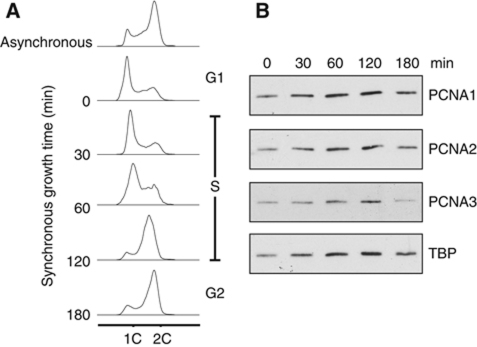
Previous biochemical and structural analyses have demonstrated that the S. solfataricus PCNA subunits PCNA1, PCNA2 and PCNA3 assemble into a stable heterotrimer in vitro and in vivo. Furthermore, all three PCNA genes are essential for viability in the related archaeon Sulfolobus islandicus (Zhang et al, 2010). We sought to confirm if this unique assembly does indeed support DNA replication in S. solfataricus cells. By adapting a cell-cycle synchronisation method previously developed for Sulfolobus acidocaldarius (Duggin et al, 2008) we synchronised S. solfataricus cells in the G1-phase of the cell cycle and followed synchronous growth through S-phase and into G2 (Figure 1A). Protein expression was analysed during synchronous growth, and demonstrated that PCNA1, PCNA2 and PCNA3 are all present throughout S-phase (Figure 1B). We therefore conclude that S. solfataricus heterotrimeric PCNA does indeed facilitate DNA replication in this organism. With reference to the loading control TATA-box binding protein (TBP), we note that there does not appear to be any significant cell-cycle modulation of PCNA protein levels across the time course tested.
Figure 1.
S. solfataricus heterotrimeric PCNA is expressed throughout S-phase. (A) Cell-cycle synchronisation of S. solfataricus cells. An asynchronous culture was applied to the ‘Baby machine’ apparatus, G1-phase cells were collected at time 0 and subsequently grown synchronously at 75°C. The DNA content of cells was analysed at the indicated time points by FACS; ‘1C’ and ‘2C’ indicate fluorescent signal corresponding to 1 or 2 copies of genomic DNA, respectively. (B) Analysis of PCNA expression throughout the cell cycle by western blotting. Samples were analysed at the indicated time points during synchronous growth. TBP was analysed as a loading control.
The replicative polymerase, PolB1, performs strand displacement synthesis on a model lagging strand substrate
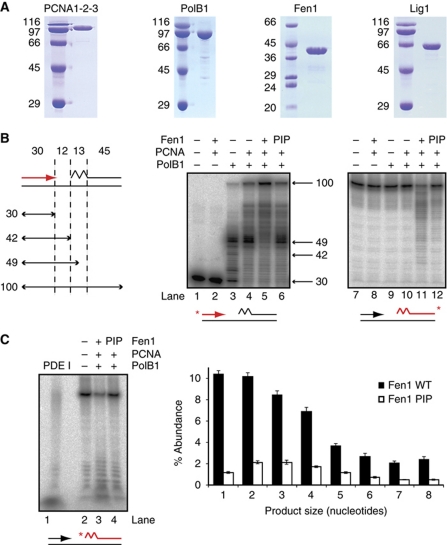
In order to analyse the process of Okazaki fragment maturation in vitro, we utilised a synthetic lagging strand substrate consisting of a 100-nt template strand annealed to two complementary strands mimicking Okazaki fragments: an upstream strand with a free 3′ end available for DNA synthesis and a downstream strand primed by 13 nt of RNA, consistent with the known structure of archaeal Okazaki fragments (Matsunaga et al, 2003). We then analysed the activity on this substrate of six recombinantly purified candidate proteins from S. solfataricus; PolB1, Fen1, Lig1 and PCNA1, PCNA2 and PCNA3 (Figure 2A). We confirmed the enzymes PolB1, Fen1 and Lig1 were catalytically active on their respective optimal substrates (Supplementary Figure S1). For simplicity, we utilised a fully functional version of PCNA in which the three subunits are fused in a single polypeptide (Dionne et al, 2008).
Figure 2.
PCNA-dependent coupling of PolB1 and Fen1 activities in vitro. (A) Analysis of purified recombinant proteins by SDS–PAGE visualised by Coomassie staining. Molecular weight markers are labelled on the left, sizes are in kDa. (B) Activity of purified PolB1, Fen1 and PCNA on an in vitro lagging strand substrate. A schematic of the substrate is illustrated on the left, with section sizes indicated in nucleotides (for sequences of oligonucleotides see Supplementary Table S1). The jagged region of the substrate denotes a ribonucleotide primer. Reactions contained 0.1 pmol PolB1, 0.1 pmol Fen1 and 20 pmol PCNA. Reaction products were separated by denaturing PAGE. The sizes and migration of notable DNA synthesis products are indicated. ‘PIP’ denotes the substitution of a Fen1 mutated in the PIP motif that is defective in PCNA binding (see Supplementary Figure S2). Substrates are also indicated below gels, with the radiolabelled strand indicated in red. Note that the upstream strand was labelled on its 5′ end and the downstream strand on its 3′ end, indicated with asterisks. (C) Analysis of CPs generated by WT and a PIP mutant version (PIP) of Fen1. Reactions were performed as in (B). Substrate is indicated below gel, labelling of the downstream strand is indicated in red, asterisk indicates 5′ end labelling. PDE I indicates substrate digestion to completion by snake phosphodiesterase I to indicate the migration of monoribonucleotides. Quantification of product abundance as a percentage of total DNA after subtraction of background degradation (lane 2) is illustrated on the right. Values are mean±s.e.m. (n=3).
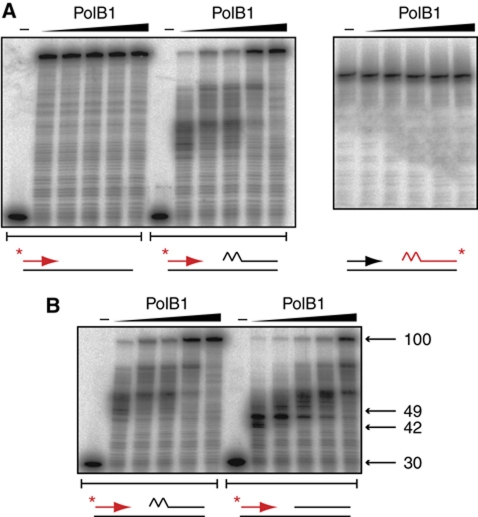
Bioinformatic and biochemical analysis suggests that PolB1 is the only replicative polymerase in S. solfataricus (Rogozin et al, 2008; Choi et al, 2011), and presumably operates as the primary DNA polymerase on both the leading and lagging strands. We therefore first measured the activity of PolB1 on our lagging strand substrate in the presence of dNTPs, separating reaction products on the basis of size by denaturing gel electrophoresis (Figure 2B). As expected, PolB1 catalyses DNA synthesis on the substrate, extending the upstream Okazaki fragment across the region of unreplicated template (lane 3). In the presence of PCNA, PolB1 activity is stimulated by 50% (as measured by full-length DNA synthesis), further extending the primer into the region occupied by the downstream Okazaki fragment (lane 4). The fact that this is not accompanied by any degradation of the downstream fragment (lane 10) suggests that the mechanism of strand displacement synthesis is operating, with the downstream strand excluded into a flap structure. This strand displacement synthesis activity, while stimulated by association with PCNA, is intrinsic to PolB1, and interestingly appears to be more active on an RNA-primed Okazaki fragment than a fragment composed entirely of DNA (Figure 3). PolB1 therefore possesses the properties required to displace unwanted RNA primers on the lagging strand in a manner significantly stimulated by PCNA. To our knowledge this is the first time such activity has been reported for S. solfataricus PolB1.
Figure 3.
Characterisation of PolB1 strand displacement activity. (A) Comparison of PolB1 activity on substrates possessing or lacking a downstream Okazaki fragment. Reactions contained 0, 15, 30, 60, 120, 240 fmol PolB1, assays were performed as described in Materials and methods with the exception that reactions contained 50 mM KCl and MnCl2 was omitted. Reaction products were separated by denaturing PAGE. Substrates are indicated below gels, the radiolabelled strand is coloured red and the position of the label is indicated with an asterisk. The jagged region of the substrate denotes a 13-ribonucleotide primer. (B) Comparison of strand displacement by PolB1 on substrates containing Okazaki fragments possessing or lacking RNA primers. Experiments were performed as in (A). The sizes and migration of notable DNA synthesis products are indicated on the right.
PolB1 coordinates with Fen1 in a PCNA-dependent manner to excise RNA primers from Okazaki fragments
Strand displacement by PolB1 is predicted to give rise to a downstream flap structure and we therefore investigated the activity of the flap endonuclease, Fen1, on the lagging strand substrate (Figure 2B). While the addition of Fen1 and PCNA alone has no effect (lanes 2 and 8), the addition of Fen1 to PolB1 and PCNA results in cleavage of the downstream Okazaki fragment (lane 11), suggesting that flap structures generated by PolB1 during strand displacement synthesis are indeed a substrate for subsequent Fen1 activity. Interestingly, downstream cleavage by Fen1 is accompanied by a further 15% increase in DNA synthesis of the upstream fragment (lane 5). This suggests that following flap cleavage, PolB1 subsequently re-engages the upstream primer to initiate further RNA primer displacement and excision, the activities of PolB1 and Fen1 becoming coupled to perform nick translation. Such activity is ideally suited to the requirement of replacing initiating RNA with error-free DNA.
To further investigate the importance of PCNA to these activities, we generated mutants to disrupt the interaction between PCNA and its binding partners, Fen1 and Lig1. Guided by previous studies (Gomes and Burgers, 2000; Dionne et al, 2003b; Pascal et al, 2006), we generated point mutants in the aromatic residues of Fen1 and Lig1 PIP motifs, which disrupted interaction with PCNA, while having no effect on the intrinsic catalytic activity of the proteins (Supplementary Figure S2). Substitution of the Fen1 PIP mutant into the primer excision assay (Figure 2B, lanes 6 and 12) resulted in a four-fold reduction in downstream flap cleavage and eliminated enhanced polymerase activity, demonstrating that the interaction of Fen1 with PCNA is crucial to coupling its activity to that of PolB1, which in turn is significantly stimulated by PCNA.
The coupled activities of PolB1 and Fen1 resulted in a range of cleaved products, with up to 30 nt of the downstream Okazaki fragment removed. To distinguish if these were the result of multiple rounds of flap creation and cleavage or whether they represent single flap cleavage events with a broad size distribution, we measured initial flap cleavage products (CPs) by labelling the downstream Okazaki fragment at the 5′ end and performing maturation assays as before (Figure 2C). Most CPs are relatively short (<4 nt), in agreement with measurements on budding yeast proteins (Garg et al, 2004; Rossi and Bambara, 2006), suggesting that the RNA primer excision observed is the result of multiple cycles of coordinated PolB1 and Fen1 activity. Interestingly, when this analysis is performed in the presence of the Fen1 PIP mutant, together with the expected decrease in the magnitude of flap cleavage, there is also an increase in the peak flap cleavage length from 1–2 nt to 2–3 nt. This supports a mechanistic basis for PCNA stimulation of Fen1 activity whereby colocalisation of PolB1 and Fen1 facilitates more rapid flap cleavage as it emerges from the strand displacing polymerase.
It has previously been reported that the lagging strand DNA polymerase in budding yeast, DNA polymerase δ, can maintain its position at the nick between two Okazaki fragment by undergoing a process termed ‘idling’; multiple cycles of nucleotide addition and accompanying strand displacement followed by exonuclease-mediated backtracking and nucleotide removal. This process has been proposed to prevent excessive strand displacement synthesis and maintain a suitable nick structure for subsequent Okazaki fragment ligation. Interestingly, using the same methodology as Garg et al (2004), we observe a specific increase in nucleotide turnover by PolB1 in the presence of a downstream Okazaki fragment, suggesting that PolB1 is also able to undergo idling at a nick, albeit at a highly reduced rate (almost three orders of magnitude lower) relative to that observed with DNA polymerase δ (Supplementary Figure S3). Consistent with the above analysis, this idling is reduced by 30% in the presence of Fen1, demonstrating the tight coupling between PolB1 and Fen1 as strand displaced RNA is cleaved by Fen1, allowing PolB1 to continue further DNA synthesis rather than backtracking.
Lig1 coordinates with PolB1 and Fen1 in a PCNA-dependent manner to ligate processed Okazaki fragments
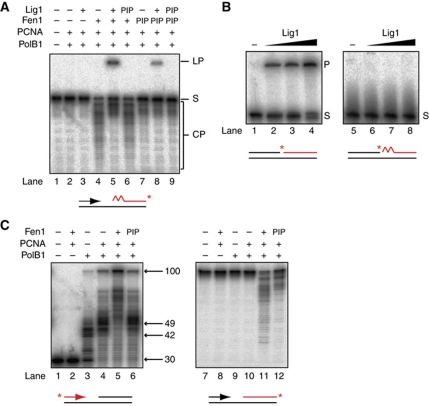
Multiple rounds of strand displacement and RNA cleavage by PolB1 and Fen1 are predicted to generate a DNA nick. We therefore investigated whether Lig1 is active on the lagging strand substrate (Figure 4A). Incubation of Lig1 with the full complement of proteins required for RNA primer excision (PolB1, PCNA and Fen1, lane 4) results in the production of a slower migrating larger DNA molecule which, under the denaturing conditions of the electrophoresis, can only arise through covalent joining of the labelled downstream Okazaki fragment to the upstream Okazaki fragment (lane 5). Notably, omission of Fen1 eliminates not only RNA primer excision but also ligation in the presence of Lig1 (lane 3). Consistent with previous measurements performed on other DNA ligases (Pascal et al, 2004), we confirmed that S. solfataricus Lig1, while demonstrating robust DNA ligase activity, is unable to ligate an RNA-containing nick intermediate, which would arise as a result of PolB1 gap filling on our lagging strand substrate (Figure 4B). This discrimination is believed to act as a control mechanism to prevent incorporation of RNA into the nascent lagging strand. This conclusion is further supported by the fact that in our reconstituted system, only downstream fragments cleaved beyond the 13 nt of initiating RNA are converted to the ligated form (Figure 4A, compare lanes 4 and 5). Thus, we observe fully coupled Okazaki fragment processing in vitro with ligation absolutely dependent on prior RNA removal, which in turn is absolutely dependent on prior strand displacement synthesis. This analysis therefore demonstrates that the catalytic activity of three proteins, PolB1, Fen1 and Lig1 are necessary and sufficient for Okazaki fragment processing in this system reconstituted from S. solfataricus.
Figure 4.
Completion of Okazaki fragment maturation in vitro by PolB1, Fen1 and Lig1. (A) Activity of PolB1, Fen1, Lig1 and PCNA on an in vitro lagging strand substrate. Reactions contained 0.1 pmol PolB1, 0.1 pmol Fen1, 1 pmol Lig1 and 20 pmol PCNA. Reaction products were separated by denaturing PAGE. The migration of substrate (S), CPs and the ligation product (LP) are indicated. Substrate is indicated below gel, with radiolabelled strand indicated in red and label position with a red asterisk. The jagged region of the substrate denotes a 13-ribonucleotide primer. ‘PIP’ denotes the substitution of a Fen1 and/or Lig1, which are mutated in the PIP motif and are defective in PCNA binding (see also Supplementary Figure S2). (B) Substrate specificity of Lig1. Reactions contained 0, 0.625, 1.25, 2.5 pmol Lig1. Reaction products were separated by denaturing PAGE. Substrates (S) are indicated below gel, 5′ end labelling of the downstream strand is indicated in red and label position with a red asterisk. The ligated product is labelled (P). (C) Activity of purified PolB1, Fen1 and PCNA on an in vitro lagging strand substrate lacking a downstream RNA primer. Reactions contained 0.1 pmol PolB1, 0.1 pmol Fen1 and 20 pmol PCNA. Reaction products were separated by denaturing PAGE. The sizes and migration of notable DNA synthesis products are indicated on the right. ‘PIP’ denotes the substitution of a Fen1 mutant defective in PCNA binding. Substrates are indicated below gels, with radiolabelled strand indicated in red and label position with a red asterisk.
Substitution of the Fen1 PIP mutant into this reaction results in reduced RNA primer excision, and thus a 5.5-fold reduction in final ligation (Figure 4, lanes 7 and 8). Furthermore, substitution of a Lig1 PIP mutant either alone (lane 6) or in combination with Fen1 PIP (lane 9) completely abolishes final ligation of Okazaki fragments. Thus, an interaction of all three essential enzymes—PolB1, Fen1 and Lig1—with PCNA is crucial to the efficiency of Okazaki fragment maturation. We confirmed that a non-fused PCNA complex assembled from PCNA1, PCNA2 and PCNA3 also supports coordinated PolB1, Fen1 and Lig1 maturation activity and thus our observations are not affected by using a fused heterotrimeric PCNA (Supplementary Figure S4).
While several different pathways have been proposed to mediate Okazaki fragment maturation in eukaryotic systems, the central role of these three enzymes in S. solfataricus appears to correlate with their orthologues constituting the core pathway for maturation in eukaryotes (Ayyagari et al, 2003). Additional factors shown to regulate this process in eukaryotes, such as the Dna2 helicase/endonuclease and the Pif1 helicase which are important in resolving large displaced flaps (Rossi et al, 2008), appear to be absent from S. solfataricus and indeed largely from archaea; a Dna2 orthologue for example has only been identified in a single sequenced archaeal genome to date (Higashibata et al, 2003). Such additional factors may therefore have evolved after the divergence of archaea and eukaryotes to function redundantly with the core DNA polymerase–Fen1–Lig1 pathway.
Type 2 RNaseH enzymes have also been implicated in RNA removal during Okazaki fragment maturation and furthermore have been shown to interact with PCNA similarly to the core factors in mammalian cells and archaea (Qiu et al, 1999; Meslet-Cladiére et al, 2007; Chon et al, 2009; Bubeck et al, 2011). S. solfataricus possesses a predicted RNaseH2 gene but we were unable to produce soluble recombinant protein to analyse in our reconstituted system. However, we note that PolB1, PCNA and Fen1 can remove the 5′ end of RNA-primed Okazaki fragments equivalently to Okazaki fragments consisting only of DNA (Figure 4C, compare with Figure 2B), arguing against an essential requirement for RNaseH2, or indeed any other additional factors, in facilitating RNA removal from Okazaki fragments. Furthermore, genetic studies in the euryarchaeal species Haloferax volcanii have proposed the dominant role of RNaseH2 to be in resolution of aberrant structures arising from DNA damage rather than during Okazaki fragment maturation (Meslet-Cladiére et al, 2007) and consistent with this, biochemical studies of Archaeoglobus fulgidus RNaseH2 demonstrate robust activity in removing single ribonucleotides embedded in DNA duplexes (Bubeck et al, 2011).
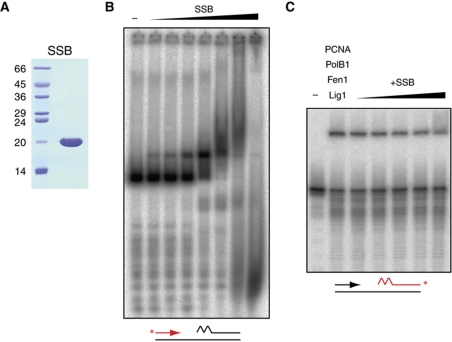
SSB binds to a lagging strand template but does not significantly affect Okazaki fragment maturation
In vivo, single-stranded DNA is bound by single-stranded binding proteins, which provide protection from DNA damage and resolve potential secondary structures. In S. solfataricus, this function is performed by the single-strand binding protein, SSB (Wadsworth and White, 2001; Haseltine and Kowalczykowski, 2002). We therefore wanted to determine if SSB has an influence on lagging strand replication, where Okazaki fragments are separated by single-stranded DNA. We purified recombinant SSB (Figure 5A), and first determined if it was able to bind to the lagging strand substrate under the same reaction conditions used to measure Okazaki fragment maturation (Figure 5B). By electrophoretic mobility shift assay, SSB was demonstrated to bind to the substrate across a range of concentrations; the binding pattern observed is broadly consistent with previous estimates of a dissociation constant in the low nanomolar range (Wadsworth and White, 2001). At lower concentrations, a discrete band is observed, presumably representing coating of the entire 12 nt region of single-stranded DNA by 2–3 molecules of SSB binding non-cooperatively (Wadsworth and White, 2001; Haseltine and Kowalczykowski, 2002). However, at higher concentrations, more heterogeneous protein–DNA complexes are observed, suggesting that SSB extends its single-stranded binding footprint and melts duplex DNA, as has been observed previously (Cubeddu and White, 2005; Marsh et al, 2006). Indeed at the highest concentrations of SSB, a decrease in the size of protein–DNA complexes relative to the starting substrate is observed, indicating complete SSB-mediated dissociation of the labelled Okazaki fragment from the template.
Figure 5.
Effect of SSB on Okazaki fragment maturation in vitro. (A) Analysis of purified SSB by SDS–PAGE visualised by Coomassie staining. Molecular weight markers are labelled on the left, sizes are in kDa (B) SSB binding to a lagging strand substrate. Substrate, indicated below the gel, was incubated with 0, 0.14, 5.6, 22.3, 89.1, 356, 1425, 5700 fmol of SSB and protein–DNA complexes separated by native gel electrophoresis. Radiolabelling of the upstream strand is indicated in red and label position with a red asterisk. (C) Effect of SSB on Okazaki fragment maturation. Reactions contained 0.1 pmol PolB1, 0.1 pmol Fen1, 1 pmol Lig1, 20 pmol PCNA, and 0, 0.14, 5.6, 22.3, 89.1, 356 fmol of SSB. Reaction products were separated by denaturing PAGE. Substrate is indicated below gel, 3′ end labelling (asterisk) of the downstream strand is indicated in red.
We next tested whether SSB has an effect on full maturation by PolB1, Fen1, Lig1 and PCNA across the range of concentrations at which we see substrate binding but not full melting (Figure 5C). Addition of SSB has a very modest inhibitory effect on processing (two-fold reduction in ligation), suggesting minimal effect on the enzymes responsible for maturation. Because our substrate is designed to study the final stages of Okazaki fragment maturation only, the amount of single-stranded DNA present is relatively small compared with that expected in vivo, where synthesis of Okazaki fragments will lag behind template production by the replicative helicase, MCM. We therefore speculate that SSB may have a greater role in lagging strand replication involving larger regions of single-stranded DNA where resolution of secondary structure may be important in facilitating progress of the replication machinery. Indeed, consistent with this, a recent study has shown a stimulatory effect of SSB on PolB1 when replicating longer stretches of single-stranded template (Choi et al, 2011). Given the negligible effect of SSB under our assay conditions, it was omitted from all subsequent experiments.
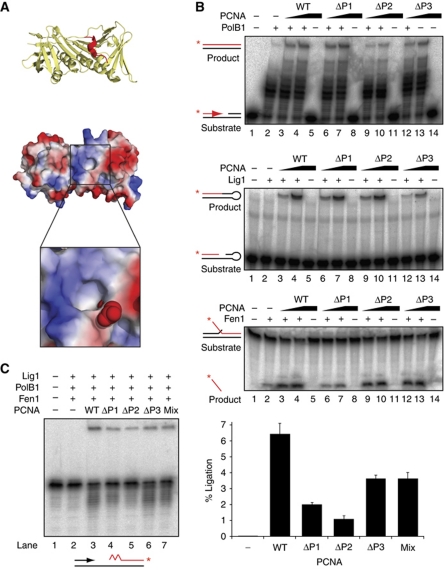
Okazaki fragment maturation is most efficient when PolB1, Fen1 and Lig1 are coordinated around a single PCNA molecule
We wanted to determine whether the importance of PCNA binding for PolB1, Fen1 and Lig1 in Okazaki fragment maturation is due to coordination of all three enzymes around a single heterotrimeric PCNA ring. To address this, we tested the effect of separating the PolB1, Fen1 and Lig1 binding sites across different PCNA rings. We utilised our fused PCNA heterotrimer to generate mutant rings in which one of the three enzyme binding sites is disrupted at a time. By using the fused heterotrimer, we eliminate the possibility of subunit exchange to generate a fully wild-type (WT) heterotrimer when different mutants are mixed. A conserved alanine residue lining the hydrophobic binding site of each PCNA was replaced with a glutamic acid to sterically and electrostatically exclude PIP motif binding (Figure 6A). These mutants were validated by comparing their ability to stimulate PolB1, Fen1 or Lig1 individually on their respective optimal model substrates (Figure 6B). Each PCNA mutant is defective in stimulating the intended enzyme activity (ΔP1=Fen1, ΔP2=PolB1, ΔP3=Lig1) but retains its ability to stimulate the remaining two enzymes, eliminating the possibility that the mutants have disrupted neighbouring binding sites or the overall ring architecture of the protein. This requirement restricted us to using relatively subtle mutations and therefore the impact of the mutated binding site is relatively small. For example, mutation of the PIP motif in Lig1 completely abrogates PCNA-stimulated activity (Figure 4A), in contrast to the 4.5-fold reduction in Lig1 stimulation exerted by the ΔP3 mutant in PCNA. The effects of these PCNA mutations are nonetheless significant and reproducible.
Figure 6.
Separating binding of PolB1, Fen1 and Lig1 across multiple PCNA rings impairs coupled activity. (A) Structural basis of PCNA mutagenesis. Upper panel shows cartoon representation of PCNA1 (yellow) in complex with the PIP peptide motif of Fen1 (red). Lower panel shows electrostatic surface view of the same PCNA1 molecule highlighting the boxed hydrophobic binding pocket. The Fen1 PIP motif is omitted for clarity. The magnified section models a mutation of Alanine 246 to glutamate, rendered in space-filling mode. Images were generated in PyMOL (http://www.pymol.org) from PDB file 2IZO. (B) Effect of PCNA mutants on stimulating the activities of PolB1, Fen1 and Lig1 on optimal model substrates in vitro. Substrates and products for each reaction are indicated on the left, with radiolabelled strands indicated in red and label position by a red asterisk. ‘WT’ PCNA is a covalent fusion of PCNA1, PCNA2 and PCNA3. ‘ΔP1’ possess an A246E mutation in the PCNA1 subunit of the fusion. ‘ΔP2’ possesses an A242E mutation in the PCNA2 subunit of the fusion. ‘ΔP3’ possesses an A241E mutation in the PCNA3 subunit of the fusion. In all, 30 fmol PolB1/65 fmol Fen1/10 pmol Lig1 were incubated with 0, 0.77 and 7.7 pmol of each PCNA protein and reaction products were separated by denaturing PAGE (C) Effect of PCNA mutants on Okazaki fragment maturation. Reactions contained 0.1 pmol PolB1, 0.1 pmol Fen1, 1 pmol Lig1 and 20 pmol PCNA. Reaction products were separated by denaturing PAGE. The substrate is indicated below the gel, with radiolabelled strand indicated in red and label position with a red asterisk. Quantification of the upper ligated band—a measure of completed Okazaki fragment maturation—is shown on the right. Values are mean±s.e.m. (n=3).
The mutant PCNAs were then substituted into the Okazaki fragment maturation assay and the amount of fully processed Okazaki fragments quantified (Figure 6C). Substitution of each mutant into the system individually results in a significant reduction in activity compared with WT PCNA, consistent with the preceding analysis in which an interaction of PolB1, Fen1 and Lig1 with their respective PCNA-binding sites is essential to the efficiency of maturation. Indeed, in the absence of PCNA, processing activity is undetectable. Crucially, when a mix of all three mutants is substituted into the system, such that the molarity of PCNA1, PCNA2 and PCNA3 binding sites is equal to WT but all three sites are never on the same PCNA molecule, processing activity is not restored to WT levels, rather the maximal activity we observe is the same as the activity of the least impaired mutant PCNA ring (ΔP3). This indicates a requirement for the three distinct enzymes PolB1, Fen1 and Lig1 to be coordinated around a single PCNA molecule for maximum efficiency of processing.
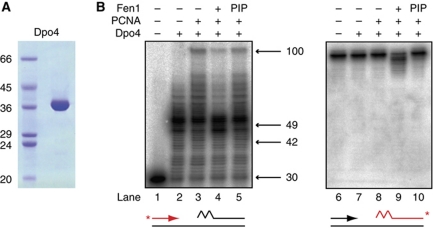
Efficient coordination of DNA polymerase and Fen1 activities requires simultaneous access to PCNA
We subsequently wanted to determine whether coordination around a single PCNA is a dynamic process, involving sequential access of PolB1, Fen1 and Lig1 to their respective binding sites, or whether this coordination involves simultaneous binding of the three enzymes to PCNA. To address this question, we analysed the effect on RNA primer excision of forcing the required enzymatic activities to alternate in their access to PCNA. To achieve this, we used as a tool the S. solfataricus translesion DNA polymerase Dpo4, which interacts with the same PCNA1 binding site as Fen1 (Dionne et al, 2008; Xing et al, 2009). Dpo4 was purified (Figure 7A) and substituted into the maturation assay (Figure 7B). Dpo4 possesses similar catalytic activity to PolB1 on our lagging strand substrate, performing PCNA-stimulated strand displacement synthesis (lanes 2 and 3). Consistent with its known lack of 3′–5′ exonuclease activity, Dpo4 is unable to undergo any significant idling at a nick (Supplementary Figure S3) and therefore potentially undergoes strand displacement by utilising a different mechanism. However, because idling activity even by PolB1 is very low, together with the very similar pattern of strand displacement observed for both enzymes (compare Figure 2B, lane 4 with Figure 7B, lane 3), we suggest that both enzymes generate the same displaced RNA flap structure under these experimental conditions. However, addition of Fen1 to Dpo4 fails to result in the coordinated enzyme activity observed with PolB1; downstream fragment cleavage is minimal (lane 9, compare with Figure 2, lane 11) while strand displacement by Dpo4 is actually decreased rather than increased in the presence of Fen1 (lane 4, compare with Figure 2, lane 5). Dpo4 does still exhibit some PCNA-stimulated strand displacement in the presence of Fen1, demonstrating that Dpo4 access to PCNA is decreased rather than totally blocked. Similarly, elimination of Dpo4 disruption by substituting the Fen1 PIP mutant (lanes 5 and 10) confirms that WT Fen1 also is still gaining access to PCNA. Therefore, despite Dpo4 and Fen1 both being able to access their shared binding site on PCNA and possessing the appropriate catalytic activities, alternating access to PCNA is not sufficient for effective coupling of their activities. We note that this antagonism between Dpo4 and Fen1 may provide a mechanism to minimise the length of DNA synthesised by this error-prone polymerase in vivo.
Figure 7.
Preventing DNA polymerase and Fen1 from simultaneously binding the same PCNA ring impairs coupled activity. (A) Analysis of purified Dpo4 by SDS–PAGE visualised by Coomassie staining. Molecular weight markers are labelled on the left, sizes are in kDa (B) Activity of purified Dpo4, Fen1 and PCNA on an in vitro lagging strand substrate. Reactions contained 50 fmol Dpo4, 0.1 pmol Fen1 and 20 pmol PCNA. Reaction products were separated by denaturing PAGE. The sizes and migration of notable DNA synthesis products are indicated. Substrates are indicated below gels, with radiolabelled strand indicated in red and label position with a red asterisk. The jagged region of the substrate denotes a 13-ribonucleotide primer. ‘PIP’ denotes the substitution of a Fen1 mutated in the PIP motif that is defective in PCNA binding (see also Supplementary Figure S2).
Discussion
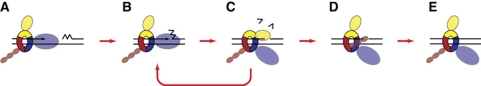
In this report, we describe the in vitro reconstitution of coupled processing of discontinuous RNA-primed Okazaki fragments into a single continuous daughter strand, with proteins derived from the crenarchaeon S. solfataricus. In conjunction with our previous demonstration that PCNA can act as a bridge between Fen1 and PolB1 and/or Lig1 (Dionne et al, 2003b), our current data support a model whereby the essential enzymes PolB1, Fen1 and Lig1 are coordinated around a single PCNA ring, with simultaneous binding to this ring supporting the most efficient coupling of their interdependent activities. Such coordination tightly couples PolB1-mediated flap creation to Fen1-mediated flap cleavage. Furthermore, colocalisation of Lig1 allows it to interrogate CPs generated by Fen1, enabling it to ligate Okazaki fragments as soon as initiating RNA is fully removed; thus providing a mechanism by which covalent integrity of the genome is efficiently maintained (Figure 8).
Figure 8.
Schematic illustration of Okazaki fragment maturation in S. solfataricus. DNA is illustrated by black lines, RNA primer is illustrated by the jagged region. (A) PolB1 (blue), Fen1 (yellow) and Lig (red) bind to their specific PCNA1, PCNA2 or PCNA3 binding sites on heterotrimeric PCNA (blue, yellow and red, respectively). PolB1 engages the template to perform DNA synthesis while Fen1 and Lig1 are carried by PCNA. (B) Upon encountering a downstream Okazaki fragment, PolB1 initiates strand displacement synthesis, displacing downstream RNA into a flap structure. (C) Fen1 engages the generated flap and cleaves it, producing a nick. Multiple rounds of PCNA-coordinated PolB1 and Fen1 coupled activity enables complete removal of RNA primers. (D) Lig1 interrogates nick structures emerging from Fen1, and, following RNA removal, encircles the DNA nick and catalyses Okazaki fragment ligation. (E) Efficient coupling of PolB1, Fen1 and Lig1 activities mediated by PCNA ensures covalent integrity for nascent lagging strands. Adapted from Beattie and Bell (2011b).
An increasing body of structural evidence has revealed how multiple proteins can be coordinated around a single PCNA. Crystallographic studies have directly visualised three molecules of human Fen1 bound to PCNA (Sakurai et al, 2005), and three molecules of A. fulgidus RNase H2 bound to PCNA (Bubeck et al, 2011). A key finding of these structural studies, together with others investigating single protein-sliding clamp interactions (Bunting et al, 2003; Nishida et al, 2009; Xing et al, 2009; Mayanagi et al, 2011), is the ability of many proteins to adopt different conformations when bound to PCNA, often mediated by a flexible linker located between the anchoring PIP motif and the core of the protein. Such conformational flexibility between apparent ‘carrier’ and ‘active’ states may provide a mechanism for ensuring accommodation of multiple different enzymes around a single PCNA, while preventing competitive access to DNA substrates. One of the most striking examples of such conformational switching is that of DNA ligase I, which in its active DNA-bound form has been shown to encircle DNA in a conformation which would presumably prevent access of other PCNA-bound factors to DNA (Pascal et al, 2004; Mayanagi et al, 2009). However, in the absence of DNA, S. solfataricus Lig1 adopts an elongated form radiating from PCNA (Pascal et al, 2006), thus providing an elegant explanation for how this particular protein may be ‘carried’ by PCNA without interfering with the action of PolB1 and Fen1 until an appropriate nick substrate arises.
The heterotrimeric PCNA of crenarchaea has provided us with a unique opportunity to investigate the architectural basis of multi-enzyme coordination during lagging strand DNA replication. The orthologous factors to PolB1, Fen1 and Lig1 all interact with eukaryotic homotrimeric PCNA, and it is possible they may adopt the same higher order arrangement. Indeed, biochemical analysis supports the simultaneous binding of two different proteins to the homotrimeric human PCNA (Doré et al, 2009), and furthermore, biophysical analysis has demonstrated simultaneous binding of distinct proteins to the homodimeric Escherichia coli β clamp (Indiani et al, 2005) and the homotrimeric euryarchaeal PCNA (Kiyonari et al, 2009). It is unclear, however, what might drive the formation of productive complexes of enzymes when the binding sites provided by the sliding clamp are identical. It is possible that crenarchaea, in which a relatively small number of PCNA-interacting proteins have been identified to date, may have evolved distinct PCNA subunits as the simplest mechanism to drive the formation of complexes composed of different enzymes. Eukaryotic PCNA, however, with a vast number of interacting proteins, may have evolved more regulated mechanisms to determine which proteins may bind PCNA in which combination at different times; in this respect, it is interesting that a number of post-translational modifications have been identified on both PCNA and PCNA-binding proteins that can modulate the interaction between the two (Henneke et al, 2003; Moldovan et al, 2007; Guo et al, 2010). It is worth noting that, as discussed, S. solfataricus PCNA has been shown to interact with additional factors outside of those involved in lagging strand DNA replication investigated here (see Introduction), and thus as with homo-oligomeric sliding clamps, there are presumably mechanisms that ensure the appropriate enzyme binds to each site at the correct time. Key questions for the future will therefore be whether DNA-sliding clamps can assemble other multi-enzyme complexes outside of lagging strand replication, such as during DNA repair, and how appropriate sets of enzymes are bound at the correct time.
Materials and methods
Constructs
SsPCNA1-2-3, SsPCNA1, SsPCNA2, SsPCNA3, SsPolB1, SsFen1, SsLig1, SsDpo4 and SsSSB expression constructs were generated previously (Boudsocq et al, 2001; Wadsworth and White, 2001; Dionne et al, 2003b, 2008). All mutations were introduced into the above constructs by single oligonucleotides using a modified QuikChange protocol (Stratagene).
Cell-cycle synchronisation
S. solfataricus P2 cells were synchronised as described previously (Duggin et al, 2008), except cells were grown in media lacking tryptone and G1 cells were collected after 5 h. At each indicated time point, 400 μl of cells were withdrawn for FACS analysis, performed as described previously (Duggin et al, 2011), and 1.5 ml of cells were analysed by western blotting with PCNA1, PCNA2, PCNA3 or TBP antisera (Dionne et al, 2003b; Duggin et al, 2008).
Protein purification
All proteins were overexpressed in E. coli Rosetta (DE3) pLysS (Novagen). Cells were grown to A600=0.4–0.6 before induction with 1 mM IPTG for 3 h at 37°C. Cells were harvested by centrifugation at 4000 r.p.m. for 15 min.
Lig1 and Lig1 PIP mutant were purified identically. Cells were ground in liquid nitrogen, resuspended in 20 mM Tris pH 8, 300 mM NaCl and complete EDTA-free protease inhibitors (Roche) and incubated for 25 min at 75°C before centrifugation at 17 000 r.p.m. for 30 min. The supernatant was loaded onto 2 ml Ni-NTA agarose resin (Qiagen) and eluted with 500 mM imidazole. Eluted fractions were loaded onto a HiLoad Superdex 200 gel-filtration column (GE Healthcare) pre-equilibrated in 10 mM Tris pH 8, 150 mM NaCl, 14 mM β-mercaptoethanol. Protein containing fractions were diluted two-fold in 10 mM Tris pH 8, 14 mM β-mercaptoethanol, loaded onto a MonoQ column (GE Healthcare) pre-equilibrated in 10 mM Tris pH 8, 75 mM NaCl, 14 mM β-mercaptoethanol, and eluted over a 15-ml linear gradient of 75–1000 mM NaCl. Protein containing fractions were pooled and stored at −80°C.
All PCNA1-2-3 variants were purified identically. Cells were resuspended in 20 mM Tris pH 8, 300 mM NaCl and protease inhibitors. Extract was lysed by French press at 20 000 psi before centrifugation at 17 000 r.p.m. for 30 min. Supernatant was heated to 75°C for 25 min before further centrifugation. The supernatant was purified by passage through Ni-NTA agarose, and eluted fractions were loaded onto a HiLoad Superdex 200 gel-filtration column pre-equilibrated in 20 mM Tris pH 8, 300 mM NaCl. Protein containing fractions were pooled and stored at −80°C.
Non-covalent heterotrimeric PCNA was prepared from individual PCNA subunits as described previously (Pascal et al, 2006) taking advantage of the resistance of the assembled heterotrimer but not individual PCNA1 and PCNA3 components to heat treatment at 85°C for 25 min.
Fen1 and Fen1 PIP mutant were purified broadly as described previously (Hutton et al, 2008). Extracts were lysed by French Press, clarified by centrifugation before incubation for 15 min at 75°C and further centrifugation. The supernatant was purified by passage through a HiTrap Heparin column (GE Healthcare) and a HiLoad Superdex 75 gel-filtration column (GE Healthcare). Protein containing fractions were pooled and stored at −80°C.
Cells expressing PolB1 were resuspended in buffer A (10 mM HEPES pH 7.5, 100 mM NaCl, 1 mM DTT) with protease inhibitors, lysed by French press and centrifuged. Supernatant was incubated for 20 min at 65°C before further centrifugation. The supernatant was loaded onto a HiTrap Heparin column pre-equilibrated in buffer A, and eluted over a 75-ml linear gradient of 100–1000 mM NaCl. Protein containing fractions were loaded onto a HiLoad Superdex 200 column pre-equilibrated in 10 mM HEPES pH 7.5, 500 mM NaCl, 1 mM DTT. Protein containing fractions were diluted five-fold in 10 mM HEPES pH 7.5, 1 mM DTT, loaded onto a MonoS column (GE Healthcare) pre-equilibrated in buffer A and eluted over a 15-ml linear gradient of 100–1000 mM NaCl. Protein containing fractions were pooled and stored at −80°C.
Cells expressing Dpo4 were resuspended in buffer A (10 mM Tris pH 8, 150 mM NaCl, 1 mM DTT) with protease inhibitors, lysed by French press and centrifuged. Supernatant was incubated at 75°C for 25 min before further centrifugation. Supernatant was loaded onto a HiTrap Heparin column pre-equilibrated in buffer A and eluted over a 75-ml linear gradient of 150–1000 mM NaCl. Protein containing fractions were loaded onto a HiLoad Superdex 200 column pre-equilibrated in 10 mM HEPES pH 7.5, 150 mM NaCl, 1 mM DTT. Protein containing fractions were pooled and stored at −80°C.
SSB was purified exactly as described previously (Wadsworth and White, 2001).
Substrate preparation
Oligonucleotides were synthesised by Eurogentec (Supplementary Table S1). Oligonucleotides were either 5′ end labelled with 32P-γ-ATP (Perkin-Elmer) using T4 polynucleotide kinase (NEB) or 3′ end labelled with 32P-α-cordycepin (Perkin-Elmer) using Terminal Transferase (NEB) as indicated. Labelled oligonucleotides were annealed to two-fold excess Template and four-fold excess secondary oligonucleotide (if present) by incubating at 95°C for 10 min before gradual cooling to room temperature. Lagging strand substrates were generated by annealing Template to Upstream and Downstream_RNA or Downstream_DNA. Fen1 substrate was generated by annealing Template to Upstream_1nt and Downstream_flap. Lig1 substrate was generated by annealing Nick 1 to two-fold excess Nick2. Unincorporated nucleotide was removed by passing through a MicroSpin G-25 column (GE Healthcare) before storage at –20°C. RNase inhibitor (NEB) was included when generating RNA-containing substrates.
Electrophoretic mobility shift assay
Reaction mixtures (20 μl) containing 0.1 pmol 32P-labelled substrate, and the indicated amount of SSB were incubated for 15 min at 50°C in 50 mM Tris pH 7.5, 100 mM KCl, 10 mM MgCl2, 100 μM dNTPs, 4 mM MnCl2, 1 mM ATP, 100 μg/ml BSA, 10 mM DTT, 5% glycerol. Protein–DNA complexes were resolved by electrophoresis through a 6% polyacrylamide, 0.5 × TBE gel run at 175 V. The gel was dried and visualised by autoradiography.
DNA polymerase assays
Reaction mixtures (20 μl) contained 50 mM Tris pH 7.5, 100 mM KCl, 10 mM MgCl2, 100 μM dNTPs, 4 mM MnCl2, 1 mM ATP, 100 μg/ml BSA, 10 mM DTT, 0.1 pmol 32P-labelled substrate, and the indicated amounts of PolB1 and PCNA. Reactions were incubated at 50°C for 15 min before being quenched by the addition of 20 μl loading dye (8 M urea, 1 × TBE, 0.05% Bromophenol Blue) and incubated for 5 min at 99°C. Reaction products were resolved by electrophoresis through a 12% polyacrylamide, 8 M urea, 1 × TBE gel. Gels were dried and visualised by autoradiography.
Flap cleavage assays
Reaction mixtures (20 μl) contained 70 mM Tris pH 7.6, 50 mM KCl, 10 mM MgCl2, 100 μg/ml BSA, 5 mM DTT, 0.1 pmol 32P-labelled substrate, and the indicated amounts of Fen1 and PCNA. Reactions were incubated at 50°C for 15 min before being quenched by the addition of 20 μl loading dye (8 M urea, 1 × TBE, 0.05% Bromophenol Blue) and incubated for 5 min at 99°C. Reaction products were resolved by electrophoresis through a 12% polyacrylamide, 8 M urea, 1 × TBE gel. Gels were dried and visualised by autoradiography.
DNA ligation assays
Reaction mixtures (20 μl) contained 50 mM Tris pH 7.5, 50 mM KCl, 10 mM MgCl2, 1 mM ATP, 100 μg/ml BSA, 10 mM DTT, 0.1 pmol 32P-labelled substrate and indicated amounts of Lig1 and PCNA. Reactions were incubated at 50°C for 15 min before being quenched by the addition of 20 μl loading dye (8 M urea, 1 × TBE, 0.05% Bromophenol Blue) and incubated for 5 min at 99°C. For PCNA-independent assays, reactions were performed in 50 mM Tris pH 7.5, 10 mM MgCl2, 1 mM ATP, 10 mM DTT, and reactions were incubated for 30 min at 70°C. Reaction products were resolved by electrophoresis through a 12% polyacrylamide, 8 M urea, 1 × TBE gel. Gels were dried and visualised by autoradiography.
Maturation assays
Reaction mixtures (20 μl) contained 50 mM Tris pH 7.5, 100 mM KCl, 10 mM MgCl2, 100 μM dNTPs, 4 mM MnCl2, 1 mM ATP, 100 μg/ml BSA, 10 mM DTT, 0.1 pmol 32P-labelled substrate, 0.1 pmol PolB1 (or 50 fmol Dpo4), 0.1 pmol Fen1, 1 pmol Lig1 and 20 pmol PCNA as appropriate. Reactions were incubated at 50°C for 15 min before being quenched by the addition of 20 μl loading dye (95% formamide, 5 mM EDTA, 0.05% Bromophenol Blue) and incubated for 5 min at 99°C. Reaction products were resolved by electrophoresis through a 12% polyacrylamide, 8 M urea, 1 × TBE gel. Gels were dried and visualised by autoradiography.
To analyse flap cleavage size, reaction products were separated on a 20% polyacrylamide, 8 M urea, 1 × TBE gel and visualised by autoradiography. For reference, 0.1 pmol 32P-labelled Downstream_RNA oligonucleotide was digested with 10 μg Phosphodiesterase I (Sigma) for 5 min at 37°C in reaction buffer before quenching.
Supplementary Material
Acknowledgments
We thank Malcolm White (St Andrews, UK) for the SSB expression plasmid. TRB was funded by a studentship from the Biotechnology and Biological Sciences Research Council, UK. Research in SDB's laboratory was also supported by the Wellcome Trust.
Author contributions: TRB and SDB designed the experiments and wrote the manuscript; TRB performed the experiments.
Footnotes
The authors declare that they have no conflict of interest.
References
- Ayyagari R, Gomes XV, Gordenin DA, Burgers PM (2003) Okazaki fragment maturation in yeast. I. Distribution of functions between FEN1 AND DNA2. J Biol Chem 278: 1618–1625 [DOI] [PubMed] [Google Scholar]
- Beattie TR, Bell SD (2011a) The role of the DNA sliding clamp in Okazaki fragment maturation in archaea and eukaryotes. Biochem Soc Trans 39: 70–76 [DOI] [PubMed] [Google Scholar]
- Beattie TR, Bell SD (2011b) Molecular machines in archaeal DNA replication. Curr Opin Chem Biol 15: 614–619 [DOI] [PubMed] [Google Scholar]
- Boudsocq F, Iwai S, Hanaoka F, Woodgate R (2001) Sulfolobus solfataricus P2 DNA polymerase IV (Dpo4): an archaeal DinB-like DNA polymerase with lesion-bypass properties akin to eukaryotic poleta. Nucleic Acids Res 29: 4607–4616 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bubeck D, Reijns MAM, Graham SC, Astell KR, Jones EY, Jackson AP (2011) PCNA directs type 2 RNase H activity on DNA replication and repair substrates. Nucleic Acids Res 39: 3652–3666 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bunting KA, Roe SM, Pearl LH (2003) Structural basis for recruitment of translesion DNA polymerase Pol IV/DinB to the beta-clamp. EMBO J 22: 5883–5892 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burgers PM (2009) Polymerase dynamics at the eukaryotic DNA replication fork. J Biol Chem 284: 4041–4045 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Choi JY, Eoff RL, Pence MG, Wang J, Martin MV, Kim EJ, Folkmann LM, Guengerich FP (2011) Roles of the four DNA polymerases of the crenarchaeon Sulfolobus solfataricus and accessory proteins in DNA replication. J Biol Chem 286: 31180–31193 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chon H, Vassilev A, DePamphilis ML, Zhao Y, Zhang J, Burgers PM, Crouch RJ, Cerritelli SM (2009) Contributions of the two accessory subunits, RNASEH2B and RNASEH2C, to the activity and properties of the human RNase H2 complex. Nucleic Acids Res 37: 96–110 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cubeddu L, White MF (2005) DNA damage detection by an archaeal single-stranded DNA-binding protein. J Mol Biol 353: 507–516 [DOI] [PubMed] [Google Scholar]
- Dionne I, Bell SD (2005) Characterization of an archaeal family 4 uracil DNA glycosylase and its interaction with PCNA and chromatin proteins. Biochem J 387: 859–863 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dionne I, Brown NJ, Woodgate R, Bell SD (2008) On the mechanism of loading the PCNA sliding clamp by RFC. Mol Microbiol 68: 216–222 [DOI] [PubMed] [Google Scholar]
- Dionne I, Nookala RK, Jackson SP, Doherty AJ, Bell SD (2003b) A heterotrimeric PCNA in the hyperthermophilic archaeon Sulfolobus solfataricus. Mol Cell 11: 275–282 [DOI] [PubMed] [Google Scholar]
- Dionne I, Robinson NP, McGeoch AT, Marsh VL, Reddish A, Bell SD (2003a) DNA replication in the hyperthermophilic archaeon Sulfolobus solfataricus. Biochem Soc Trans 31: 674–676 [DOI] [PubMed] [Google Scholar]
- Doré AS, Kilkenny ML, Jones SA, Oliver AW, Roe SM, Bell SD, Pearl LH (2006) Structure of an archaeal PCNA1-PCNA2-FEN1 complex: elucidating PCNA subunit and client enzyme specificity. Nucleic Acids Res 34: 4515–4526 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Doré AS, Kilkenny ML, Rzechorzek NJ, Pearl LH (2009) Crystal structure of the rad9-rad1-hus1 DNA damage checkpoint complex--implications for clamp loading and regulation. Mol Cell 34: 735–745 [DOI] [PubMed] [Google Scholar]
- Duggin IG, Dubarry N, Bell SD (2011) Replication termination and chromosome dimer resolution in the archaeon Sulfolobus solfataricus. EMBO J 30: 145–153 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Duggin IG, McCallum SA, Bell SD (2008) Chromosome replication dynamics in the archaeon Sulfolobus acidocaldarius. Proc Natl Acad Sci USA 105: 16737–16742 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garg P, Stith CM, Sabouri N, Johansson E, Burgers PM (2004) Idling by DNA polymerase delta maintains a ligatable nick during lagging-strand DNA replication. Genes Dev 18: 2764–2773 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gomes XV, Burgers PM (2000) Two modes of FEN1 binding to PCNA regulated by DNA. EMBO J 19: 3811–3821 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guo Z, Zheng L, Xu H, Dai H, Zhou M, Pascua MR, Chen QM, Shen B (2010) Methylation of FEN1 suppresses nearby phosphorylation and facilitates PCNA binding. Nat Chem Biol 6: 766–773 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haseltine CA, Kowalczykowski SC (2002) A distinctive single-strand DNA-binding protein from the Archaeon Sulfolobus solfataricus. Mol Microbiol 43: 1505–1515 [DOI] [PubMed] [Google Scholar]
- Henneke G, Koundrioukoff S, Hübscher U (2003) Phosphorylation of human Fen1 by cyclin-dependent kinase modulates its role in replication fork regulation. Oncogene 22: 4301–4313 [DOI] [PubMed] [Google Scholar]
- Higashibata H, Kikuchi H, Kawarabayasi Y, Matsui I (2003) Helicase and nuclease activities of hyperthermophile Pyrococcus horikoshii Dna2 inhibited by substrates with RNA segments at 5′-end. J Biol Chem 278: 15983–15990 [DOI] [PubMed] [Google Scholar]
- Hlinkova V, Xing G, Bauer J, Shin YJ, Dionne I, Rajashankar KR, Bell SD, Ling H (2008) Structures of monomeric, dimeric and trimeric PCNA: PCNA-ring assembly and opening. Acta Crystallogr D Biol Crystallogr 64: 941–949 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hutton RD, Roberts JA, Penedo JC, White MF (2008) PCNA stimulates catalysis by structure-specific nucleases using two distinct mechanisms: substrate targeting and catalytic step. Nucleic Acids Res 36: 6720–6727 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Indiani C, McInerney P, Georgescu RE, Goodman MF, O’Donnell M (2005) A sliding-clamp toolbelt binds high- and low-fidelity DNA polymerases simultaneously. Mol Cell 19: 805–815 [DOI] [PubMed] [Google Scholar]
- Kiyonari S, Tahara S, Shirai T, Iwai S, Ishino S, Ishino Y (2009) Biochemical properties and base excision repair complex formation of apurinic/apyrimidinic endonuclease from Pyrococcus furiosus. Nucleic Acids Res 37: 6439–6453 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marsh VL, McGeoch AT, Bell SD (2006) Influence of chromatin and single strand binding proteins on the activity of an archaeal MCM. J Mol Biol 357: 1345–1350 [DOI] [PubMed] [Google Scholar]
- Matsunaga F, Norais C, Forterre P, Myllykallio H (2003) Identification of short ‘eukaryotic’ Okazaki fragments synthesized from a prokaryotic replication origin. EMBO Rep 4: 154–158 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mayanagi K, Kiyonari S, Nishida H, Saito M, Kohda D, Ishino Y, Shirai T, Morikawa K (2011) Architecture of the DNA polymerase B-proliferating cell nuclear antigen (PCNA)-DNA ternary complex. Proc Natl Acad Sci USA 108: 1845–1849 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mayanagi K, Kiyonari S, Saito M, Shirai T, Ishino Y, Morikawa K (2009) Mechanism of replication machinery assembly as revealed by the DNA ligase-PCNA-DNA complex architecture. Proc Natl Acad Sci USA 106: 4647–4652 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meslet-Cladiére L, Norais C, Kuhn J, Briffotaux J, Sloostra JW, Ferrari E, Hübscher U, Flament D, Myllykallio H (2007) A novel proteomic approach identifies new interaction partners for proliferating cell nuclear antigen. J Mol Biol 372: 1137–1148 [DOI] [PubMed] [Google Scholar]
- Moldovan G-L, Pfander B, Jentsch S (2007) PCNA, the maestro of the replication fork. Cell 129: 665–679 [DOI] [PubMed] [Google Scholar]
- Nishida H, Mayanagi K, Kiyonari S, Sato Y, Oyama T, Ishino Y, Morikawa K (2009) Structural determinant for switching between the polymerase and exonuclease modes in the PCNA-replicative DNA polymerase complex. Proc Natl Acad Sci USA 106: 20693–20698 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pascal JM, O’Brien PJ, Tomkinson AE, Ellenberger T (2004) Human DNA ligase I completely encircles and partially unwinds nicked DNA. Nature 432: 473–478 [DOI] [PubMed] [Google Scholar]
- Pascal JM, Tsodikov OV, Hura GL, Song W, Cotner EA, Classen S, Tomkinson AE, Tainer JA, Ellenberger T (2006) A flexible interface between DNA ligase and PCNA supports conformational switching and efficient ligation of DNA. Mol Cell 24: 279–291 [DOI] [PubMed] [Google Scholar]
- Qiu J, Qian Y, Frank P, Wintersberger U, Shen B (1999) Saccharomyces cerevisiae RNase H(35) functions in RNA primer removal during lagging-strand DNA synthesis, most efficiently in cooperation with Rad27 nuclease. Mol Cell Biol 19: 8361–8371 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roberts JA, Bell SD, White MF (2003) An archaeal XPF repair endonuclease dependent on a heterotrimeric PCNA. Mol Microbiol 48: 361–371 [DOI] [PubMed] [Google Scholar]
- Rogozin IB, Makarova KS, Pavlov YI, Koonin EV (2008) A highly conserved family of inactivated archaeal B family DNA polymerases. Biol Direct 3: 32. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rossi ML, Bambara RA (2006) Reconstituted Okazaki fragment processing indicates two pathways of primer removal. J Biol Chem 281: 26051–26061 [DOI] [PubMed] [Google Scholar]
- Rossi ML, Pike JE, Wang W, Burgers PM, Campbell JL, Bambara RA (2008) Pif1 helicase directs eukaryotic Okazaki fragments toward the two-nuclease cleavage pathway for primer removal. J Biol Chem 283: 27483–27493 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sakurai S, Kitano K, Yamaguchi H, Hamada K, Okada K, Fukuda K, Uchida M, Ohtsuka E, Morioka H, Hakoshima T (2005) Structural basis for recruitment of human flap endonuclease 1 to PCNA. EMBO J 24: 683–693 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vivona JB, Kelman Z (2003) The diverse spectrum of sliding clamp interacting proteins. FEBS Lett 546: 167–172 [DOI] [PubMed] [Google Scholar]
- Wadsworth RIM, White MF (2001) Identification and properties of the crenarchaeal single-stranded DNA binding protein from Sulfolobus solfataricus. Nucleic Acids Res 29: 914–920 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Williams GJ, Johnson K, Rudolf J, McMahon SA, Carter L, Oke M, Liu H, Taylor GL, White MF, Naismith JH (2006) Structure of the heterotrimeric PCNA from Sulfolobus solfataricus. Acta Crystallogr F 62: 944–948 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xing G, Kirouac K, Shin YJ, Bell SD, Ling H (2009) Structural insight into recruitment of translesion DNA polymerase Dpo4 to sliding clamp PCNA. Mol Microbiol 71: 678–691 [DOI] [PubMed] [Google Scholar]
- Zhang C, Guo L, Deng L, Wu Y, Liang Y, Huang L, She Q (2010) Revealing the essentiality of multiple archaeal pcna genes using a mutant propagation assay based on an improved knockout method. Microbiology 156: 3386–3397 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.