Abstract
Post-embryonic growth in plants depends on the continuous supply of undifferentiated cells within meristems. Proliferating cells maintain their competence for division by active repression of differentiation and the associated endocycle entry. We show by upregulation and downregulation of E2FA that it is required for maintaining proliferation, as well as for endocycle entry. While E2FB–RBR1 (retinoblastoma-related protein 1) complexes are reduced after sucrose addition or at elevated CYCD3;1 levels, E2FA maintains a stable complex with RBR1 in proliferating cells. Chromatin immunoprecipitation shows that RBR1 binds in the proximity of E2F promoter elements in CCS52A1 and CSS52A2 genes, central regulators for the switch from proliferation to endocycles. Overexpression of a truncated E2FA mutant (E2FAΔRB) lacking the RBR1-binding domain interferes with RBR1 recruitment to promoters through E2FA, leading to decreased meristem size in roots, premature cell expansion and hyperactivated endocycle in leaves. E2F target genes, including CCS52A1 and CCS52A2, are upregulated in E2FAΔRB and e2fa knockout lines. These data suggest that E2FA in complex with RBR1 forms a repressor complex in proliferating cells to inhibit premature differentiation and endocycle entry. Thus, E2FA regulates organ growth via two distinct, sequentially operating pathways.
Keywords: Arabidopsis , cell proliferation, endocycle, E2F, retinoblastoma
Introduction
Post-embryonic growth in plants depends on meristematic activity, which is regulated by developmental programmes and environmental conditions. Meristematic cells proliferate through cell division and delay terminal differentiation. How cell differentiation is repressed within meristems, including the founder stem cells, and how it is activated when cells leave the meristem is a basic question in plant developmental biology (Bogre et al, 2008; Doonan and Sablowski, 2010).
Differentiating plant cells may enter into a modified cell cycle, called the endocycle, in which DNA synthesis is repeatedly activated without intervening mitosis, leading to an increase in the DNA content. The molecular players underlying the switch from the mitotic cell cycle into an endocycle are emerging (Lee et al, 2009; Nieuwland et al, 2009). The endocycle plays an important role in cell expansion (Breuer et al, 2009) and in cell fate acquisition and maintenance (Bramsiepe et al, 2010). The current molecular model for endocycle regulation suggests that it shares components with the proliferative cell cycle, such as the A-type cyclin-dependent kinase CDKA;1 (Leiva-Neto et al, 2004; Verkest et al, 2005a), whereas its onset is achieved through a selective inactivation of M phase-promoting factors, such as the B1-type CDK (CDKB1;1) through proteolytic destruction of its cyclin partner, CYCA2;3 (Boudolf et al, 2004, 2009). Correspondingly, Arabidopsis relatives of the animal fizzy-related activators of the anaphase-promoting complex (APC), CCS52A1 and CCS52A2, stimulate the switch from mitosis to endocycle (Larson-Rabin et al, 2009; Vanstraelen et al, 2009). In part, the expression of CCS52A2 is confined to cells engaged in endocycle by the atypical E2F, DEL1/E2FE (Lammens et al, 2008).
The retinoblastoma-related protein 1 (RBR1) and its targets, the E2F transcription factors are known to take part in the decision between cell proliferation and differentiation (Wildwater et al, 2005; Wyrzykowska et al, 2006). Arabidopsis has a single RBR1 gene with an essential function in plant development, gamete formation and meiosis (Ebel et al, 2004; Park et al, 2005; Wildwater et al, 2005; Desvoyes et al, 2006; Jordan et al, 2007; Lageix et al, 2007; Chen et al, 2009, 2011; Borghi et al, 2010; Johnston et al, 2010; Gutzat et al, 2011), while it holds three RBR1 interacting E2F transcription factors, E2FA, E2FB and E2FC. These E2Fs require association with one of the two DIMERISATION PARTNER proteins, DPA or DPB for DNA binding (Inze and De Veylder, 2006; Magyar, 2008). The transcription factor activity of the E2F-DP dimer is regulated by RBR1 binding, although in plants only indirect evidence supports this model, including resemblance of overexpression line phenotypes of E2FA, E2FB and CYCD3;1 with those of RBR1-RNAi plants (De Veylder et al, 2002; Rossignol et al, 2002; Magyar et al, 2005; Wildwater et al, 2005) and regulation of E2F targets by overexpression of CYCD3;1, RBR1, E2F and DP genes (Ramirez-Parra et al, 2003; Vandepoele et al, 2005; de Jager et al, 2009). According to current models, CYCD3;1 in complex with CDKA;1 regulates cell-cycle entry by phosphorylation of RBR1, leading to the release of RBR1-bound E2F transcription factors to drive the expression of genes required for the cell-cycle phase transitions (Nakagami et al, 1999, 2002; Uemukai et al, 2005). In accordance, the triple mutant cycd3;1 cycd3;2 cycd3;3 has smaller organs with fewer cells (Dewitte et al, 2007), whereas ectopic expression of CYCD3;1 inhibits organ growth by repressing differentiation, further supporting its role in maintaining the balance between cell proliferation and differentiation (Dewitte et al, 2003). The CDK inhibitor proteins, called KIP-related proteins (KRPs) oppose CYCD–CDK activities and inhibit cell-cycle progression (Verkest et al, 2005b).
Functional characterization of E2Fs has been mostly restricted to ectopic overexpression studies: lines co-transformed with E2FA and DPA results in the activation of both mitotic and endocycle (De Veylder et al, 2002), whereas overexpression of E2FB induces mitosis but represses the endocycle (Magyar et al, 2005; Sozzani et al, 2006). On the other hand, silencing of E2FC leads to cell proliferation and compromised endocycle, suggesting that E2FC would be analogous to the repressor-type animal E2Fs (del Pozo et al, 2006). Based on these data, E2FB and E2FC are antagonistic transcription factors, while E2FA has dual functionality (Magyar, 2008).
Here, we investigated how E2FA can regulate both cell proliferation and differentiation-associated endocycle; two processes that are spatially separated during plant development. We demonstrate that E2FA forms a stable complex with RBR1 in proliferating cells and suggest that this repressor complex plays a role in maintaining the meristematic state. We addressed the dual function of E2FA by analysing e2fa knockout mutant, E2FA silenced lines and lines with elevated levels of E2FA within its own expression domains. We show that E2FA promotes the maintenance of cells in the proliferative state while stimulates endocycle later during leaf development.
Results
E2FA and RBR1 are co-regulated in proliferating cells
Because RBR1 regulates the E2F/DP dimer, we investigated whether they are co-regulated by analysing publicly available microarray data. We found that only E2FA co-expressed with RBR1 with a 0.7 expression correlation coefficient (Supplementary Table S1). In addition, both E2FA and RBR1 showed a highly similar co-expression neighbourhood. GO overrepresentation analysis by BINGO (Maere et al, 2005) on the list of genes that show correlated expression with E2FA or RBR1 using thresholds between 0.5–0.7 correlation coefficients yielded overlapping GO categories for these two genes, and contained GO categories of DNA replication, chromosome organisation (Supplementary Table S2).
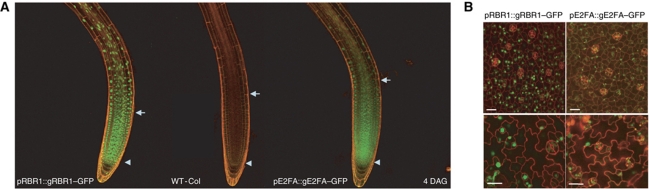
To investigate the spatial overlap in the accumulation patterns of the E2FA and RBR1 proteins in developing organs, we analysed Arabidopsis plants carrying constructs with the green fluorescence protein (GFP) marker fused to the C-terminus of the E2FA and RBR1 genes under the control of their own promoters (pE2FA::gE2FA–GFP and pRBR1::gRBR1–GFP). The pRBR1::gRBR1–GFP construct is functional, since it complemented the rbr1 mutant phenotype (data not shown). The E2FA–GFP fusion retained its ability to bind RBR1 (Figure 4C), DPA and DPB (Supplementary Figure S7). Furthermore, the elevated expression of E2FA–GFP led to phenotypes of overproliferation, increased endocycle (Figure 4; see later) that are similar to what was published for the overexpression of wild-type (WT) E2FA (De Veylder et al, 2002). E2FA–GFP accumulated within the root meristem, becoming gradually weaker, but still detectable at the transition zone, where cells leave proliferation and start elongation (Figure 1A; Supplementary Figure S1A). RBR1–GFP was also expressed in the meristem, and remained present at the elongation zone and in differentiated columella cells (Figure 1A; Supplementary Figure S1A). Interestingly, elevated E2FA expression in the pE2FA::gE2FA–GFP line 81 resulted in a larger root meristem compared with the WT, while elevated RBR1 expression in the pRBR1::gRBR1–GFP line had a smaller meristem (Supplementary Figure S1B and C). In young leaves, both E2FA–GFP and RBR1–GFP were abundant in proliferating cells (Figure 1B) but still detectable in differentiated pavement cells (Figure 1B). In summary, we detected E2FA–GFP and RBR1–GFP proteins in cells that undergo mitotic cycles, and to a lesser extent in cells that differentiate and undergo endocycles, such as enlarged and lobed leaf cells.
Figure 1.
Both RBR1 and E2FA are present in proliferating cells in Arabidopsis roots and leaves. (A) Confocal microscopy images of the root tip of Arabidopsis plants 4 DAG expressing either the translational GFP fusion of E2FA (pE2FA::gE2FA–GFP—left) or RBR1 (pRBR1::gRBR1–GFP—right) and the WT Columbia as control (WT-Col—middle). Arrows indicate the cortex transition zone, arrowheads mark the quiescent centre. (B) Confocal microscopy images of adaxial leaf surfaces from the first leaf pair of the same transgenic lines as in (A) 8 days and 12 DAG (upper and lower images, respectively). GFP signal (green) is counterstained for cell wall with propidium iodide (red). Scale bar is 100 μM.
E2FA is required both for proliferation and for endocycle
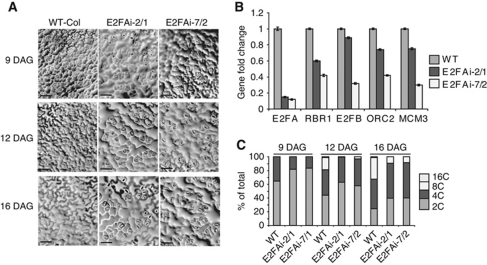
Dual functions of E2FA were proposed from overexpression studies together with its DPA dimerization partner (De Veylder et al, 2002; Kosugi and Ohashi, 2003), but so far awaiting validation by loss-of-function genetics. At the start of this work, the only publicly available T-DNA insertion mutant line for E2FA (SALK_034842) revealed to have no insert at the expected position. Therefore, we generated E2FA silencing lines in order to address its physiological role in cell proliferation and endocycle. We tested the specificity of silencing by transiently expressing 35SCaMV promoter driven E2FA and E2FB-RNAi construct with E2FA–HA and E2FB–HA tagged constructs in protoplasts. The E2FA-RNAi construct specifically targeted E2FA but not E2FB, while E2FB-RNAi effectively eliminated E2FB, but showed some minor reduction also in E2FA (Supplementary Figure S2A). Because the E2FA-RNAi construct was shown to be specific, we stably transformed Arabidopsis plants with this construct, and selected two independent lines that showed a strong reduction in E2FA levels (below 20% of the endogenous mRNA), though we found some variation in the silencing efficiency from experiment to experiment (Supplementary Figure S2B). The E2FA-RNAi plants were smaller, displaying shorter roots with foreshortened meristem size (Supplementary Figure S3A–C). Epidermal pavement cells in young leaves (9 DAG) of E2FA-RNAi lines were enlarged, when compared with control plants, indicative of premature exit from the proliferation phase (Figure 2A). These cells were abnormally large compared with WT also at later time points (12 and 16 DAG), while cells around the stomata remained small.
Figure 2.
E2FA silencing leads to compromised cell proliferation and endocycle. (A) Representative images from the adaxial epidermal cell layer of the first leaf pairs at three developmental time points (DAG) from the WT, and the E2FA-RNA-interference lines (E2FAi line 2/1 and 7/2) as indicated. Bar=100 μM. (B) The expression levels of E2FA, RBR1, E2FB, ORC2 and MCM3 were determined by Q-RT–PCR in 1-week-old seedlings of E2FAi lines and expressed as fold changes compared with WT. (C) The percentage of DNA ploidy levels were determined by flow cytometry in samples taken at three time points (9, 12, 16 DAG) of developing first leaf pairs (L1–2) of WT and two independent E2FAi lines (2/1, 7/2).
To investigate how the silencing of E2FA affected the expression of cell-cycle regulators, we analysed the transcript levels of E2FB, RBR1, MCM3, ORC2 mRNAs in the two selected lines. All of these genes are putative E2F targets based on their transcriptional upregulation in E2FA/DPA overexpression lines in microarray experiments and on the presence of E2F cis-acting elements in their promoters (Vandepoele et al, 2005). We found that these selected E2F target genes were all downregulated in the E2FA silencing line (Figure 2B). As shown previously, the E2FA-RNAi construct has no effect on the 35CaMV promoter driven E2FB expression, and therefore the diminishing of E2FB mRNA in the E2FA-RNAi plants represents a cross-regulation between E2FA and E2FB, most likely through the E2F element in the promoter of E2FB, as has been published before (Sozzani et al, 2006). Suggestive of a complex regulatory network among E2Fs, we found that a number of them are deregulated in the e2fa knockout mutant (see later). In parallel to E2FA silencing, RBR1 mRNA and protein levels were also reduced, in accordance to the co-regulation of these genes across many experimental conditions (Supplementary Table S1). CDKB1;1 level is linked to cell proliferation (Boudolf et al, 2004), and in accordance with compromised proliferation, we found a reduced CDKB1;1 protein level in the E2FA-RNAi plants (Supplementary Figure S3D).
In Arabidopsis leaves when cells exit cell proliferation and start cell expansion, the endocycle is activated. To measure the switch from proliferation to endocycle, we determined the DNA content of nuclei from leaf 9–16 DAG by flow cytometry. In WT leaves, 9 DAG the proliferating cells alternate between G1 (65%) and G2 (35%). In E2FA-RNAi lines, the proportion of cells in G2 was reduced with an accompanied increase in G1. Together with the data that cells became larger in the E2FA-RNAi leaves at this stage, compared with WT (Figure 2A), the flow cytometry data suggest a G1 arrest rather than a shortened G2 phase in these cells. At 12–16 DAG, 8C and 16C nuclei were detected in the WT leaf, indicative of endocycle onset. In contrast, in the E2FA-RNAi lines the switch from proliferation to endocycle was delayed (Figure 2C), showing reduced levels of 8C nuclei and absence of 16C nuclei. Similarly, the ploidy level was reduced in fully mature leaves (Supplementary Figure S3E). Although endocycle has been correlated with cell enlargement (Breuer et al, 2010), in the E2FA-RNAi this appears not to be the case, since leaf epidermal cells enlarged with only a modest increase in their DNA content. This shows that endocycle at this stage of leaf pavement cell differentiation is not essential for cell expansion. The endocycle is also compromised in the cotyledons of the E2FA-RNAi plants, as well as in the recently published e2fa-1 knockout line (Berckmans et al, 2011b).
Taken together, these results suggest that E2FA is required both for cell proliferation and for endocycle.
The amount of E2FA–RBR1 complex correlates with the level of proliferation
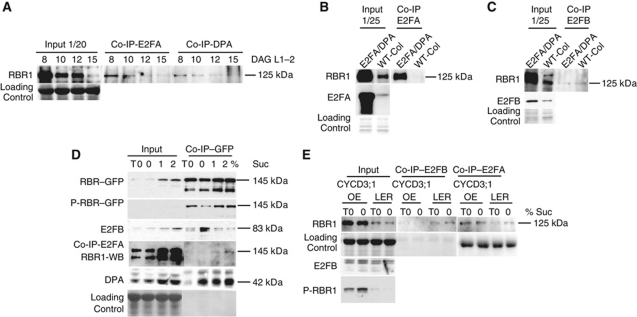
It is generally assumed that the cell proliferation and endocycle-promoting activities of E2FA is restrained by RBR1; however, there are no experimental data to show when and how RBR1 regulates E2FA during plant development. To study the interaction between RBR1 and E2FA over a developmental time course, we dissected the first two emerging leaves at 8 days after germination (DAG), when large proportion of cells proliferate, and at 10, 12 and 15 DAG, when cells gradually exit proliferation (Beemster et al, 2006), and performed co-immunoprecipitation (co-IP) with E2FA- or DPA-specific antibodies with these samples. E2FA levels in these samples were too low in crude extracts to be detected with our E2FA-specific antibody. However, we detected the presence of RBR1 in the immunoprecipitate (Figure 3A). RBR1 levels were the highest at 8 DAG, and gradually diminishing as leaves develop. This is in accordance to previous reports that RBR1 is most abundant in proliferating tissues in Arabidopsis (Wildwater et al, 2005). However, this is not necessarily the case in all plants, as in maize leaves RB increases as cells exit proliferation (Huntley et al, 1998). RBR1–E2F association is primarily regulated by phosphorylation and disrupted through CDK–CYCD activities in proliferating tissues. However, we found that the amount of RBR1 in complex with E2FA and DPA mirrored RBR1 abundance, being highest in proliferating leaf cells (Figure 3A).
Figure 3.
RBR1 makes complex with E2FA in proliferating cells while it associates with E2FB in cells leaving cell proliferation. (A) Interaction of E2FA with RBR1 during leaf development. IP with anti-E2FA or anti-DPA antibodies from protein extracts prepared from the juvenile first leaf pair (L1–2) of WT Arabidopsis plant at the indicated time points (DAG). Co-IP of RBR1 with E2FA and DPA was probed on western blot with anti-RBR1 antibody. In all, 1/20 of the IP from the extract was loaded as input. Coomassie staining of the same membrane was used for loading control. (B, C) Interaction of E2FA or E2FB with RBR1 in Arabidopsis Columbia (WT-Col) or E2FA/DPA transgenic Arabidopsis lines. IP with anti-E2FA (B) or anti-E2FB (C) antibodies from protein extracts of WT-Col and E2FA/DPA seedlings 1 week after germination. The co-IP of RBR1 was tested with anti-RBR1 on western blot. In all, 1/25 of the IP from the extract was loaded as input to determine RBR1, E2FA or E2FB levels as indicated. Coomassie-stained proteins on the same membranes are shown as loading control. (D) Interaction of RBR1–GFP with E2FA and E2FB is differently regulated by sucrose. Six days old pRBR::gRBR1–GFP seedlings grown on plates in the presence of 1% sucrose (T0) was treated with liquid media with 0, 1 or 2% sucrose for 6 hours. IP with anti-GFP or anti-E2FA from these seedlings in (D) as indicated. The co-IP experiments with specific antibodies and the input levels as well as the corresponding molecular masses are indicated. (E) Interaction of E2FA or E2FB with RBR1 in WT Arabidopsis Landsberg (Ler) or CYCD3;1 overexpression line (CYCD3;1-OE). Six days old seedlings (T0) grown on plate in the presence of 1% sucrose were treated with liquid medium without sucrose (0%) for 6 hours. IP from protein extracts of these seedlings with anti-E2FA or anti-E2FB antibodies in (E) as indicated. The co-IP of RBR1 was probed with anti-RBR1 antibody in western blots. Input levels of RBR1, P-RBR1 and E2FB proteins and loading controls are indicated. The quantitation of all western blots are provided in Supplementary Figure S13.
E2FA and DPA ectopic co-overexpression leads to overproliferation in root columella cells and cotyledon, whereas in cortical root cells it induces endocycle (De Veylder et al, 2002). It was proposed that the elevated E2FA levels escapes from RBR1 repression and promote both cell proliferation and endocycle, dependent on the tissue-specific availability of mitosis inducing factors. Unexpectedly, we find that the E2FA/DPA overexpression plants showed a strongly upregulated RBR1 amount, and had a correspondingly elevated level of RBR1–E2FA complex (Figure 3B). E2FB level is slightly upregulated in the E2FA/DPA overexpression line (Figure 3C), in agreement to what we have found in the pE2FA::gE2FA–GFP lines (Figure 4A) and was published before by Sozzani et al (2006). Despite the increased E2FB and RBR1 amounts in the E2FA/DPA overexpression line, there was less E2FB/RBR1 complex present compared with WT (Figure 3C), suggesting a distinct regulation of RBR1 interaction with E2FA than with E2FB.
Figure 4.
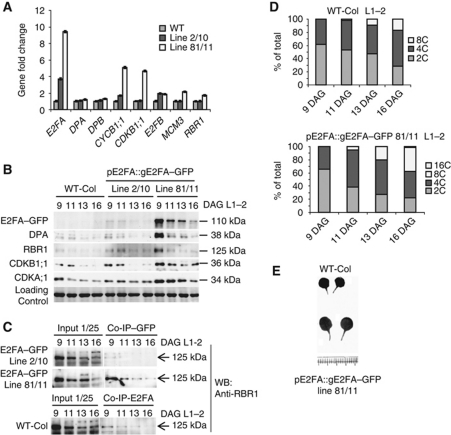
The pE2FA::gE2FA–GFP lines have increased expression of E2FA within its own expression domain that leads to more proliferation and endocycle compared with WT. (A) Two independent pE2FA::gE2FA–GFP lines (line 2/10 and line 81/11) were tested for the expression levels of E2FA, E2FA interacting proteins; DPA, DPB as well as cell-cycle markers and selected E2F target genes; CYCB1;1, CDKB1;1, E2FB, MCM3 and RBR1 by Q-RT–PCR in samples derived from proliferating first leaf pairs (9 DAG). (B) E2FA–GFP, DPA, RBR1, CDKB1;1 and CDKA;1 protein levels in samples of the first leaf pairs (L1–2) of WT and the two independent pE2FA::gE2FA–GFP lines at four time point (DAG) detected by specific antibodies on western blots as indicated. Molecular masses (kDa) are indicated. (C) Interaction of E2FA–GFP with RBR1 in WT and in the pE2FA::gE2FA–GFP lines using samples representing a developmental series of leaf pairs (L1–2) at the indicated time points (9, 11, 13, 16 DAG). IPs used anti-GFP or anti-E2FA antibodies as indicated. Co-IP of RBR1 was tested by anti-RBR1 antibody on western blot (RBR1-WB), the RBR1 protein is indicated with arrow. In all, 1/25 of the IP from the extract was loaded as input. (D) Ploidy levels of pE2FA::gE2FA–GFP line 81/11 compared with WT in samples of the first leaf pair—L1–2 taken at 9, 11, 13, 16 DAG. (E) Representative images of the first leaf pair from pE2FA::gE2FA–GFP line 81/11 shows an increase in leaf size compared with WT.
In summary, the amount of RBR1–E2FA complex correlates with the extent of proliferation in WT, E2FA/DPA overexpression line, pE2FA::gE2FA–GFP lines (see below), while there is less RBR1–E2FB complex in the E2FA/DPA line.
The amount of RBR1–E2FB complex is decreased whereas RBR1–E2FA complex is elevated when CYCD3;1 is overexpressed
One possible explanation for the elevated level of RBR1–E2FA complex in samples when a high proportion of cells are in proliferation is that in cycling cells proportionally more cells would go through the G1 phase, where RBR1 is predicted to interact with E2Fs. D-type cyclins promote cell proliferation notably in G1 phase through phosphorylation of RBR1, and thereby through the disruption of RBR1–E2F complex (van den Heuvel and Dyson, 2008). First we set out to test whether this is indeed the case during Arabidopsis leaf development by using a phospho-RB (P-RB) antibody that recognizes two conserved CDK phosphorylation sites (Ser807/811) present in RBR1 protein. This commercial antibody was shown to cross-react only with the phosphorylated RBR1 form in plants (Abraham et al, 2011). To further ascertain that the P-RB antibody recognises a genuine RBR1, we immunoprecipitated with P-RB from WT and pRBR1::RBR1–GFP plants and probed the immunoprecipitate with the Arabidopsis-specific RBR1 or GFP antibodies and found a 125 and 145 kDa protein corresponding to the expected RBR1 and RBR1–GFP sizes, respectively (Supplementary Figure S4A–C and F). We found RBR1 phosphorylation mostly in young leaves (9 DAG; Supplementary Figure S4A). Furthermore, overexpression of CYCD3;1 led to RBR1 hyperphosphorylation (Supplementary Figure S4B), while overexpression of KRP2 reduced its phosphorylation (Supplementary Figure S4C).
It is known that sugar availability regulates the plant cell cycle by influencing the expression of D-type cyclins (Menges et al, 2006). Because RBR1 is a pivotal target for the CYCD–CDKA complex (Nakagami et al, 2002), we monitored RBR1 phosphorylation in Arabidopsis seedlings in response to short-term (6 h) treatment with 2% sucrose or no sucrose in the media (Supplementary Figure S5A). The level of RBR1 phosphorylation became high in the presence of sucrose and was reduced when WT seedlings were incubated in the absence of sucrose. We also tested how the sugar-induced phosphorylation of RBR1 is regulated in CYCD3;1 and KRP2 overexpression lines and found it to be low in KRP2 and high in CYCD3;1, irrespective of the sucrose levels (Supplementary Figure S5A). These results show that RBR1 phosphorylation on the conserved Ser807/811 sites is sucrose dependent and rely on a CDK regulated by CYCD3;1 and KRP2.
Next, we tested whether the change in RBR1 phosphorylation after sucrose treatments is linked to an altered cell proliferation at different sucrose levels by monitoring DNA content using flow cytometry and the expression of cell cycle marker genes by Q-RT–PCR (Supplementary Figure S5B and C). Addition of 2% sucrose elevated the proportion of cells in S-phase after 6 h followed by an increase in G2 cells after 12 h. In contrast, in sucrose-free media, the proportion of S-phase decreased while the G1 increased (Supplementary Figure S5B). The expression of cell-cycle genes also changed, though in a rather complex manner, indicating that both the G1 and G2 control points are targets for sugar signalling pathways. Depletion of sucrose first reduced the CYCA2;3 level at 6 h and then CDKB1;1 at 24 h while addition of sucrose induced the CYCD3;1 and CYCB1;1 expression at 6 h. CYCA2;3 peaked at 12 h and diminished at 24 h, suggestive for a synchronous cell proliferation (Supplementary Figure S5B and C).
The next question was how RBR1 association with E2FA and E2FB is regulated by sucrose. We utilised the RBR1–GFP line and performed IP with GFP-specific antibody and detected RBR1, P-RBR1, E2FB and DPA, while for the interaction of E2FA with RBR1 we immunoprecipitated with E2FA-specific antibody and detected RBR1 on the western blot (Figure 3D). In agreement to the previous experiment in WT Arabidopsis seedlings (Supplementary Figure S5A), both RBR1–GFP protein and the P-RBR1 levels increased parallel with the elevated sucrose concentration but only P-RBR1 level was reduced in sucrose-free condition. The majority of E2FB protein was in complex with RBR1 in sucrose-free condition (Figure 3D). Though E2FB protein level was increased in the presence of sucrose, its RBR1 association greatly diminished. Thus, sucrose regulates the level of RBR1-free E2FB, which potentially can go on and trigger cell proliferation. Contrary to E2FB, the E2FA–RBR1 complex was the most abundant in the presence of sucrose, and paralleled the RBR1 protein level. DPA can form heterodimmers both with E2FA (Supplementary Figure S7) and with E2FB (Magyar et al, 2000, 2005) but not with E2FC (Gutierrez, 2009). In agreement, DPA could be detected in complex with RBR1 at both the presence and absence of sucrose, when E2FA and E2FB forms complex with RBR1, respectively (Figure 3D). All these data indicate that RBR1 represses cell-cycle progression through E2FB in sucrose-limited condition, while RBR1 forms complex with E2FA when sucrose is present and cells proliferate.
We also have found above that CYCD3;1 overexpression can increase and overcome the sucrose-dependent RBR1 phosphorylation. Therefore, we investigated how CYCD3;1 level affects the association of RBR1 with E2FA and E2FB. We took advantage of sucrose-depleted condition, which we know to increase E2FB but decrease E2FA interaction with RBR1. We immunoprecipitated E2FA and E2FB from WT Landsberg erecta and CYCD3;1 overexpression lines and tested the level of RBR1 in the complex. In agreement to our results above there is an increased RBR1 amount in the CYCD3;1 overexpression line compared with WT. The RBR1–E2FA complex also increased in parallel to RBR1 levels in the CYCD3;1 line (Figure 3E; Supplementary Figure S6A). In agreement that DPA is the dimerisation partner of E2FA, the DPA association with RBR1 also increases in the CYCD3;1 overexpression line (Supplementary Figure S6A). These results are surprising, because the majority of RBR1 is phosphorylated upon CYCD3;1 overexpression, and according to the current models this should lead to the dissociation of RBR1–E2F complex. This was indeed the case for E2FB, which showed a decreased association with RBR1 in the CYCD3;1 overexpression line compared with WT (Figure 3E).
Cells in cotyledons exit proliferation and enter into endocycle early during development, which is strongly inhibited by CYCD3;1 overexpression (Dewitte et al, 2003). An extended proliferation of cotyledons in the CYCD3;1 overexpression line is also indicated by the elevated levels of E2FB, DPA and CDKB1;1 cell-cycle proteins (Supplementary Figure S6B). We found that RBR1 forms a complex with E2FA but not with E2FB in the cotyledon of CYCD3;1 overexpressor line (Supplementary Figure S6C). These data further substantiate that E2FA–RBR1 and E2FB–RBR1 complexes are distinct in their regulation and E2FA–RBR1 abundance correlates with cell proliferation.
Since both CYCD3;1 and E2FA/DPA overexpression lines show an increase in proliferation and RBR1 levels we compared their RBR1 phosphorylation status. CYCD3;1 line had vastly more P-RBR1 than E2FA/DPA, though both had increased amounts compared with the WT. The ratio of unphosphorylated to phosphorylated RBR1 forms was substantially higher for E2FA/DPA compared with CYCD3;1 overexpression line, suggesting that the mechanism for overproliferation for these two lines are distinct; for CYCD3;1 this is due to RBR1 phosphorylation, while for E2FA/DPA this could relate to increased amount of RBR1–E2FA complex (Supplementary Figure S4B).
To investigate whether the phosphorylated or non-phosphorylated RBR1 forms a complex with E2FA in proliferating cells, we have used two independent pE2FA::gE2FA–GFP lines with lower and higher expression levels, lines 2 and 81, respectively. This enabled us to immunoprecipitate E2FA with GFP antibody and to probe the immunoprecipitates for phospho-RB, RBR1 and GFP. In agreement with the above-described results, line 81, which has more E2FA–GFP, also contained a higher amount of RBR1 (Supplementary Figure S4D). However, no dramatic difference in the P-RBR1 level was observed between the two lines and we could not detect the P-RBR1 form in complex with E2FA (Supplementary Figure S4D). This result was confirmed by a reciprocal IP with the P-RB antibody; from line 81; we could detect RBR1 in complex with E2FA, but there was no detectable association between E2FA- and P-RBR1 (Supplementary Figure S4E). Because we detect E2FA–RBR1 complexes in the CYCD3;1 overexpression line, a pool of RBR1, possibly the one that form a complex with E2FA, appears to be protected from CYCD3;1-CDKA;1 phosphorylation.
Elevated E2FA level results in more E2FA–RBR1 complex and increased cell proliferation
To further investigate how E2FA level affect cell proliferation and endocycle, we analysed the pE2FA::gE2FA–GFP plants (lines 2 and 81) that had different E2FA expression levels (Figure 4A), but the developmental regulation remained unaltered; more E2FA protein was present in the young proliferating leaves (9 DAG) and it diminished as leaves develop (11–16 DAG; Figure 4B), correlating with the expression levels of the mitotic regulators; CYCB1;1 and CDKB1;1 (Figure 4A). Although we did not detect a significant increase in DPA and RBR1 transcripts, their protein amount increased proportionally to the level of E2FA–GFP (Figure 4B). This effect could be due to the stabilization of RBR1 and DPA when complexed with E2FA. To test this hypothesis, we performed a co-IP experiment with a GFP antibody and determined the presence of RBR1 (Figure 4C). The increased E2FA levels indeed resulted in a corresponding increase in the E2FA–RBR1 complex (Figure 4C).
Confirming the upregulation of cell proliferation, we found increased and sustained CDKB1;1 and CDKA;1 protein levels in the pE2FA::gE2FA–GFP line 81/11 compared with the WT (Figure 4B). Moreover, endocycle was also stimulated, as illustrated by flow cytometric measurements on leaves from 11 DAG onwards, when proliferation starts to decrease, resulting in an increased percentage of 8C and 16C nuclei (Figure 4D). These results illustrate that the outcome of E2FA overexpression is determined by the proliferative status of the tissue, and correlates with the amount of E2FA–RBR1 complex. This dual effect of E2FA on cell proliferation and endocycle resulted in overall larger leaf size (Figure 4E).
To investigate how RBR1 levels affect mitosis and endocycle, we measured DNA content of developing leaves in RBR1–GFP lines (Supplementary Figure S6E) that we know to have around 10 times higher RBR1 level (data not shown). We analysed the effect of increased RBR1 level on leaf development at two time points; at 7 DAG when most leaf cells are dividing and at 16 DAG when endocycle is more prominent. RBR1 was found to repress the progression of both mitotic cell cycle and endocyle (Supplementary Figure S6E). From this experiment it is not clear whether the elevated RBR1 at distinct time points could act through the same or different E2Fs.
Blocking RBR1 recruitment to promoters through E2FA by overexpression of an E2FA form with a deleted RBR1-binding domain triggers premature exit from proliferation and hyperactivate endocycles
E2FA was suggested to regulate the proliferative cell cycle and endocycle through its ability to bind to promoter sequences and transactivate E2F target genes that are involved in DNA synthesis (De Veylder et al, 2002). To unravel the role of transactivation versus the recruitment of a repressor RBR1 complex to promoters specifically through E2FA, we constructed a truncated form of E2FA in which the last 65 C-terminal amino acids responsible for both RB binding and transactivation were removed (E2FAΔRB; Supplementary Figure S8A), and expressed it in Arabidopsis plants under the control of CaMV35S promoter. A similar deletion mutation was described for the animal E2F1, and was shown to confer growth factor independent cell proliferation, suggesting that not the masking of E2F transactivation capacity upon RB binding, but the recruitment of RB to promoters is required for repression of cell division (Zhang et al, 1999). Our biochemical analysis demonstrated that E2FAΔRB is still able to associate with E2F cis-acting elements and to dimerise with DPA (Supplementary Figure S8B–D), whereas it was impaired in RBR1 association (Supplementary Figure S8E and F) and transactivation, tested by its ability to activate E2F target promoters (Supplementary Figure S8G).
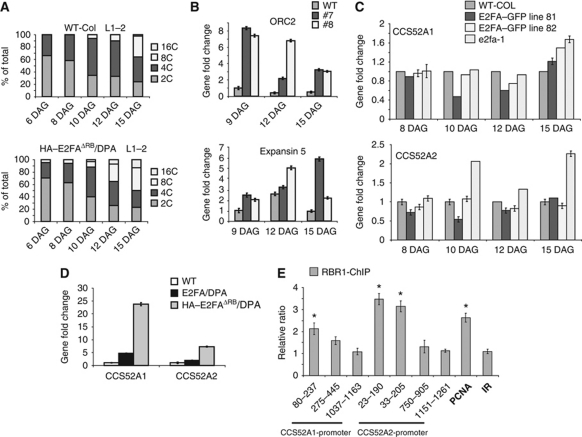
Arabidopsis 35S::E2FAΔRB lines had no obvious macroscopic phenotypes, and the mutant E2FAΔRB protein was hardly detectable through its HA-tag. However, when E2FAΔRB was co-expressed with DPA, the stability and the DNA-binding activity of E2FAΔRB was improved (Supplementary Figures S8B–D and S9A). The 35S::E2FAΔRB/DPA plants had smaller-sized cotyledons and leaves than the WT (Supplementary Figure S9B). The size of the root meristem was also compromised in the 35S::E2FAΔRB/DPA plants in comparison to the WT root (Supplementary Figure S9C). Leaf epidermal cells enlarged prematurely in the 35S::E2FAΔRB/DPA plants compared with the WT, indicating that cells exited the proliferative stage earlier than WT cells (Supplementary Figure S9D). Consequently, leaf size was significantly smaller in the 35S::E2FAΔRB/DPA plants compared with the WT (Supplementary Figure S9E). Trichomes were over branched, containing up to 10 branches compared with the typical three in the WT (Supplementary Figure S9F and G). The over branched trichomes indicated a strong positive effect on the endocycle, which we confirmed by DNA content analysis using flow cytometry. The ploidy levels were significantly higher; up to 32C in the mature leaves of 35S::E2FAΔRB/DPA lines compared with the single 35S::E2FAΔRB and DPA overexpressors, as well as the WT control, where the maximum DNA amount was 8C (Supplementary Figure S9H). Moreover, we detected an 8C ploidy level as early as 6 DAG in the E2FAΔRB/DPA line, whereas in WT leaves, cells remain diploid up to 10 DAG, demonstrating that the endocycle was prematurely activated (Figure 5A). This phenotype required the formation of the E2FAΔRB/DPA dimer, as the single transformants were indistinguishable from WT (Supplementary Figure S9G and H).
Figure 5.
E2FA interaction with RBR1 is required for the repression of premature cell elongation and endocycle. (A) DNA content was determined in the first leaf pairs by flow cytometry and it shows a premature increase in ploidy in the 35S::HA–E2FAΔRB/DPA overexpressor line in comparison to the WT. (B) Expression analysis of ORC2 and Expansin 5 were determined by Q-RT–PCR from the first leaf pairs at three developmental time points (DAG) from the WT and two 35S::HA–E2FAΔRB/DPA lines. (C) Expression analysis of CCS52A1 and CCS52A2 were determined by Q-RT–PCR from the developing first leaf pair of WT, and two pE2FA::gE2FA–GFP lines (the high expressor line 81 and the low expressor line 82) and the T-DNA insertion line e2fa-1 in the indicated time points (DAG). (D) Expression analysis of CCS52A1 and CCS52A2 were determined by Q-RT–PCR from seedlings of WT, 35S::E2FA/DPA and 35S::E2FAΔRB/DPA overexpressor lines 1 week after germination. (E) ChIP analysis of CCS52A1 and CCS52A2 promoters showing RBR1 association around their consensus E2F sites were carried out on 5 days old roots of Arabidopsis Columbia seedlings. PCNA1 (At1g07370; Kosugi and Ohashi, 2002) promoter was used as a positive control while a random intergenic region from chromosome 3 (IR; between At3g03660-70) was used as a negative control. Asterisks show the presence of consensus cis-acting E2F element in the promoter fragments amplified by using Q-RT–PCR primers in distance from the translational start codon as indicated.
To demonstrate the requirement of E2FA binding to DNA for its dual effect in regulation of proliferation and endocycle, we mutated the DNA-binding domain of DPA (DPAΔDB) and co-expressed it with E2FA (Supplementary Figure S10A). We did not find any phenotypes that were described for 35S::E2FA/DPA line, and there was no significant alteration in ploidy levels neither in the 35S::E2FA/DPAΔDB plants nor in the 35S::E2FAΔRB/DPAΔDB plants and remained similar to WT and to the singly transformed lines (Supplementary Figure S10B and C). These results suggest that E2FA/DPA or E2FAΔRB/DPA dimer needs to associate with DNA for its phenotypic effects.
We also determined the expression of S-phase regulatory genes in the 35S::E2FAΔRB/DPA seedlings, including ORC2, MCM3 and RNR2 (Figure 5B; Supplementary Figure S11B and C). We found all these genes to be upregulated. We do not know whether these genes are direct targets for the E2FA–RBR1 repressor complex, or their upregulation is a consequence of hyperactivated endocycle.
We conclude that the overexpressed E2FAΔRB/DPA binds to DNA at E2F sites and blocks the formation of the repressor RBR1 complex at these sites leading to hyperactivated endocycle and compromised meristem maintenance.
CCS52 genes are repressed by the recruitment of RBR1 through E2FA to their promoters
In proliferating cells, genes required for endocycle onset need to be repressed. These include the CCS52 genes that play important regulatory roles in the transition from mitosis to endocycle by stimulating the degradation of mitotic cyclins (Vanstraelen et al, 2009). To test the impact of E2FA levels on the expression of CCS52 genes, we followed their expression during the development of the first leaf pairs in lines with increased E2FA expression in its own expression domain (strong and weak expressor E2FA–GFP lines; 81 and 82, respectively) and in the e2fa-1 knockout mutant (Berckmans et al, 2011b). Both CCS52 genes were repressed in the leaves with elevated expression of E2FA in E2FA–GFP lines; 81 and 82 at 10 DAG when leaf cells normally begin to exit proliferation and enter into endocycle (Figure 5C). In contrast, CCS52A2 expression was doubled in the e2fa-1 mutant leaf at 10 DAG and later at 15 DAG (Figure 5C). On the basis of these data, we suggest that E2FA function as a repressor on the CCS52A genes possibly through the recruitment of RBR1 to their promoters.
To study whether the E2FAΔRB/DPA binding to DNA blocks the recruitment of innate E2FA to promoters for transactivation, or prevents the recruitment of the repressor RBR1 complex to promoters through E2FA, we tested the expression of various putative E2FA target genes (Vandepoele et al, 2005). In the first scenario, target gene expression is expected to be downregulated, while in the second they would be upregulated. We found that both CCS52 genes (CCS52A1 and CCS52A2) were largely upregulated in the 35S::E2FAΔRB/DPA lines (Figure 5D). Similarly, genes described before to be repressed in 35S::E2FA/DPA overexpression lines with a role in cell enlargement and differentiation (Vandepoele et al, 2005), such as EXPANSIN 5 (Park et al, 2010) and ECERIFERUM 1 (CER1; Lai et al, 2007) were upregulated in the 35S::E2FAΔRB/DPA lines, compared with the WT, supporting the notion that both endocycle and cell expansion are accelerated in the leaves of the 35S::E2FAΔRB/DPA lines (Figure 5A and B; Supplementary Figures S9D and S11A).
To confirm that RBR1 indeed present on the promoters of the CCS52 genes where E2Fs can bind in vivo, we performed chromatin IP (ChIP) in 5 days old Arabidopsis roots by using RBR1-specific antibody. Both CCS52A1 and A2 gene contains exactly the same overlapping E2F-binding sites in close distance to their translational start codon (159–170 bp and 110–122 bp, respectively; Lammens et al, 2008). To test whether RBR1 binds to the E2F sites, we analysed three different regions in these promoters. As it is shown in Figure 5E, enrichment was detected by ChIP in a region spanning from 23 to 237 bp upstream of the ATG start codon in both CCS52 promoters correlating with the presence of putative E2F-binding sites. Interestingly, more CCS52A2 was precipitated by RBR1 than CCS52A1, suggesting that the level of RBR1 repression could be different on these genes in the root. In agreement, CCS52A2 expression is restricted only to few cells located in the root cap (Vanstraelen et al, 2009).
We conclude that E2FA, together with RBR1 forms a repressor complex required for inhibiting the activity of genes involved in, endocycle; CCS52A1 and A2 and cell expansion, differentiation, such as EXPANSIN 5, CER1, in order to maintain the diploid state and proliferation potential in meristems.
E2FA is part of an RBR1-dependent and -independent E2F regulatory network involved in defining the balance between proliferation and endocycle
Previously, it was shown that DEL1/E2FE represses the expression of CCS52A2 in proliferating cells, and thereby restrain CCS52A2 expression to cells exiting proliferation and undergoing endocycle (Lammens et al, 2008). This provides an RBR1-independent mechanism to repress the CCS52A genes in the meristem. E2FC was also shown to repress genes in G2 phase control, such as CYCB1;1, and to promote endocycle (del Pozo et al, 2006). How these genes are linked in a regulatory network to regulate the transition from cell proliferation to endocycle is little understood. Therefore, we investigated how the expression of other E2Fs during leaf development is effected in the e2fa-1 knockout mutant. We found a decreased DEL1/E2FE expression at 8 DAG and an increased level of DEL1–3/E2FD-F at 15 DAG in the e2fa-1 mutant compared with WT, indicating that E2FA is required for DEL1/E2FE expression in proliferating cells, while as cells leave the proliferative state during leaf development E2FA represses the expression of all DELs (Supplementary Figure S12). E2FC expression is elevated at all time points during development in the e2fa-1 mutant, suggesting that E2FA suppresses E2FC expression. Interestingly, in the e2fa-1 mutant E2FB expression changes relative to WT but this is developmental stage dependent. In summary, E2FA levels can influence the expression of other E2Fs and thereby these RBR1-dependent and -independent regulatory mechanisms are connected in a regulatory network.
Discussion
Plant development is largely post-embryonic, and relies on a pool of undifferentiated cells within the meristems. Several regulators are required for stem cell maintenance and this is the case for RBR1 that has a pivotal function in roots (Wildwater et al, 2005), and in the leaf (Borghi et al, 2010). Part of the RBR1 function depends on downstream targets such as E2FA that regulates cell proliferation within the meristem, whereas in post-mitotic cells it promotes endocycle (De Veylder et al, 2002; Inze and De Veylder, 2006). E2FA is most abundant in proliferating cells, but both E2FA transcripts (De Veylder et al, 2002) and protein can clearly be detected in differentiated cells known to be engaged in endocycle. The effect of E2FA on these two processes is dose-dependent; modest E2FA ectopic overexpression only boosts the endocycle, as indicated by the selective upregulation of S-phase related genes (de Jager et al, 2009), while strong ectopic overexpression disrupts tissue organisation by deregulating both cell proliferation and endocycle (De Veylder et al, 2002). By manipulating E2FA level within its own expression domain, we show that E2FA is necessary and sufficient for both processes and thus it coordinates growth through maintaining cell proliferation within the meristems and promoting endocycle and cell enlargement outside the meristems.
RBR1 forms a stable complex with E2FA that is not dissociated when CYCD3;1 level is high
RB is a tumour suppressor gene, involved in the repression of cell proliferation in animals (van den Heuvel and Dyson, 2008). In plants, the rbr1 knockout mutant and RBR1 silencing lines show cell type-specific overproliferation (Ebel et al, 2004; Park et al, 2005; Wildwater et al, 2005; Desvoyes et al, 2006; Jordan et al, 2007; Lageix et al, 2007; Chen et al, 2009, 2011; Borghi et al, 2010; Johnston et al, 2010; Gutzat et al, 2011), while ectopic overexpression of RBR1 induces cell differentiation (Wildwater et al, 2005; Wyrzykowska et al, 2006). However, the function of the different RBR1–E2F complexes is still not well understood. Three E2F transcription factors have the ability to form a complex with RBR1 in Arabidopsis, E2FA, E2FB and E2FC (Van Leene et al, 2010). Here, we show that in proliferating cells, the complexes between RBR1 and the two ‘activator-type’ E2Fs, E2FA and E2FB are differently regulated by CYCD3;1. The abundance of the RBR1–E2FB complex is responsive to CYCD3;1 levels and in agreement with the model of E2F control by RBR1, disrupted upon RBR1 phosphorylation (Figure 6A). CYCD3;1 overexpression results in overproliferation, whereas its knockout compromises cell division (Dewitte et al, 2003, 2007), suggesting that CYCD3;1 acts on RBR1–E2FB. We show that sucrose availability is linked to RBR1 phosphorylation dependent on CYCD3;1 and KRP2 levels. When sucrose is abundant, the free E2FB is high leading to proliferation, while in sucrose limiting conditions, E2FB is associated with RBR1. In contrast, the abundance of the RBR1–E2FA complex was found to be increased upon CYCD3;1 overexpression and the amount of this complex correlated with the extent of proliferation, and was high in the presence rather than in the absence of sucrose. These data suggest that either the RBR1–E2FA complex is not disrupted upon RBR1 phosphorylation by the CYCD3;1/CDKA;1 or that the RBR1–E2FA complex is protected from the CYCD3;1-CDK phosphorylation activity. The first hypothesis would be compatible with recent findings in the unicellular alga Chlamydomonas, where the phosphorylated MAT3/RBR remains bound to the E2F1/DP1 heterodimers during all stages of the cell cycle (Olson et al, 2010). Also in animal cells, DNA damage promotes the formation of stable pRB–E2F1 complex in proliferating cells regardless of the RB phosphorylation status (Ianari et al, 2009). Our results, however, show that in Arabidopsis, RBR1 phosphorylated by CYCD–CDK on the conserved Ser807/811 sites cannot be detected in association with E2FA. Therefore, we suggest that the RBR1 bound to E2FA escapes from CYCD–CDK phosphorylation. Elevating E2FA levels proportionally increases the RBR1–E2FA complex and leads to a higher proportion of unphosphorylated RBR1, providing additional evidence that this RBR1–E2FA complex is resistant to phosphorylation by CYCD3;1-CDK, probably due to its participation in a large multi-subunit complex in which phosphorylation sites are hidden.
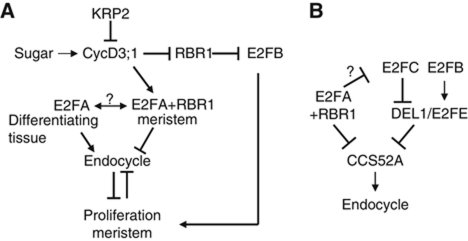
Figure 6.
Model explaining the functions of E2FA and E2FB in proliferating and in endocycling cells. (A) RBR1 represses E2FB activity, which is released by CYCD3;1- and KRP2-regulated RBR1 kinase in a sucrose-dependent manner. The RBR1-free E2FB stimulates cell proliferation by activating genes involved in cell-cycle progression. E2FA–RBR1 complex, however, is more stable in proliferating cells present in the meristem and represses genes involved in the switch from mitosis to endocycle (in B). E2FA is released from its RBR1-bound form by an unknown mechanism and stimulate endocycle in cells committed for differentiation. (B) E2FA–RBR1 complex inhibit endocycle in actively dividing cells by directly repressing the expression of CCS52A genes. E2FC is also repressed by E2FA, though it is not yet clear whether this is direct. E2FC is known to promote endocycle, and opposing E2FB in this activity. Recently, it was shown that the balance of E2FB/E2FC is measured by direct transcriptional regulation of DEL1/E2FE (Berckmans et al, 2011a). Thus, endocycle is regulated by CCS52A through an interlinked RBR1-dependent and RBR1-independent E2F transcription factors.
The identification of evolutionary conserved pocket protein/E2F complexes in Drosophila has provided new insight into E2F-mediated gene-regulation (Dimova et al, 2003; van den Heuvel and Dyson, 2008). Moreover, the animal multi-subunit E2F–RB complexes alternatively called dREAM (named after the complex of Drosophila melanogaster RBF, E2F and MYB) or Myb-MuvB (MMB) are present in actively dividing cells regardless of the presence of active RB-kinases and their major functions are to repress developmental genes and regulate G2- and M-phase of the cell cycle (Korenjak et al, 2004; Korenjak and Brehm, 2005; Litovchick et al, 2007; Knight et al, 2009). Components of putative dREAM complexes appear to be conserved in plants. There are five related Arabidopsis MYB3R genes; two of them were shown to regulate mitotic gene expression (Haga et al, 2007). Also in plants, E2FA and RBR1 have been reported to cause chromosome instability (Henriques et al, 2010; Johnston et al, 2010; Chen et al, 2011). Therefore, E2FA and RBR1 could participate in the plant homologue of the dREAM complex to regulate different aspects of cell division and development, although further studies are required to test this hypothesis.
RBR1–E2FA complex functions in maintaining proliferation through repression of cell differentiation
Overexpression of E2FA together with DPA dramatically represses cellular differentiation and promotes stem cell maintenance (De Veylder et al, 2002; Wildwater et al, 2005). It was suggested that the largely overexpressed E2FA can escape from RBR1 repression leading to uncontrolled cell proliferation and delayed differentiation. Similar maintenance of the undifferentiated state has been found with overexpression of a number of other positive cell-cycle regulators, such as CYCD3;1, CYCA3;2 and CYCA2;3, which also simultaneously inhibits the entry into endocycle (Dewitte et al, 2003; Yu et al, 2003; Imai et al, 2006). How cell cycle promotion impinges on differentiation is not well understood. In animal cells, differentiation was suggested to be a default pathway that must be inhibited in stem cells and in cells maintaining their proliferative state (Orford and Scadden, 2008).
In animal systems, the E2F and RB family members are known to provide a broadly utilised switch that not only controls the temporal expression of genes for cell proliferation, but also represses developmentally regulated genes independent from cell proliferation (van den Heuvel and Dyson, 2008). Genome-wide expression studies and in silico promoter analysis for the presence of E2F elements revealed a battery of Arabidopsis genes that are repressed by E2FA (Vlieghe et al, 2003; Vandepoele et al, 2005; de Jager et al, 2009). Surprisingly, none of the downregulated genes has cell cycle related functions, but they are involved in cell elongation, development and metabolism. Recently, it was shown that RBR1 not only regulates cell-cycle genes, but is required to repress late embryonic genes during seedling development (Gutzat et al, 2011). We have tested the regulation of a selective set among these genes, such as the CCS52A1, CCS52A2, CER1 and EXPANSIN5, and found all to be upregulated in the 35S::E2FAΔRB/DPA line, suggesting that the E2FA–RBR1 complex, formed in proliferating cells, is involved in the repression of these differentiation genes. Supporting the role of RBR1 in the repression of endocycle is the increased ploidy level found in cells where RBR1 function is inhibited through the expression and binding of viral proteins (Desvoyes et al, 2006; Jordan et al, 2007). However, reduced RBR1 level can also lead to elevated ploidy through endomitosis (Henriques et al, 2010; Johnston et al, 2010). We show that increasing RBR1 levels within its own expression domain suppresses the onset of endocycle. Endocycle is also constrained in Drosophila follicle cells via repression of genes for origin recognition complex by RBF1, E2F1 and E2F2, and thus appears to represent an evolutionary conserved mechanism present in animals and plants (Cayirlioglu et al, 2003).
CCS52A1 and CCS52A2 are two activators of the APC, whose functions are to regulate the developmental switch between mitosis and endocycle by stimulating the degradation of mitotic cyclins (Fulop et al, 2005). CCS52A1 expression is excluded from the root meristem; accumulates from the first elongating cell upwards and the ccs52a1 mutant shows a delayed exit from mitosis (Larson-Rabin et al, 2009; Vanstraelen et al, 2009). Both the expression and the phenotypic consequences of E2FA silencing and overexpression are opposite to those of CCS52A1, suggesting that CCS52A1 could be an important target for the E2FA–RBR1 repressor complex. Both CCS52A1 and CCS52A2 promoters contain E2F-binding sites, and we show by ChIP that RBR1 is present on these promoters in the vicinity of E2F elements. Interestingly, the atypical E2F repressor protein, DEL1/E2FE only regulates CCS52A2 during leaf development (Lammens et al, 2008). Elevated E2FA levels repress both CCS52A1 and CCS52A2, while in the e2fa-1 knockout mutant both CCS52As are elevated, though CCS52A2 at an earlier time point during leaf development. These suggest that there are both RBR1-dependent and RBR1-independent repressor complexes that play roles to regulate the onset of endocycle through the regulation of CCS52A genes (Figure 6B).
E2FA promotes cell growth through stimulating endocycle
The proliferation and the endocycle functions of E2FA are confined to spatially distinct tissues, and the 35S::E2FAΔRB/DPA line allowed us to separate these roles. The compromised maintenance of the proliferative state in the 35S::E2FAΔRB/DPA line suggests that RBR1 binding is required for proliferation, while for endocycle E2FA needs to escape from RBR1 control to bind DNA. Though RBR1 itself is most abundant in proliferating cells, it is more ubiquitous than E2FA in differentiating cells. The exact mechanism by which E2FA function is uncoupled from RBR1 control to regulate endocycle is not clear. One possibility would be the preferential association of RBR1 with another E2F, as cells leave the meristem and E2FA–RBR1 complex diminish. This transition coincides with the decrease of CYCD3;1 and consequently the dephosphorylation of RBR1, that favours the interaction of RBR1 with E2FB. Additionally, other regulators, such as the Arabidopsis S6K1, could play a role at the transition zone to promote the RBR1–E2FB interaction and cell-cycle repression (Henriques et al, 2010). Alternatively, E2FA might promote endocycle by direct interaction with the ORC (origin of replication complex), as it was found in Drosophila (Royzman et al, 1999; Bosco et al, 2001; Wells et al, 2003).
In Arabidopsis, there are 6 E2F transcription factors; E2FA, E2FB and E2FC are able to interact with RBR1, while DEL1/E2FE, DEL2/E2FD and DEL3/E2FF are RBR1 independent. The promoters of all except E2FA contain putative E2F elements, and indeed it was shown that they can cross-regulate each others expression and thereby constitute a gene regulatory network. It was shown that E2FA can induce E2FB expression (Sozzani et al, 2006), while DEL2/E2FD increases the expression of E2FA, E2FB and DEL1/E2FE (Sozzani et al, 2010). We show that E2FA regulates DEL1/E2FE and E2FC expression (Figure 6B). Recently, we have shown that E2FB and E2FC oppositely regulates DEL1/E2FE expression, while E2FA does not bind to the DEL1 promoter (Berckmans et al, 2011a). It is possible that E2FA therefore indirectly regulate DEL1 through modifying the expression of E2FC and E2FB. In agreement, E2FC expression was increased in the e2fa-1 knockout plant. Thus, there is an interconnected RBR1–E2F gene regulatory network that regulates the balance between cell proliferation and endocycle, cell differentiation (Figure 6).
In conclusion, E2FA promotes cell proliferation indirectly, through maintaining the dedifferentiated state of cells in complex with RBR1. In differentiated cells, there is a distinct mechanism where E2FA is required to promote endocycle that regulates cellular growth. Thus, E2FA impinges on the two major mechanisms that determine plant organ growth.
Materials and methods
Plant material and growth conditions
Arabidopsis thaliana ecotype Columbia (WT-Col) or Landsberg erecta (WT-Ler) was used as control plants. For all analysis, plants were grown under 16 h light/8 h dark photoperiod at 22°C in vitro on germination medium or on soil as described before (Henriques et al, 2010). The 35S::CYCD3;1 overexpressor line was kindly provided by James AH Murray (Cardiff University, Wales, UK). The cotyledons and the first leaf pairs of Arabidopsis WT-Col or the transgenic Arabidopsis lines (pE2FA::gE2FA–GFP; pRBR1::gRBR1–GFP; 35S::HA–E2FAΔRB/DPA; CYCD3;1-OE; e2fa-1-ko) were harvested from 6 to 16 days post-germination grown in vitro, and were flash frozen and stored at −80°C. Protoplasts were isolated from Arabidopsis cell suspension and the transformation for transient expression was carried out as described before (Magyar et al, 2005; Henriques et al, 2010). The T-DNA insertion mutant of E2FA (e2fa-1) has recently been described by Berckmans et al (2011b).
Plasmid constructs, generation of transgenic Arabidopsis plants
The construct of the pE2FA::gE2FA–GFP translational fusion has been described before (Henriques et al, 2010). To construct the pRBR1::gRBR1–GFP translational fusion, the RBR1 promoter and the genomic open reading frame fragments of RBR1 were amplified from Col-0 genomic DNA using the primer combinations described in Supplementary Table S2, and RBR1 genomic sequence was fused at the 3′ end with the coding sequence of GFP in a pGreenII-based pGII0125 destination vector (Galinha et al, 2007), by using the Invitrogen 3way gateway system (Invitrogen, USA). The HA epitope tagged C-terminal deletion mutant of E2FA (E2FAΔRB) missing a 65-amino-acid long region containing the conserved RB-binding motif was described (Magyar et al, 2000). E2FA and E2FB have been cloned into pDON-201 plasmid (Magyar et al, 2005) and they were further cloned into pK7GWIWG2(I), a gateway RNA-interference binary destination vector (Karimi et al, 2002). The HA-tagged E2FA and E2FB as well as the c-myc-tagged DPA has been constructed previously (Magyar et al, 2005).
Transgenic Arabidopsis plants were generated by using the floral-dip method for Agrobacterium-mediated transformation as described (Zhang et al, 2006) and mutants were selected on the presence of the appropriate antibiotic. A single T-DNA insertion line containing the 35S::HA–E2FAΔRB insert was identified and homozygous T2 segregation was selected on kanamycin-containing medium. Homozygous 35S::HA–E2FAΔRB and the 35S::DPA (De Veylder et al, 2002) was crossed and double overexpressor lines were selected on medium containing kanamycin and hygromycin. Two T2 lines were selected later named as 7 and 8; line 7 was heterozygous for both E2FAΔRB, and DPA while line 8 was heterozygous for E2FAΔRB but homozygous for DPA. Double overexpressor 35S::HA–E2FAΔRB/35S::Myc–DPAΔDB transgenic lines were generated by crossing the homozygous lines. The construct for the RNR2 promoter-GUS was described (Horvath et al, 2006).
RNA extraction and quantitative RT–PCR
Whole seedlings or the first leaf pair was snap frozen in liquid nitrogen. All samples were ground to fine powder using a chilled (with liquid nitrogen) pestle and mortar. RNA was extracted using the RNeasy Plant Mini kit (Qiagen, UK). The concentration of each RNA sample was calculated using the nanodrop 100 (Labtech, UK). Each sample was normalised to a concentration of 0.5 μg RNA and ran on a 0.8% agarose gel to check for quality of RNA. cDNA was synthesised using 1 μg of RNA using the QuantiTect Reverse transcription kit (Qiagen). Real-time amplification in the presence of SYBR Green was performed using a BioScript PCR kit (Bioline, UK) according to the manufacturer's instructions in a Rotor-Gene 6000 apparatus (Corbet Life Science, Australia). All data were normalised to housekeeping genes (PIP2 or actin) and the calculated efficiency was added to the analysis. All reactions took place in triplicate.
Cytological analysis, confocal microscopy, flow cytometry analysis
To visualize the leaf epidermis, gel cast was made of the leaf surface (the adaxial side of the first leaf pair) and then observed under DIC light microscope Nikon Optiphot 2 as described (Horiguchi et al, 2006). Laser scanning confocal microscopy (Olympus FV1000) was used to examine roots and leaves of 4- to 12-day-old plants stained with 20 μg/ml propidium iodide. For flow cytometry measurements, the first leaf pair were collected and chopped with razor blades in nuclei extraction buffer and stained with DAPI as described before (Magyar et al, 2005). Flow cytometry data were obtained using a Partec PAS2 Particle Analysing system (Partec, Germany).
Immunoprecipitations, immunoblotting
IP and immunoblotting assays have been carried out as described (Henriques et al, 2010). The following antibodies have been used in co-IP experiments: anti-E2FA polyclonal rabbit antibody (Takahashi et al, 2008), anti-DPA (Magyar et al, 2005) and anti-DPB (Umbrasaite et al, 2010), and anti-E2FB polyclonal rabbit antibodies (Magyar et al, 2005), anti-GFP monoclonal mouse (Roche) or GFP-Trap coupled to magnetic beads (ChromoTek) or anti-GFP polyclonal rabbit antibodies (AbCam), and anti-HA monoclonal mouse (Roche) or anti-HA polyclonal rabbit (Upstate), anti-phospho-specific Rb (Ser807/811) polyclonal rabbit antibody (Cell Signaling Tech). Generally, 400–800 μg of total protein extract derived from a week or 2 weeks old seedlings or leaf samples (the first leaf pairs derived from 8 to 16 days old seedlings) were used in co-IP experiments. Precipitated material was separated on 10% SDS–PAGE together with equal loading of 20–25 μg of total protein extract as input material and blotted to PVDF membrane. Antibodies used in immunoblotting experiments: anti-RBR1 (Horvath et al, 2006), mouse monoclonal anti-PSTAIRE (CDKA;1 specific; Sigma), rabbit polyclonal antibody anti-CDKB1;1 (Magyar et al, 2005).
Chromatin immunoprecipitation
ChIP were carried out on root material of 5 days old Col-0 seedlings. IP was performed on sonicated chromatin fragments of 500–700 bp in the presence and absence (negative control) of antibody, specific for AtRBR1 protein as described in Horvath et al (2006). To assay the in vivo binding of the AtRBR1 protein to the promoter regions of the CCS52A1 and CCS52A2, genes using Q-RT–PCRs primers were designed to amplify fragments between 100–200 bps that span along 1.4 and 1.5 kb regions, respectively. The primers tiling along the relevant promoter regions and primers used as control showed in each cases same amplification efficiency. The primers are listed in Supplementary Table S3. To calculate the enrichment on a promoter region, the relative ratio of the amplified DNA coming from chromatin samples after IP in the presence and absence of antibody was taken. As a negative control, a random intergenic region (IR; between At3g03660-70) and as a positive control PCNA1 (At1g07370) was used (Kosugi and Ohashi, 2003).
Supplementary Material
Acknowledgments
We thank JM Murray, Cardiff, UK for the CYCD3;1 overexpression line. This work was supported by the BBSRC BB/D017599/1 to LB and ZM at RH; as well as by the young investigator award to ZM at BRC, Szeged, Hungary. RH was funded by the Fundação para a Ciência e Tecnologia (SFRH/BPD/7164/2001).
Author contributions: ZM designed and performed the majority of the experiments in Figures 1, 2, 3, 4 and 5 and Supplementary Figures S1–S13; BH produced the pRBR1::gRBR1–GFP lines that was instrumental in Figure 1 and Supplementary Figures S1, S4 and S6 contributed in the construction of pE2FA::gE2FA–GFP line used in Figures 1, 4 and 5 and Supplementary Figures S1, S4 and S5 and performed the chromatin immunoprecipitation (ChIP) experiment presented in Figure 5E, discussed the data, commented and contributed to the paper; SK performed the Q-RT–PCR experiments for Figures 2, 4 and 5 and Supplementary Figures S2 and S11. BM has conducted experiments for Figure 5C and Supplementary Figures S5 and S11D. RH contributed to generate the 35S::E2FAΔRB/DPA crosses used in Figure 5, Supplementary Figures S9–S11. LBakó together with BH has established and performed the ChIP. LDV participated in the initial design of the experiments, contributed materials and plant lines. BS provided valuable ideas on analysing root development, discussed the data, commented and contributed to the paper. LB jointly with ZM conceived the study, designed the experiments, performed flow cytometry measurements and wrote the manuscript.
Footnotes
The authors declare that they have no conflict of interest.
References
- Abraham E, Miskolczi P, Ayaydin F, Yu P, Kotogany E, Bako L, Otvos K, Horvath GV, Dudits D (2011) Immunodetection of retinoblastoma-related protein and its phosphorylated form in interphase and mitotic alfalfa cells. J Exp Bot 62: 2155–2168 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beemster GT, Vercruysse S, De Veylder L, Kuiper M, Inze D (2006) The Arabidopsis leaf as a model system for investigating the role of cell cycle regulation in organ growth. J Plant Res 119: 43–50 [DOI] [PubMed] [Google Scholar]
- Berckmans B, Lammens T, Van Den Daele H, Magyar Z, Bogre L, De Veylder L (2011a) Light-dependent regulation of DEL1 is determined by the antagonistic action of E2Fb and E2Fc. Plant Physiol 157: 1440–1451 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Berckmans B, Vassileva V, Schmid SP, Maes S, Parizot B, Naramoto S, Magyar Z, Kamei CL, Koncz C, Bogre L, Persiau G, De Jaeger G, Friml J, Simon R, Beeckman T, De Veylder L (2011b) Auxin-dependent cell cycle reactivation through transcriptional regulation of Arabidopsis E2Fa by lateral organ boundary proteins. Plant Cell 23: 3671–3683 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bogre L, Magyar Z, Lopez-Juez E (2008) New clues to organ size control in plants. Genome Biol 9: 226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Borghi L, Gutzat R, Futterer J, Laizet Y, Hennig L, Gruissem W (2010) Arabidopsis retinoblastoma-related is required for stem cell maintenance, cell differentiation, and lateral organ production. Plant Cell 22: 1792–1811 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bosco G, Du W, Orr-Weaver TL (2001) DNA replication control through interaction of E2F-RB and the origin recognition complex. Nat Cell Biol 3: 289–295 [DOI] [PubMed] [Google Scholar]
- Boudolf V, Lammens T, Boruc J, Van Leene J, Van Den Daele H, Maes S, Van Isterdael G, Russinova E, Kondorosi E, Witters E, De Jaeger G, Inze D, De Veylder L (2009) CDKB1;1 forms a functional complex with CYCA2;3 to suppress endocycle onset. Plant Physiol 150: 1482–1493 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boudolf V, Vlieghe K, Beemster GT, Magyar Z, Torres Acosta JA, Maes S, Van Der Schueren E, Inze D, De Veylder L (2004) The plant-specific cyclin-dependent kinase CDKB1;1 and transcription factor E2Fa-DPa control the balance of mitotically dividing and endoreduplicating cells in Arabidopsis. Plant Cell 16: 2683–2692 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bramsiepe J, Wester K, Weinl C, Roodbarkelari F, Kasili R, Larkin JC, Hulskamp M, Schnittger A (2010) Endoreplication controls cell fate maintenance. PLoS Genet 6: e1000996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Breuer C, Ishida T, Sugimoto K (2010) Developmental control of endocycles and cell growth in plants. Curr Opin Plant Biol 13: 654–660 [DOI] [PubMed] [Google Scholar]
- Breuer C, Kawamura A, Ichikawa T, Tominaga-Wada R, Wada T, Kondou Y, Muto S, Matsui M, Sugimoto K (2009) The trihelix transcription factor GTL1 regulates ploidy-dependent cell growth in the Arabidopsis trichome. Plant Cell 21: 2307–2322 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cayirlioglu P, Ward WO, Silver Key SC, Duronio RJ (2003) Transcriptional repressor functions of Drosophila E2F1 and E2F2 cooperate to inhibit genomic DNA synthesis in ovarian follicle cells. Mol Cell Biol 23: 2123–2134 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen Z, Hafidh S, Poh SH, Twell D, Berger F (2009) Proliferation and cell fate establishment during Arabidopsis male gametogenesis depends on the Retinoblastoma protein. Proc Natl Acad Sci USA 106: 7257–7262 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen Z, Higgins JD, Hui JT, Li J, Franklin FC, Berger F (2011) Retinoblastoma protein is essential for early meiotic events in Arabidopsis. EMBO J 30: 744–755 [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Jager SM, Scofield S, Huntley RP, Robinson AS, den Boer BG, Murray JA (2009) Dissecting regulatory pathways of G1/S control in Arabidopsis: common and distinct targets of CYCD3;1, E2Fa and E2Fc. Plant Mol Biol 71: 345–365 [DOI] [PubMed] [Google Scholar]
- De Veylder L, Beeckman T, Beemster GT, de Almeida Engler J, Ormenese S, Maes S, Naudts M, Van Der Schueren E, Jacqmard A, Engler G, Inze D (2002) Control of proliferation, endoreduplication and differentiation by the Arabidopsis E2Fa-DPa transcription factor. EMBO J 21: 1360–1368 [DOI] [PMC free article] [PubMed] [Google Scholar]
- del Pozo JC, Diaz-Trivino S, Cisneros N, Gutierrez C (2006) The balance between cell division and endoreplication depends on E2FC-DPB, transcription factors regulated by the ubiquitin-SCFSKP2A pathway in Arabidopsis. Plant Cell 18: 2224–2235 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Desvoyes B, Ramirez-Parra E, Xie Q, Chua NH, Gutierrez C (2006) Cell type-specific role of the retinoblastoma/E2F pathway during Arabidopsis leaf development. Plant Physiol 140: 67–80 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dewitte W, Riou-Khamlichi C, Scofield S, Healy JM, Jacqmard A, Kilby NJ, Murray JA (2003) Altered cell cycle distribution, hyperplasia, and inhibited differentiation in Arabidopsis caused by the D-type cyclin CYCD3. Plant Cell 15: 79–92 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dewitte W, Scofield S, Alcasabas AA, Maughan SC, Menges M, Braun N, Collins C, Nieuwland J, Prinsen E, Sundaresan V, Murray JA (2007) Arabidopsis CYCD3 D-type cyclins link cell proliferation and endocycles and are rate-limiting for cytokinin responses. Proc Natl Acad Sci USA 104: 14537–14542 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dimova DK, Stevaux O, Frolov MV, Dyson NJ (2003) Cell cycle-dependent and cell cycle-independent control of transcription by the Drosophila E2F/RB pathway. Genes Dev 17: 2308–2320 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Doonan JH, Sablowski R (2010) Walls around tumours—why plants do not develop cancer. Nat Rev Cancer 10: 794–802 [DOI] [PubMed] [Google Scholar]
- Ebel C, Mariconti L, Gruissem W (2004) Plant retinoblastoma homologues control nuclear proliferation in the female gametophyte. Nature 429: 776–780 [DOI] [PubMed] [Google Scholar]
- Fulop K, Tarayre S, Kelemen Z, Horvath G, Kevei Z, Nikovics K, Bako L, Brown S, Kondorosi A, Kondorosi E (2005) Arabidopsis anaphase-promoting complexes: multiple activators and wide range of substrates might keep APC perpetually busy. Cell Cycle 4: 1084–1092 [PubMed] [Google Scholar]
- Galinha C, Hofhuis H, Luijten M, Willemsen V, Blilou I, Heidstra R, Scheres B (2007) PLETHORA proteins as dose-dependent master regulators of Arabidopsis root development. Nature 449: 1053–1057 [DOI] [PubMed] [Google Scholar]
- Gutierrez C (2009) The Arabidopsis cell division cycle. The Arabidopsis Book 7: e0120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gutzat R, Borghi L, Futterer J, Bischof S, Laizet Y, Hennig L, Feil R, Lunn J, Gruissem W (2011) Retinoblastoma-related protein controls the transition to autotrophic plant development. Development 138: 2977–2986 [DOI] [PubMed] [Google Scholar]
- Haga N, Kato K, Murase M, Araki S, Kubo M, Demura T, Suzuki K, Muller I, Voss U, Jurgens G, Ito M (2007) R1R2R3-Myb proteins positively regulate cytokinesis through activation of KNOLLE transcription in Arabidopsis thaliana. Development 134: 1101–1110 [DOI] [PubMed] [Google Scholar]
- Henriques R, Magyar Z, Monardes A, Khan S, Zalejski C, Orellana J, Szabados L, de la Torre C, Koncz C, Bogre L (2010) Arabidopsis S6 kinase mutants display chromosome instability and altered RBR1-E2F pathway activity. EMBO J 29: 2979–2993 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Horiguchi G, Fujikura U, Ferjani A, Ishikawa N, Tsukaya H (2006) Large-scale histological analysis of leaf mutants using two simple leaf observation methods: identification of novel genetic pathways governing the size and shape of leaves. Plant J 48: 638–644 [DOI] [PubMed] [Google Scholar]
- Horvath BM, Magyar Z, Zhang Y, Hamburger AW, Bako L, Visser RG, Bachem CW, Bogre L (2006) EBP1 regulates organ size through cell growth and proliferation in plants. EMBO J 25: 4909–4920 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huntley R, Healy S, Freeman D, Lavender P, de Jager S, Greenwood J, Makker J, Walker E, Jackman M, Xie Q, Bannister AJ, Kouzarides T, Gutierrez C, Doonan JH, Murray JA (1998) The maize retinoblastoma protein homologue ZmRb-1 is regulated during leaf development and displays conserved interactions with G1/S regulators and plant cyclin D (CycD) proteins. Plant Mol Biol 37: 155–169 [DOI] [PubMed] [Google Scholar]
- Ianari A, Natale T, Calo E, Ferretti E, Alesse E, Screpanti I, Haigis K, Gulino A, Lees JA (2009) Proapoptotic function of the retinoblastoma tumor suppressor protein. Cancer Cell 15: 184–194 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Imai KK, Ohashi Y, Tsuge T, Yoshizumi T, Matsui M, Oka A, Aoyama T (2006) The A-type cyclin CYCA2;3 is a key regulator of ploidy levels in Arabidopsis endoreduplication. Plant Cell 18: 382–396 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Inze D, De Veylder L (2006) Cell cycle regulation in plant development. Annu Rev Genet 40: 77–105 [DOI] [PubMed] [Google Scholar]
- Johnston AJ, Kirioukhova O, Barrell PJ, Rutten T, Moore JM, Baskar R, Grossniklaus U, Gruissem W (2010) Dosage-sensitive function of retinoblastoma related and convergent epigenetic control are required during the Arabidopsis life cycle. PLoS Genet 6: e1000988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jordan CV, Shen W, Hanley-Bowdoin LK, Robertson DN (2007) Geminivirus-induced gene silencing of the tobacco retinoblastoma-related gene results in cell death and altered development. Plant Mol Biol 65: 163–175 [DOI] [PubMed] [Google Scholar]
- Karimi M, Inze D, Depicker A (2002) GATEWAY vectors for Agrobacterium-mediated plant transformation. Trends Plant Sci 7: 193–195 [DOI] [PubMed] [Google Scholar]
- Knight AS, Notaridou M, Watson RJ (2009) A Lin-9 complex is recruited by B-Myb to activate transcription of G2/M genes in undifferentiated embryonal carcinoma cells. Oncogene 28: 1737–1747 [DOI] [PubMed] [Google Scholar]
- Korenjak M, Brehm A (2005) E2F-Rb complexes regulating transcription of genes important for differentiation and development. Curr Opin Genet Dev 15: 520–527 [DOI] [PubMed] [Google Scholar]
- Korenjak M, Taylor-Harding B, Binne UK, Satterlee JS, Stevaux O, Aasland R, White-Cooper H, Dyson N, Brehm A (2004) Native E2F/RBF complexes contain Myb-interacting proteins and repress transcription of developmentally controlled E2F target genes. Cell 119: 181–193 [DOI] [PubMed] [Google Scholar]
- Kosugi S, Ohashi Y (2003) Constitutive E2F expression in tobacco plants exhibits altered cell cycle control and morphological change in a cell type-specific manner. Plant Physiol 132: 2012–2022 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lageix S, Catrice O, Deragon JM, Gronenborn B, Pelissier T, Ramirez BC (2007) The nanovirus-encoded Clink protein affects plant cell cycle regulation through interaction with the retinoblastoma-related protein. J Virol 81: 4177–4185 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lai C, Kunst L, Jetter R (2007) Composition of alkyl esters in the cuticular wax on inflorescence stems of Arabidopsis thaliana cer mutants. Plant J 50: 189–196 [DOI] [PubMed] [Google Scholar]
- Lammens T, Boudolf V, Kheibarshekan L, Zalmas LP, Gaamouche T, Maes S, Vanstraelen M, Kondorosi E, La Thangue NB, Govaerts W, Inze D, De Veylder L (2008) Atypical E2F activity restrains APC/CCCS52A2 function obligatory for endocycle onset. Proc Natl Acad Sci USA 105: 14721–14726 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Larson-Rabin Z, Li Z, Masson PH, Day CD (2009) FZR2/CCS52A1 expression is a determinant of endoreduplication and cell expansion in Arabidopsis. Plant Physiol 149: 874–884 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee HO, Davidson JM, Duronio RJ (2009) Endoreplication: polyploidy with purpose. Genes Dev 23: 2461–2477 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leiva-Neto JT, Grafi G, Sabelli PA, Dante RA, Woo YM, Maddock S, Gordon-Kamm WJ, Larkins BA (2004) A dominant negative mutant of cyclin-dependent kinase A reduces endoreduplication but not cell size or gene expression in maize endosperm. Plant Cell 16: 1854–1869 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Litovchick L, Sadasivam S, Florens L, Zhu X, Swanson SK, Velmurugan S, Chen R, Washburn MP, Liu XS, DeCaprio JA (2007) Evolutionarily conserved multisubunit RBL2/p130 and E2F4 protein complex represses human cell cycle-dependent genes in quiescence. Mol Cell 26: 539–551 [DOI] [PubMed] [Google Scholar]
- Maere S, Heymans K, Kuiper M (2005) BiNGO: a Cytoscape plugin to assess overrepresentation of gene ontology categories in biological networks. Bioinformatics 21: 3448–3449 [DOI] [PubMed] [Google Scholar]
- Magyar Z (2008) Keeping the balance between proliferation and differentiation by the E2F transcriptional regulatory network is central to plant growth and development. In Plant Growth Signaling, Bogre L, Beemster GT (eds), 10. Berlin Heidelberg: Springer [Google Scholar]
- Magyar Z, Atanassova A, De Veylder L, Rombauts S, Inze D (2000) Characterization of two distinct DP-related genes from Arabidopsis thaliana. FEBS Lett 486: 79–87 [DOI] [PubMed] [Google Scholar]
- Magyar Z, De Veylder L, Atanassova A, Bako L, Inze D, Bogre L (2005) The role of the Arabidopsis E2FB transcription factor in regulating auxin-dependent cell division. Plant Cell 17: 2527–2541 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Menges M, Samland AK, Planchais S, Murray JA (2006) The D-type cyclin CYCD3;1 is limiting for the G1-to-S-phase transition in Arabidopsis. Plant Cell 18: 893–906 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakagami H, Kawamura K, Sugisaka K, Sekine M, Shinmyo A (2002) Phosphorylation of retinoblastoma-related protein by the cyclin D/cyclin-dependent kinase complex is activated at the G1/S-phase transition in tobacco. Plant Cell 14: 1847–1857 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakagami H, Sekine M, Murakami H, Shinmyo A (1999) Tobacco retinoblastoma-related protein phosphorylated by a distinct cyclin-dependent kinase complex with Cdc2/cyclin D in vitro. Plant J 18: 243–252 [DOI] [PubMed] [Google Scholar]
- Nieuwland J, Scofield S, Murray JA (2009) Control of division and differentiation of plant stem cells and their derivatives. Semin Cell Dev Biol 20: 1134–1142 [DOI] [PubMed] [Google Scholar]
- Olson BJ, Oberholzer M, Li Y, Zones JM, Kohli HS, Bisova K, Fang SC, Meisenhelder J, Hunter T, Umen JG (2010) Regulation of the chlamydomonas cell cycle by a stable, chromatin-associated retinoblastoma tumor suppressor complex. Plant Cell 22: 3331–3347 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Orford KW, Scadden DT (2008) Deconstructing stem cell self-renewal: genetic insights into cell-cycle regulation. Nat Rev Genet 9: 115–128 [DOI] [PubMed] [Google Scholar]
- Park CH, Kim TW, Son SH, Hwang JY, Lee SC, Chang SC, Kim SH, Kim SW, Kim SK (2010) Brassinosteroids control AtEXPA5 gene expression in Arabidopsis thaliana. Phytochemistry 71: 380–387 [DOI] [PubMed] [Google Scholar]
- Park JA, Ahn JW, Kim YK, Kim SJ, Kim JK, Kim WT, Pai HS (2005) Retinoblastoma protein regulates cell proliferation, differentiation, and endoreduplication in plants. Plant J 42: 153–163 [DOI] [PubMed] [Google Scholar]
- Ramirez-Parra E, Frundt C, Gutierrez C (2003) A genome-wide identification of E2F-regulated genes in Arabidopsis. Plant J 33: 801–811 [DOI] [PubMed] [Google Scholar]
- Rossignol P, Stevens R, Perennes C, Jasinski S, Cella R, Tremousaygue D, Bergounioux C (2002) AtE2F-a and AtDP-a, members of the E2F family of transcription factors, induce Arabidopsis leaf cells to re-enter S phase. Mol Genet Genomics 266: 995–1003 [DOI] [PubMed] [Google Scholar]
- Royzman I, Austin RJ, Bosco G, Bell SP, Orr-Weaver TL (1999) ORC localization in Drosophila follicle cells and the effects of mutations in dE2F and dDP. Genes Dev 13: 827–840 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sozzani R, Maggio C, Giordo R, Umana E, Ascencio-Ibanez JT, Hanley-Bowdoin L, Bergounioux C, Cella R, Albani D (2010) The E2FD/DEL2 factor is a component of a regulatory network controlling cell proliferation and development in Arabidopsis. Plant Mol Biol 72: 381–395 [DOI] [PubMed] [Google Scholar]
- Sozzani R, Maggio C, Varotto S, Canova S, Bergounioux C, Albani D, Cella R (2006) Interplay between Arabidopsis activating factors E2Fb and E2Fa in cell cycle progression and development. Plant Physiol 140: 1355–1366 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takahashi N, Lammens T, Boudolf V, Maes S, Yoshizumi T, De Jaeger G, Witters E, Inze D, De Veylder L (2008) The DNA replication checkpoint aids survival of plants deficient in the novel replisome factor ETG1. EMBO J 27: 1840–1851 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Uemukai K, Iwakawa H, Kosugi S, de Uemukai S, Kato K, Kondorosi E, Murray JA, Ito M, Shinmyo A, Sekine M (2005) Transcriptional activation of tobacco E2F is repressed by co-transfection with the retinoblastoma-related protein: cyclin D expression overcomes this repressor activity. Plant Mol Biol 57: 83–100 [DOI] [PubMed] [Google Scholar]
- Umbrasaite J, Schweighofer A, Kazanaviciute V, Magyar Z, Ayatollahi Z, Unterwurzacher V, Choopayak C, Boniecka J, Murray JA, Bogre L, Meskiene I (2010) MAPK phosphatase AP2C3 induces ectopic proliferation of epidermal cells leading to stomata development in Arabidopsis. PLoS One 5: e15357. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van den Heuvel S, Dyson NJ (2008) Conserved functions of the pRB and E2F families. Nat Rev Mol Cell Biol 9: 713–724 [DOI] [PubMed] [Google Scholar]
- Van Leene J, Hollunder J, Eeckhout D, Persiau G, Van De Slijke E, Stals H, Van Isterdael G, Verkest A, Neirynck S, Buffel Y, De Bodt S, Maere S, Laukens K, Pharazyn A, Ferreira PC, Eloy N, Renne C, Meyer C, Faure JD, Steinbrenner J et al. (2010) Targeted interactomics reveals a complex core cell cycle machinery in Arabidopsis thaliana. Mol Syst Biol 6: 397. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vandepoele K, Vlieghe K, Florquin K, Hennig L, Beemster GT, Gruissem W, Van de Peer Y, Inze D, De Veylder L (2005) Genome-wide identification of potential plant E2F target genes. Plant Physiol 139: 316–328 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vanstraelen M, Baloban M, Da Ines O, Cultrone A, Lammens T, Boudolf V, Brown SC, De Veylder L, Mergaert P, Kondorosi E (2009) APC/C-CCS52A complexes control meristem maintenance in the Arabidopsis root. Proc Natl Acad Sci USA 106: 11806–11811 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Verkest A, Manes CL, Vercruysse S, Maes S, Van Der Schueren E, Beeckman T, Genschik P, Kuiper M, Inze D, De Veylder L (2005a) The cyclin-dependent kinase inhibitor KRP2 controls the onset of the endoreduplication cycle during Arabidopsis leaf development through inhibition of mitotic CDKA;1 kinase complexes. Plant Cell 17: 1723–1736 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Verkest A, Weinl C, Inze D, De Veylder L, Schnittger A (2005b) Switching the cell cycle. Kip-related proteins in plant cell cycle control. Plant Physiol 139: 1099–1106 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vlieghe K, Vuylsteke M, Florquin K, Rombauts S, Maes S, Ormenese S, Van Hummelen P, Van de Peer Y, Inze D, De Veylder L (2003) Microarray analysis of E2Fa-DPa-overexpressing plants uncovers a cross-talking genetic network between DNA replication and nitrogen assimilation. J Cell Sci 116: 4249–4259 [DOI] [PubMed] [Google Scholar]
- Wells J, Yan PS, Cechvala M, Huang T, Farnham PJ (2003) Identification of novel pRb binding sites using CpG microarrays suggests that E2F recruits pRb to specific genomic sites during S phase. Oncogene 22: 1445–1460 [DOI] [PubMed] [Google Scholar]
- Wildwater M, Campilho A, Perez-Perez JM, Heidstra R, Blilou I, Korthout H, Chatterjee J, Mariconti L, Gruissem W, Scheres B (2005) The RETINOBLASTOMA-RELATED gene regulates stem cell maintenance in Arabidopsis roots. Cell 123: 1337–1349 [DOI] [PubMed] [Google Scholar]
- Wyrzykowska J, Schorderet M, Pien S, Gruissem W, Fleming AJ (2006) Induction of differentiation in the shoot apical meristem by transient overexpression of a retinoblastoma-related protein. Plant Physiol 141: 1338–1348 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu Y, Steinmetz A, Meyer D, Brown S, Shen WH (2003) The tobacco A-type cyclin, Nicta;CYCA3;2, at the nexus of cell division and differentiation. Plant Cell 15: 2763–2777 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang HS, Postigo AA, Dean DC (1999) Active transcriptional repression by the Rb-E2F complex mediates G1 arrest triggered by p16INK4a, TGFbeta, and contact inhibition. Cell 97: 53–61 [DOI] [PubMed] [Google Scholar]
- Zhang X, Henriques R, Lin SS, Niu QW, Chua NH (2006) Agrobacterium-mediated transformation of Arabidopsis thaliana using the floral dip method. Nat Protoc 1: 641–646 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.