Abstract
Control of DNA replication initiation is essential for normal cell growth. A unifying characteristic of DNA replication initiator proteins across the kingdoms of life is their distinctive AAA+ nucleotide-binding domains. The bacterial initiator DnaA assembles into a right-handed helical oligomer built upon interactions between neighbouring AAA+ domains, that in vitro stretches DNA to promote replication origin opening. The Bacillus subtilis protein Soj/ParA has previously been shown to regulate DnaA-dependent DNA replication initiation; however, the mechanism underlying this control was unknown. Here, we report that Soj directly interacts with the AAA+ domain of DnaA and specifically regulates DnaA helix assembly. We also provide critical biochemical evidence indicating that DnaA assembles into a helical oligomer in vivo and that the frequency of replication initiation correlates with the extent of DnaA oligomer formation. This work defines a significant new regulatory mechanism for the control of DNA replication initiation in bacteria.
Keywords: AAA+, DnaA, helix, ParA, Soj
Introduction
Successful replication and segregation of genetic information prior to cell division is essential for all living organisms. Loss of replication control can dramatically reduce an organism's competiveness in its environment, and in extreme cases can lead to unchecked cell proliferation or cell death. Throughout the kingdoms of life, chromosome duplication is instigated by DNA replication initiator protein complexes (Mott and Berger, 2007; Wigley, 2009; Kawakami and Katayama, 2010). A unifying characteristic of initiator proteins is their AAA+ nucleotide-binding domain, which is critical for their structure and function (Tucker and Sallai, 2007; Kawakami and Katayama, 2010).
Bacterial chromosomes are typically replicated bi-directionally from a single origin (oriC); an event orchestrated by the multi-domain initiator protein DnaA (Supplementary Figure S1; for review see Mott and Berger, 2007; Leonard and Grimwade, 2010). At the C-terminus, domain IV contains the helix-turn-helix and basic loop motifs required for specific double-stranded DNA-binding activity (Erzberger et al, 2002; Fujikawa et al, 2003). Domain III contains the AAA+ motif involved in ATP binding and ATP hydrolysis, as well as residues required for coordinating single-stranded DNA (Erzberger et al, 2002; Ozaki et al, 2008; Duderstadt et al, 2011). Domain II is a poorly conserved flexible linker (Abe et al, 2007; Molt et al, 2009) connecting domains III–IV to domain I, which acts as a hub for additional protein interactions and directs loading of the replicative helicase (Sutton et al, 1998).
Initiation of DNA replication in bacteria requires stepwise structural transitions, resulting in the assembly of DnaA into an active initiation complex (for reviews see Ozaki and Katayama, 2009; Leonard and Grimwade, 2010). Through domain IV, DnaA is thought to stably bind conserved nine basepair sequences (DnaA-boxes) in the oriC region throughout the cell cycle (Cassler et al, 1995). These founding DnaA proteins recruit further DnaA molecules onto neighbouring low-affinity binding sites via dimerization of domain I (Simmons et al, 2003; Miller et al, 2009). Additional ATP-bound DnaA proteins then assemble onto this platform to form a large nucleoprotein complex observable by electron microscopy as a particle wrapped in DNA (Funnell et al, 1987). This oligomeric structure may correspond to the right-handed helix, built via interactions between neighbouring AAA+ domains, which has been observed by X-ray crystallography (Carr and Kaguni, 2001; Erzberger et al, 2006). Amino-acid substitutions in DnaA that perturb helix formation in vitro inhibit replication origin unwinding in vitro and functionality in vivo (Duderstadt et al, 2010), and it has recently been proposed that the DnaA helix destabilizes an AT-rich sequence within the origin (the DNA unwinding element; DUE) by stretching one strand of the DNA duplex to promote origin opening (Duderstadt et al, 2011). This activity appears to be accompanied by a transition in DNA-binding modes from double-stranded to single-stranded; a result of domain IV engaging the AAA+ motif of a neighbouring monomer within the helical oligomer (Erzberger et al, 2006; Duderstadt et al, 2010). This compact helix is thought to continue onto the upper strand of the now single-stranded DUE via residues in domain III, stabilizing the DUE in its unwound state (Speck and Messer, 2001; Ozaki et al, 2008). Following open complex formation, DnaA directly recruits the replicative helicase onto the single-stranded DNA via interactions with domains I and III (Sutton et al, 1998; Abe et al, 2007). The remaining replisomal components are then recruited in a stepwise manner, which culminates in an active DNA replication complex.
There are several steps during initiation at which regulatory systems have been found to control bacterial DNA replication (for review see Katayama et al, 2010). DnaA binding to oriC can be inhibited either by protein occlusion (SeqA in Escherichia coli, Spo0A in Bacillus subtilis, and CtrA in Caulobacter crescentus), by spatial sequestration (YabA in B. subtilis), or by titration (datA in E. coli and DBCs in B. subtilis). DnaA assembly at oriC can be either stimulated (DiaA in E. coli and HobA in Helicobacter pylori) or repressed (SirA in B. subtilis) by the binding of regulatory proteins to domain I. Lastly, DnaA is inactivated following replisome formation through the stimulation of its ATP hydrolysis activity (Hda in E. coli and C. crescentus).
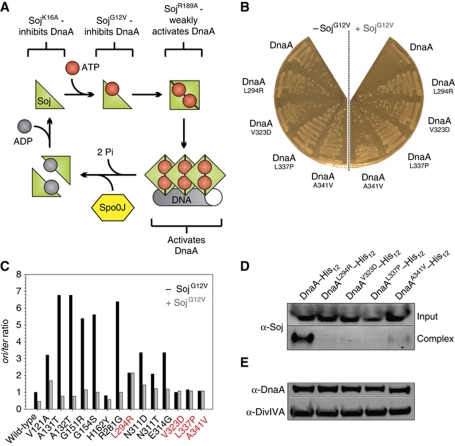
In a previous study we identified the highly conserved ParA protein (Soj) as a novel regulator of DNA replication in B. subtilis (Murray and Errington, 2008). Soj is a Walker-type ATPase that forms an ATP-dependent sandwich dimer that can bind DNA (Leonard et al, 2005). We have shown that the monomeric Soj protein inhibits DnaA, while dimerization of Soj switches the protein into an activator of DnaA (Scholefield et al, 2011). These results indicate that Soj acts as a molecular switch to control DnaA activity, with its opposing regulatory activities being dictated by its quaternary state. Detailed biochemical characterization of Soj proteins has identified amino-acid substitutions that arrest Soj quaternary changes at different steps (Figure 1A; Leonard and Grimwade, 2005; Hester and Lutkenhaus, 2007; Scholefield et al, 2011). Two separate substitutions inhibit Soj dimerization: SojK16A is unable to bind ATP, while SojG12V can bind ATP but cannot dimerize due to a steric clash in the dimerization interface; both of these proteins inhibit DnaA activity. The SojR189A substitution allows ATP-dependent dimerization but disrupts DNA-binding activity: this mutant protein is relatively inert, presumably because DNA-binding activity is required for Soj to efficiently activate DnaA (Scholefield et al, 2011).
Figure 1.
Specific mutations in dnaA either bypass or suppress the inhibition of DNA replication initiation by SojG12V. (A) Pathway of the Soj activity cycle. (B) Point mutations in DnaA introduced by error-prone PCR were found to overcome the small colony phenotype characteristic of SojG12V overexpression. Strains were grown on NA plates in the presence or absence of 1% xylose to induce sojG12V expression. Wild-type (HM524), DnaAL294R (HM527), DnaAV323D (HM528), DnaAL337P (HM529), DnaAA341V (HM530). (C) The oriC-to-terminus ratios of dnaA point mutations generated using PCR mutagenesis were determined using MFA in the presence and absence of SojG12V overexpression (1% xylose). Suppressor mutations (red) were found to be recalcitrant to SojG12V activity. Cells were grown in LB medium at 30°C. Values were normalized to the ori:ter ratio of the wild-type strain grown in the absence of xylose. DnaAV121A (HM713), DnaAA131T (HM714), DnaAA132T (HM710), DnaAG151R (HM705), DnaAG154S (HM706), DnaAH162Y (HM707), DnaAR281G (HM708), DnaAN311D (HM712), DnaAN311T (HM709), DnaAE314G (HM711). (D) The SojG12V suppressor mutations in dnaA perturb the formation of a Soj:DnaA–His12 complex in vivo. Cells were grown in LB medium at 30°C, crosslinked with formaldehyde, and the DnaA–His12 complexes were purified before the crosslinks were reversed and proteins were separated by SDS–PAGE. Soj and DnaA–His12 were detected by western blot analysis. The top panel shows the amount of Soj protein in the cell lysate (Input) and the bottom panel shows the amount of Soj found in a complex with DnaA–His12 following purification (Complex). DnaA–His12 (HM657), DnaAL294R–His12 (HM716), DnaAV323D–His12 (HM555), DnaAL337P–His12 (HM658), DnaAA341V–His12 (HM725). (E) The amount of each DnaASup–His12 protein was determined by western blot analysis. DivIVA was used as a loading control.
Here, we have investigated the negative regulation of DnaA by monomeric Soj. We have identified amino-acid substitutions in DnaA that render the protein insensitive to inhibition by monomeric Soj and that do not form a complex with Soj in vivo. Using these proteins we show that Soj directly interacts with DnaA in vitro. Importantly, we have developed a site-specific crosslinking assay that detects DnaA oligomers assembling on single-stranded and double-stranded DNA substrates, both of which appear to represent a helical conformation built upon the AAA+ domains. Using this assay we show that monomeric Soj specifically inhibits DnaA helix formation in vitro. Furthermore, we adapted our site-specific crosslinking assay to demonstrate that (i) DnaA forms oligomers in vivo, (ii) monomeric Soj inhibits DnaA oligomerization in vivo, and (iii) the extent of DnaA oligomerization in vivo correlates with the rate of DNA replication initiation. Together, these results establish the DnaA helix as an important target for regulation, as well as providing critical biochemical evidence supporting the physiological relevance of DnaA helix formation during DNA replication initiation.
Results
Specific point mutations in dnaA disrupt Soj inhibition in vivo
Previously, we have shown that monomeric Soj inhibits DnaA activity and forms a complex with DnaA in vivo (Murray and Errington, 2008). However, it remained unclear whether this regulation was mediated by a direct interaction between the proteins. To address this question, we screened for mutations in dnaA that suppress the growth inhibition caused by overexpressing monomeric SojG12V (Figure 1B). A chloramphenicol marker was integrated downstream of the dnaAN operon and genomic DNA from this strain was used as a substrate for error-prone PCR to generate point mutations in dnaA. PCR products were transformed into a strain harbouring an inducible sojG12V allele and plated under SojG12V overexpression conditions, resulting in slow growth of wild-type colonies. Genomic DNA from large colonies was backcrossed into the parent strain to confirm that the mutation conferring fast growth was linked to dnaA. DNA sequencing identified 14 distinct mutations that caused single amino-acid substitutions within DnaA.
To characterize the mutations in dnaA, marker frequency analysis (MFA) was used to measure the relative levels of origin and terminus DNA, thereby generating a measure of DNA replication initiation frequency (Figure 1C). The mutations within dnaA fell into two classes: hypermorphs that bypassed SojG12V inhibition (DnaAHyp proteins) by having a high basal rate of initiation, and suppressors that had an approximately wild-type rate of initiation but were resistant to SojG12V inhibition (DnaASup proteins: DnaAL294R, DnaAV323D, DnaAL337P, DnaAA341V). The suppressor mutations were each independently cloned into dnaA and transformed into the SojG12V overexpression strain to demonstrate that they were responsible for the large colony phenotype. The resulting strains displayed rates of DNA replication initiation and DnaA expression levels similar to wild type in the presence or absence of SojG12V overexpression (Supplementary Figure S2).
The ability of Soj to form a complex with DnaA–His12 and DnaASup–His12 proteins in vivo was investigated using nickel affinity purification following formaldehyde crosslinking. Compared with wild-type DnaA–His12, all four DnaASup–His12 proteins were defective in their ability to form a complex with Soj (Figure 1D). Western blot analysis confirmed that all DnaA–His12 proteins were expressed to a similar level as wild type (Figure 1E). Taken together, the data indicate that these amino-acid substitutions in DnaA suppress SojG12V inhibition by disrupting DnaA–Soj complex formation.
Soj interacts directly with DnaA in vitro
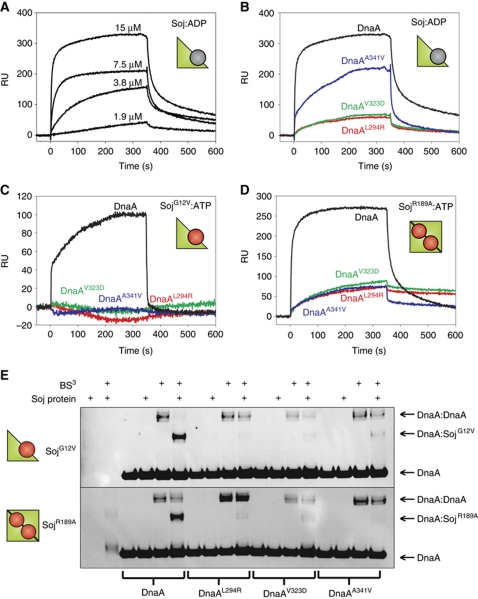
To test whether Soj and DnaA directly interact, we purified several DnaA and Soj proteins and measured binding in vitro using surface plasmon resonance (SPR). B. subtilis DnaA lacks cysteine residues, allowing for the introduction of a C-terminal cysteine following a His5-tag. Conjugation of these proteins to the sensor chip using a ligand thiol coupling technique produced a homo-orientated DnaA surface. Wild-type and mutant Soj proteins were then systematically injected over the wild-type and DnaASup surfaces. SPR analysis showed that wild-type Soj in the monomeric, ADP-bound form (Soj:ADP) binds to DnaA with an KD of ∼30 μM (Figure 2A). In contrast, the DnaAL294R and DnaAV323D proteins were severely defective in their interaction with Soj:ADP, and the DnaAA341V protein displayed an intermediate interaction profile (Figure 2B). Furthermore, all the DnaASup proteins failed to support complex formation with monomeric SojG12V (Figure 2C).
Figure 2.
Soj directly interacts with DnaA. (A–D) SPR sensorgrams. DnaA proteins were immobilized onto the SPR chip surface via a unique C-terminal cysteine residue to create a homogenous surface. Cartoon representations of Soj are shown to indicate the quaternary state of various proteins. (A) Two-fold serial dilution of wild-type Soj:ADP injected over DnaAH485C. (B–D) The indicated Soj proteins (15 μM) were injected over wild-type and mutant DnaA surfaces starting at time zero for 360 s. (E) In vitro crosslinking assay using the primary amine-specific crosslinker (BS3) in the presence of DNA (pBSoriC4; 3 nM) and ATP (2 mM). Protein complexes were separated by SDS–PAGE and the DnaA protein was visualized by western blotting. Pluses located above each lane indicate the presence of BS3 and/or Soj protein (32 μM). The identity of the DnaA proteins (3 μM) are indicated below. The identity of the Soj proteins is indicated to the left of each gel.
To substantiate the results observed by SPR, DnaA proteins were subjected to primary amine-specific crosslinking (BS3) in solution with and without SojG12V. Protein complexes were separated by SDS–PAGE and DnaA was detected by western blot analysis (Figure 2E). The appearance of a signal at a molecular weight expected for a Soj:DnaA complex (27 kDa + 54 kDa=81 kDa) was observed in the presence of SojG12V. By contrast, complex formation was dramatically reduced when the DnaASup proteins were tested. In addition, the DnaA:DnaA complex (108 kDa) was reduced in the presence of SojG12V.
Cytological analysis of GFP–SojG12V localization in B. subtilis cells suggests that it associates with origin bound DnaA (Murray and Errington, 2008). To ascertain if SojG12V is capable of interacting with a DnaA:DNA complex in vitro, a pull-down experiment was performed. His-tagged SojG12V was incubated with pBsoriC4 in the presence and absence of native DnaA (DnaAnat). Proteins and DNA were crosslinked using a concentration of formaldehyde that yielded a specific Soj:DnaA interaction (Supplementary Figure S3A). Complexes were then bound to nickel beads via the histidine tag on Soj, washed, and the crosslinks reversed. The amount of pBsoriC4 in these complexes was detected using qPCR. There was an ∼13-fold enrichment of pBsoriC4 bound to SojG12V in the presence of DnaA, indicating that Soj is capable of forming a complex with DnaA bound to DNA (Supplementary Figure S3B).
Taken together, the SPR and crosslinking assays indicate that monomeric Soj directly interacts with DnaA both in solution and bound to DNA, and that the substitutions in the DnaASup proteins disrupt complex formation.
Mapping of the DnaASup and DnaAHyp substitutions onto DnaA structures suggests a mechanism for Soj regulation
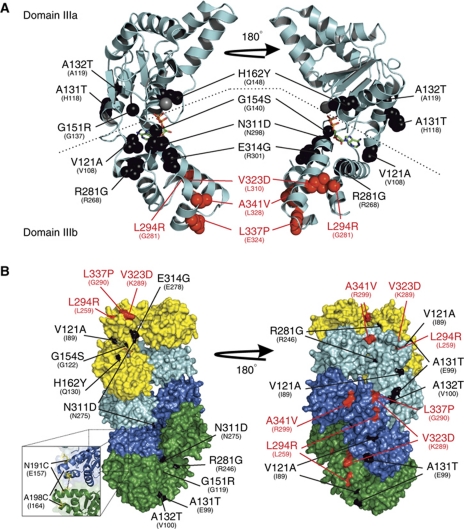
We noted that although our PCR mutagenesis strategy targeted the entire dnaA region (∼5 kb flanking either side of dnaA, including the dnaA promoter and all of oriC), all of the isolated DnaAHyp substitutions were located within the AAA+ motif of DnaA, while all of the DnaASup substitutions were located in domain IIIb (or at the border between domains IIIb and IV, depending upon the assignment of domain boundaries; see Supplementary Figure S1). Strikingly, when these amino-acid substitutions were mapped onto the crystal structure of DnaA domain III from Thermotoga maritima (Figure 3A), all four of the suppressor substitutions were found to cluster in domain IIIb, strongly suggesting that this region is the binding site for Soj.
Figure 3.
The DnaA hypermorph and suppressor substitutions are located in domain III. (A) A cartoon representation of monomeric DnaA from the T. maritima crystal structure (PDB ID: 2Z4S) bound to ADP (stick). Domains IIIa and IIIb are separated by a dashed line. The SojG12V hypermorph (black) and suppressor (red) substitutions are shown as spacefill representations. The identity and positions of the B. subtilis amino-acid substitutions are indicated above the corresponding residue of the T. maritima protein. (B) The majority of the hypermorphic substitutions are located either adjacent to, or buried within, the DnaA:DnaA interface. A surface representation of the helical DnaA structure from A. aeolicus (PDB ID: 2HCB) bound to AMP-PCP. DnaA monomers are coloured independently (yellow, cyan, blue, and green). The amino-acid substitutions are coloured and annotated as in (A) above. The inset shows the location of the two residues changed to cysteines for the crosslinking assays, with the black dashed line indicating where BMOE acts.
The DnaAHyp substitutions were found more widely distributed throughout the AAA+ motif. Three of the amino acids (G151, G154, and V121) were located around the nucleotide-binding pocket, with the backbone of the latter two residues mediating direct contacts with the terminal phosphate(s) and sugar, respectively. However, the remaining six DnaAHyp substitutions appear to be surface exposed and distal from the nucleotide-binding pocket; thus, it was unclear what effect these substitutions were having. To gain insight into how the DnaAHyp proteins might affect DnaA activity, the positions of these amino-acid substitutions were mapped onto the helical crystal structure of Aquifex aeolicus DnaA bound to AMP-PCP (Figure 3B, note A. aeolicus DnaA lacks a 14 amino-acid stretch present in all classically studied bacteria including B. subtilis, which harbours two of the four suppressor substitutions (V323D and L337P); see Supplementary Figures S1 and S4). Significantly, all of these hypermorphic substitutions were either buried inside (70%) or adjacent to the DnaA:DnaA interface and were generally solvent exposed only at each end of the helix. Since these substitutions lead to hyperactivity of DnaA, we speculate that they may promote AAA+ mediated oligomerization by increasing the affinity of the DnaA:DnaA interaction. Taken together with the observation that SojG12V disrupts DnaA:DnaA complex formation (Figure 2E), we hypothesized that monomeric Soj regulates DnaA ATP-dependent oligomerization.
B. subtilis DnaA assembles into an ATP-dependent oligomer in vitro
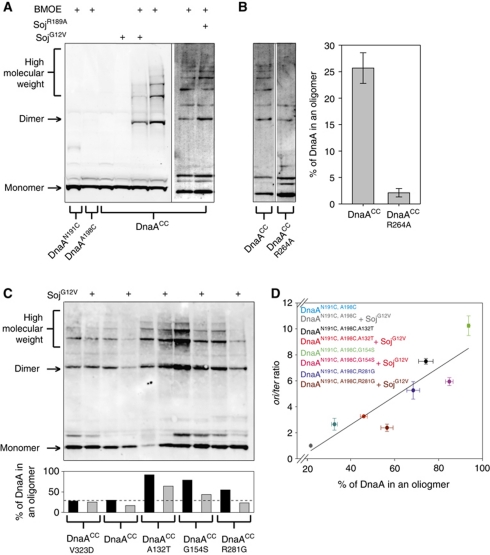
To test this model we designed a crosslinking assay to specifically detect ATP-dependent oligomerization of DnaA (Chen, 1991). Guided by the A. aeolicus structure, a pair of cysteine residues were introduced into domain IIIa at N191 and A198 (Figure 3B, inset). Within the oligomer, the N191 residue from one DnaA monomer is in close proximity (∼9 Å) to the A198 residue of the adjacent monomer. DnaAN191C,A198C (hereafter referred to as DnaAcc) was incubated with the cysteine-specific crosslinker bis(maleimido)ethane (BMOE; spacer arm 8.0 Å), protein complexes were separated by SDS–PAGE, and DnaA was detected by western blot analysis.
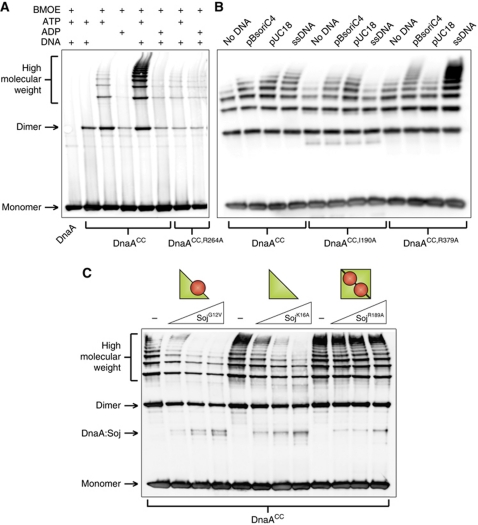
Figure 4A shows that crosslinking of DnaACC captures multiple high molecular weight complexes that run as a ladder on the gel. Formation of these DnaA oligomers was dependent on ATP and dramatically stimulated by DNA. Critically, mutation of the arginine finger residue (R264A) that coordinates the γ-phosphate of the ATP from the neighbouring DnaA molecule (Erzberger et al, 2006) greatly diminished DnaA oligomerization (lanes 7 and 8). The stimulation by DNA appeared to be non-specific as the formation of DnaA oligomers was indistinguishable when comparing plasmids with or without the B. subtilis origin of replication (pBsoriC4 versus pUC18, respectively), comparing supercoiled DNA with linear DNA, and comparing double-stranded DNA with single-stranded DNA (Figure 4B and data not shown).
Figure 4.
Monomeric Soj inhibits DnaA oligomerization in vitro. (A) In vitro oligomer formation assays using the cysteine-specific crosslinker BMOE. DnaA proteins were incubated in oligomer formation buffer for 15 min prior to the addition of BMOE. Pluses located above each lane indicate the presence of BMOE (2 mM), nucleotide (2 mM), and/or DNA (pBsoriC4; 3 nM). The identity of the DnaA proteins (3 μM) are indicated below. DnaA proteins were separated by SDS–PAGE and visualized by western blotting. (B) DnaA oligomers form on both single-stranded and double-stranded DNA. DnaA proteins (3 μM) were incubated in oligomer formation buffer with NaCl (400 mM) in the presence of ATP (2 mM), with or without DNA, for 15 min prior to the addition of BMOE. The identity of each DNA substrate is indicated above the respective lanes, and the amount of DNA is equal in each reaction (120 fmol). Both plasmids are supercoiled and the single-stranded DNA (ssDNA) is oligonucleotide oGJS159. DnaA proteins were separated by SDS–PAGE and visualized by western blotting. (C) Monomeric Soj disrupts DnaA oligomer formation in vitro. Triangles above the lanes represent an increasing concentration of various Soj proteins (12, 24, and 36 μM). DnaA proteins (3 μM) were incubated in oligomer formation buffer in the presence of ATP (2 mM) and DNA (pBsoriC4; 3 nM) for 15 min prior to the addition of BMOE. DnaA proteins were separated by SDS–PAGE and visualized by western blotting.
Previous work has suggested that the crystalized DnaA oligomer cannot accommodate binding to double-stranded DNA through the helix-turn-helix in domain IV due to substantial steric clashes (Duderstadt et al, 2010). Because we readily observed that double-stranded DNA stimulates DnaA oligomer formation, we wondered whether this stimulation was dependent upon residues required for the DNA-binding activities of domains III and IV. An amino-acid substitution was introduced into domain III (I190A) that previously was shown to disrupt single-stranded DNA-binding activity of DnaA in vitro and DNA replication in vivo (Ozaki et al, 2008). As expected, oligomerization of DnaACC,I190A was not stimulated by single-stranded DNA, although it was stimulated by double-stranded plasmids (Figure 4B). Next, to investigate the role played by domain IV in oligomer formation, arginine 379 was mutated to an alanine. In E. coli DnaA, the equivalent residue has been shown to interact with both specific basepairs and backbone phosphates in the minor groove of double-stranded DNA (Fujikawa et al, 2003). In contrast to the results obtained with DnaACC,I190A, oligomerization of DnaACC,R379A was stimulated by single-stranded DNA, but not by the double-stranded pUC18 plasmid. Interestingly, oligomerization of DnaACC,R379A was stimulated by a plasmid that harboured oriC (Figure 4B). These results correlated with the ability of DnaACC,R379A to bind to the respective plasmids as judged by an electrophoretic mobility shift assay (Supplementary Figure S5). We note that single-stranded DNA stimulated helix formation of DnaACC,R379A to a greater degree than DnaACC. This observation suggests that the arginine residue may interact with the phosphate backbone of the single-stranded substrate and inhibit the docking of domain IV into the AAA+ domain, a transition proposed to be critical for single-stranded DNA-binding activity (Duderstadt et al, 2010). Taken together, these experiments indicate that DnaA is capable of forming ATP-dependent oligomers on both single-stranded and double-stranded DNA substrates, in contrast to the apparent constraints placed upon domain IV within the structure of the DnaA oligomer (see Discussion).
Monomeric Soj specifically inhibits DnaA oligomer formation in vitro
The DnaA oligomer formation assay was then used to investigate the effect of Soj on DnaA activity. The presence of SojG12V significantly reduced both the length and abundance of the DnaA oligomers formed in the presence of plasmid DNA (Figure 4C). To determine whether the observed inhibition was specific to monomeric Soj, the effect of SojK16A (monomeric) and SojR189A (dimeric) proteins was also investigated. While SojK16A clearly disrupted DnaA oligomer formation, SojR189A had little or no effect (Figure 4C) even though it was capable of interacting with DnaA (Figure 2D and E). Thus, despite sharing apparent interaction determinants, disruption of DnaA oligomerization is a specific property of the monomeric Soj protein. These results are consistent with our previous findings that monomeric Soj specifically inhibits DnaA initiation activity in vivo (Scholefield et al, 2011).
Oligomers formed by DnaAL294R and DnaAV323D were highly resistant to SojG12V activity (Figure 5A). In comparison, oligomers formed by DnaAA341V were partially susceptible to SojG12V, consistent with the intermediate response observed between the two proteins by SPR analysis (Figures 2B and 5A). Importantly, all of the DnaASup proteins were found to form oligomers under the same conditions as wild type (Supplementary Figure S6A). These results indicate that the effect of SojG12V on DnaA oligomerization is dependent on the same residues required for the formation of a Soj:DnaA complex.
Figure 5.
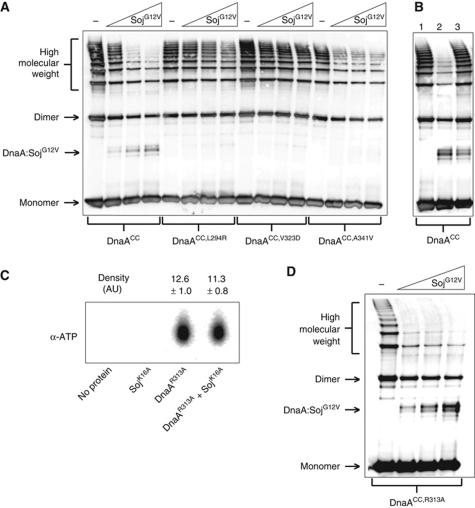
Monomeric Soj specifically prevents DnaA oligomerization in vitro. (A) In vitro oligomer formation assay with DnaASup proteins. DnaA proteins (3 μM) and SojG12V (12, 24, and 36 μM) were incubated in oligomer formation buffer in the presence of ATP (2 mM) and DNA (pBsoriC4; 3 nM) for 15 min prior to the addition of BMOE. DnaA proteins were separated by SDS–PAGE and visualized by western blotting. (B) Monomeric Soj is unable to disassemble pre-formed DnaA oligmers. Reaction conditions are the same as in (A). DnaA was crosslinked at T=15 min. Lane 1, DnaA was added to the reaction buffer at T=13. Lane 2, SojG12V was added at T=0 and DnaA added at T=13. Lane 3, DnaA was added at T=0 and SojG12V added at T=13. (C) Monomeric Soj does not affect DnaA ATP binding. DnaAR313A (3 μM) and/or SojK16A (36 μM) were incubated with α-P32 ATP before being isolated from the reaction using magnetic nickel beads and denatured with methanol. The released nucleotides were separated on a PEI cellulose TLC plate and visualized by autoradiography. Density values were measured using ImageJ (n=3). (D) Monomeric Soj disrupts DnaA oligomers independent of DnaA ATPase activity. In vitro oligomer formation assay with the ATP hydrolysis deficient DnaA protein. DnaAR313A (3 μM) and SojG12V (12, 24, and 36 μM) were incubated in oligomer formation buffer in the presence of ATP (2 mM) and DNA (pBsoriC4; 3 nM) for 15 min prior to the addition of BMOE. DnaA proteins were separated by SDS–PAGE and visualized by western blotting.
To determine whether monomeric Soj prevents DnaA oligomer formation and/or disassembles pre-formed DnaA oligomers, an order of addition experiment was performed. Figure 5B shows that DnaA oligomers formed prior to SojG12V addition are highly resistant to SojG12V inhibition (lane 3) when compared with oligomers formed after SojG12V addition (lane 2), indicating that monomeric Soj acts by preventing DnaA oligomer formation.
Since the DnaA oligomer can form on both single-stranded and double-stranded DNA, it was possible that SojG12V only acted on one of these complexes. SojG12V was found to disrupt oligomerization of both DnaACC in the presence of single-stranded DNA and DnaACC,I190A in the presence of double-stranded DNA (Supplementary Figure S6B). Thus, monomeric Soj can inhibit DnaA oligomers forming on either single-stranded or double-stranded substrates, although we note that the degree of inhibition appeared to be greater in the presence of single-stranded DNA.
Previous work has established that domain I plays an important role in DnaA oligomerization in E. coli (Felczak et al, 2005). To test whether SojG12V inhibition of DnaA oligomerization was dependent on this domain I self-interaction, we created a truncated version of DnaACC that lacked domains I and II (DnaAIII/IV,CC). As expected from the interaction analysis described above, the DnaAIII/IV protein retained the ability to interact with Soj (Supplementary Figure S7A). The truncated DnaA was readily capable of forming oligomers and, like the full-length protein, these oligomers were inhibited by monomeric SojG12V but were not affected by dimeric SojR189A (Supplementary Figure S7B). These results indicate that the oligomerization activity observed for the DnaACC protein is independent of domains I and II, and taken together with the interaction experiments, they support a model in which monomeric Soj inhibits DnaA oligomerization by specifically regulating an activity of the AAA+ domain.
SojG12V inhibition of DnaA is independent of DnaA ATP binding, ATP hydrolysis, and DNA-binding activities
Because oligomerization of DnaACC was stimulated by both ATP and DNA, we investigated whether monomeric Soj inhibits DnaA oligomerization by modulating the interaction of DnaA to either of these molecules. To test whether monomeric Soj affects ATP binding to DnaA, we incubated an ATP hydrolysis deficient DnaA protein (DnaAR313A) with α-P32 ATP in the presence and absence of SojK16A. The DnaAR313A protein was utilized to prevent hydrolysis of the bound ATP during the experiment (Supplementary Figure S8A) and SojK16A was used because it is ATP-binding deficient (Scholefield et al, 2011). Reactions were assembled and incubated in an identical manner to the oligomer formation assay before the proteins were separated from the reaction buffer using magnetic nickel beads and washed to remove unbound ATP. The bound ATP was extracted and then separated by thin layer chromatography. There was no significant difference between the amount of ATP bound to DnaAR313A in the presence or absence of SojK16A (Figure 5C), indicating that monomeric Soj does not inhibit ATP binding. Furthermore, SojG12V did not stimulate the ATPase activity of wild-type DnaA (Supplementary Figure S8A) and oligomerization of the hydrolysis deficient DnaACC,R313A protein remained sensitive to SojG12V inhibition (Figure 5D), indicating that monomeric Soj does not act by regulating the ATPase activity of DnaA.
To determine whether monomeric Soj inhibits DnaA oligomerization by inhibiting the DNA-binding activity of DnaA, we determined whether SojG12V was capable of inhibiting DnaACC oligomer formation in the absence of DNA. As before, SojG12V inhibited DnaACC oligomerization but had little effect on DnaACC,V323D (Supplementary Figure S8B). These results indicate that monomeric Soj can regulate DnaA helix formation in a manner that is independent of the ability of DnaA to bind to DNA. Taken together, the data support the model that monomeric Soj directly interacts with the AAA+ domain of DnaA to specifically prevent ATP-dependent oligomer formation.
Monomeric Soj inhibits DnaA oligomer formation in vivo
To determine if the inhibition of DnaA oligomer formation caused by monomeric Soj in vitro was physiologically relevant, the site-specific crosslinking assay was modified to detect DnaA oligomers in vivo. The endogenous dnaA gene was replaced with dnaAN191C,A198C (dnaACC) and BMOE was used to crosslink DnaACC proteins in intact cells that were harvested during exponential growth. The strain harbouring dnaACC mildly overinitated DNA replication compared with wild type, although its growth and morphology appeared normal (Supplementary Figure S9A). Following incubation with BMOE, cells containing DnaACC (but not cells with single cysteine substitutions) formed oligomers and this was dependent upon the arginine finger required for the interaction of neighbouring AAA+ domains (Figure 6A–C). Thus, the site-specific crosslinking assay captures DnaA oligomers within B. subtilis cells that likely correspond to the complexes observed in vitro.
Figure 6.
The DnaA oligomer forms in vivo and is inhibited by monomeric Soj. (A) A DnaA oligomer can be isolated in vivo and is specifically inhibited by monomeric Soj. Cells were grown in crosslinking medium at 30°C to an A600 of 0.5, washed, and resuspended in in vivo crosslinking buffer in the presence of BMOE. Where indicated, 1% xylose was added or removed (in the case of the DnaAHyp strains) at an OD600 of 0.1 to modulate Soj protein expression. The identity of DnaA and/or Soj proteins are indicated below and above each lane, respectively. Proteins were separated by SDS–PAGE and DnaA was visualized by western blotting. DnaACC SojG12V (sGJS006), DnaAN198C (sGJS021), DnaAN191C (sGJS022), DnaACC SojR189A (sGJS033). (B) The DnaA oligomer is dependent on the arginine finger (R264) in vivo. Because the arginine finger mutation (dnaAR264A) is lethal (Duderstadt et al, 2010), the allele was introduced into a strain harbouring a plasmid origin (oriN) that allows viability in the absence of DnaA and/or oriC (Hassan et al, 1997). The identity of the DnaA proteins expressed are indicated below the respective lanes. DnaACC (sGJS051), DnaACC,R264A (sGJS052). Quantification of three biological repeats is shown. Error bars represent the standard deviation of the data. (C) The hypermorphic DnaA mutants have an increased propensity to form oligomers in vivo. Reaction conditions and figure annotations are as described in (A). The bar chart below shows the quantification of the gel. DnaA was defined as being in a helix if found in a dimer or higher molecular weight complex. DnaACC (sGJS006), DnaACC,V323D (sGJS008), DnaACC,A132T (sGJS036), DnaACC,G154S (sGJS037), DnaACC,R281G (sGJS038). (D) The percentage of DnaA found in an oligomer correlates with initiation frequency. The reactions in (C) (with the exception of DnaAV323D) were repeated in triplicate, the lanes quantified using ImageJ, and the percentage of DnaA found in the helix calculated as described above. Error bars represent the s.e.m. of the data. Concurrently, the oriC-to-terminus ratios were determined by MFA. Errors bars represent the standard deviation of the data. Linear regression analysis yields an R2 value of 0.82 while the Pearson product moment correlation coefficient is 0.903 (P-value=0.00212).
Figure 6A shows that overexpression of monomeric SojG12V resulted in an ∼50% decrease in the percentage of DnaACC found in the oligomer compared with wild type, while overexpression of dimeric SojR189A had no inhibitory effect (both Soj proteins were overexpressed to the same extent; Supplementary Figure S9B). Moreover, the oligomers formed by one of the DnaASup proteins (DnaACC,V323D) were resistant to SojG12V overexpression (Figure 6C). These results suggest that monomeric Soj is capable of directly inhibiting DnaA oligomer formation in vivo as well as in vitro.
In addition to analysis of the DnaASup protein, the in vivo oligomer formation assay was used to investigate the activity of several DnaAHyp proteins. The three DnaAHyp proteins tested (A132T, G154S, and R281G) showed an ∼2–3-fold increased propensity to form oligomers compared with the wild type (Figure 6C). The helices formed by the DnaAHyp proteins remained susceptible to inhibition by SojG12V, consistent with the results from the MFA described above (Figure 1C). These observations are compatible with the model that the DnaAHyp proteins bypass SojG12V inhibition by promoting interactions between adjacent DnaA monomers, thus leading to an increase in DnaA oligomer formation and in the rate of DNA replication initiation.
Interestingly, the DNA replication initiation frequency of wild-type DnaA and each DnaAHyp protein (in the presence or absence of SojG12V) strongly correlated with the percentage of protein observed in oligomers (correlation coefficient=0.903) (Figure 6D). This result further supports the biological significance of DnaA oligomer formation, and suggests that this activity may be the rate-limiting step required for DNA replication initiation in B. subtilis.
Discussion
Monomeric Soj inhibits DnaA helix formation
In this report we show that monomeric Soj stalls DNA replication initiation by directly interacting with the initiator protein DnaA and specifically inhibiting DnaA oligomer formation. Based on the arguments outlined below, we propose that monomeric Soj regulates DnaA assembly into a right-handed helical structure. If this is correct, we have uncovered a novel mode of DnaA regulation mediated via the Soj protein. We also provide critical biochemical evidence demonstrating that the DnaA helix observed by X-ray crystallography assembles and has functional relevance in vivo, thereby supporting the physiological role of both this DnaA structure and the proposed Soj regulatory mechanism.
To study the negative regulation of DnaA by monomeric Soj, we established a cysteine crosslinking assay capable of detecting an ATP-dependent DnaA oligomer. To achieve crosslinking via BMOE, this oligomer has to fulfil two requirements: first, the arginine finger residue (R264) has to coordinate the ATP of the neighbouring monomer, and second, the two cysteines from adjacent AAA+ motifs have to be brought within 9 Å of each other. This means that there are two relatively fixed contacts between neighbouring AAA+ domains necessary for an efficient reaction. Since these dual requirements would allow minimal flexibility of the AAA+ domains relative to each other, the DnaACC site-specific crosslinking assay is most likely capturing the AAA+ domains in an orientation analogous to the published crystal structure. Therefore, we propose that our DnaACC oligomer formation assay captures a helical structure of B. subtilis DnaA.
The DnaA helix can form on single-strand and double-stranded DNA
It has been proposed that the compact DnaA helix observed in the crystal structure could only form on single-stranded DNA through interactions in domain III, since domain IV is packed tightly into the compact helix, effectively sequestering the double-stranded DNA-binding interface (Erzberger et al, 2006; Ozaki et al, 2008; Duderstadt et al, 2010). Consistent with this model and supporting the importance of the compact DnaA helix, it has been shown that amino-acid substitutions in the domain III–IV interface that affect DnaA oligomerization inhibit both open complex formation in vitro and replication origin function in vivo (Duderstadt et al, 2010). However, it should be noted that genetic analyses, electron microscopy analyses, and footprinting assays together strongly indicated that ATP-bound DnaA must first cooperatively assemble into a large nucleoprotein complex built upon defined interactions on double-stranded DNA within oriC prior to open complex formation (Mott and Berger, 2007; Rozgaja et al, 2011). Thus, it was not clear how the compact DnaA helix would be assembled at the replication origin.
We observed that an ATP-dependent DnaA helix can be built on either single-stranded or double-stranded DNA. The DnaA helix formed on single-stranded DNA was found to depend on I190 in domain III, located within the central pore of the helix. By contrast, the DnaA helix formed on non-specific double-stranded DNA was found to depend on R379 in domain IV and was independent of I190. These observations imply that during DnaA helix formation on double-stranded DNA, domain IV must be extended away from the AAA+ helical core. This also leads to the conclusion, supported by the structures of DnaA bound to ADP and AMP-PCP (Erzberger et al, 2002, 2006; Ozaki et al, 2008; Duderstadt et al, 2010), that the α-helix linking domains III and IV is a semi-flexible linker, allowing domain IV to adopt multiple conformations relative to the helical AAA+ core.
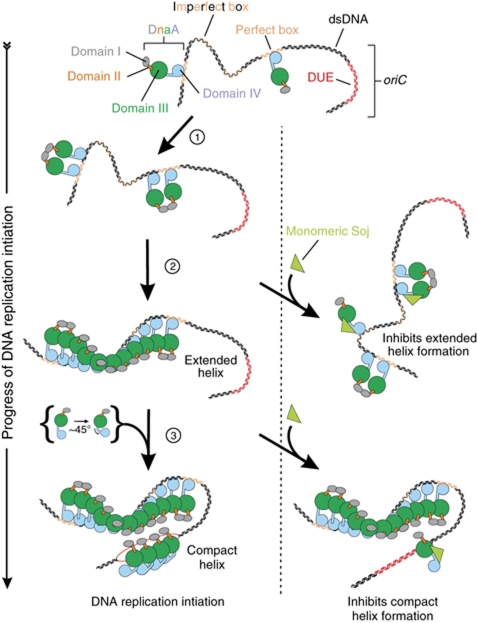
These results are consistent with the model, originally proposed by Erzberger et al (2006), whereby ATP-bound DnaA initially assembles into an extended helical structure that is engaged with specific double-stranded DNA-binding sequences through domain IV, followed by the transition to a compact helix at which point domain IV dissociates from double-stranded DNA and folds into domain III, thereby promoting the single-stranded DNA-binding activity of residues located in the central channel formed by the AAA+ motifs (Figure 7). Although the precise architecture of the DNA replication initiation complex has not yet been determined (see Ozaki and Katayama, 2011), our data help to reconcile how multiple distinct helical DnaA oligomers could be assembled specifically at bacterial replication origins.
Figure 7.
Model of monomeric Soj regulation of DnaA during DNA replication initiation. DnaA is stably bound to its high-affinity sites during most of the cell cycle. At initiation, additional DnaA molecules are recruited through a domain I interaction (step 1). At this point, Soj could interact with DnaA to prevent the structural transition necessary for it to form a helix on double-stranded DNA (step 2). This nucleoprotein complex facilitates the unwinding of the DUE and the loading of a compact helix onto ssDNA (step 3). Soj could interact with this newly recruited DnaA preventing the bending of the α-helix linking domains III and IV.
In addition to the helical DnaA oligomer built upon the AAA+ core, it is important to note that domain I plays critical roles in DnaA assembly and activity. Many bacterial DnaA proteins can directly form homo-oligomers that are dependent upon domain I (Weigel et al, 1999; Simmons et al, 2003). This domain I self-interaction is required for DnaA both to recruit additional DnaA proteins to the replication origin and to load the replicative helicase (Felczak et al, 2005; Miller et al, 2009). Although this domain I self-interaction is not necessary for either assembly or ssDNA stretching activity of the DnaA helical oligomer (Supplementry Figure S7B; Erzberger et al, 2006; Duderstadt et al, 2010, 2011), it could stimulate DnaA helix formation indirectly either by increasing the local concentration of DnaA at oriC or by stabilizing DnaA oligomers at oriC. Future studies will be needed to determine how these distinct DnaA interfaces, along with the information encoded by the DNA sequence of oriC, act in concert to construct an active DnaA initiation complex.
Potential mechanisms for Soj activity
Our genetic analysis suggests that Soj directly interacts with domain IIIB (or the region linking domains IIIb and IV) of DnaA. Comparison of the ADP-bound monomeric and the AMP-PCP-bound helical DnaA structures indicates that domain IIIB must shift and domain IV must bend to generate the space required for the arginine finger from the neighbouring monomer to engage the γ-phosphate of ATP (Erzberger et al, 2006). Furthermore, our biochemical analyses show that monomeric Soj neither inhibits ATP binding, nor stimulates ATP hydrolysis, nor disassembles pre-formed DnaA helices.
Based on these considerations, we envisage two potential mechanisms by which Soj could act to prevent helix formation (Figure 7). First, Soj could bind to domain IIIB and sterically inhibit helix assembly. Second, Soj could bind to domain IIIB and prevent DnaA undergoing the conformational changes required for it to assemble into a helix. Taking into account the observations that both monomeric and dimeric Soj proteins bind to DnaA, that the DnaASup substitutions impair the binding of both Soj conformations, and that prevention of DnaA helix formation is specific to monomeric Soj, we currently favour the latter allosteric model because binding of the larger Soj dimer would presumably create an even greater steric wedge between neighbouring DnaA molecules.
Our preliminary data suggest that dimeric Soj stimulates DnaA helix formation in vivo, consistent with its ability to increase the frequency of DNA replication initiation (GJS and HM, unpublished data). In addition, previous data indicate that Soj can positively regulate DnaA activity as a transcriptional regulator (Murray and Errington, 2008). Thus, we wonder whether Soj could allosterically regulate DnaA helix formation both negatively and positively, with the direction of regulation depending upon the quaternary state of the Soj protein.
Although the interaction studies indicate that both monomeric and dimeric Soj proteins bind to a similar region of DnaA (Figure 2C–E), we note that the dimeric Soj protein remained capable of interacting with all of the DnaASup substitutions, albeit to a lesser extent than with the wild-type protein (Figure 2D). This observation suggests that dimeric Soj may be able to interact with a second site on DnaA, and this additional contact could influence the outcome of the interaction between the two proteins. These questions regarding dichotomies between the activities and the interactions of monomeric and dimeric Soj proteins are currently under investigation.
Biological implications of regulating DnaA helix formation
The signal that switches Soj from a monomer to a dimer, and thus from an inhibitor to an activator of DnaA, is unknown. In all growth conditions tested so far, it appears that Soj dimerization is efficiently inhibited by the regulatory protein Spo0J (ParB). Therefore, Soj probably spends most of its time in the inhibitory monomeric state. Previous cytological studies have shown that GFP–SojG12V localizes as a focus at oriC in a DnaA-dependent manner (Murray and Errington, 2008), suggesting that Soj prevents DnaA helix formation at oriC. By stalling DNA replication initiation at the stage of DnaA helix formation, it would allow DnaA to remain primed for rapid activation (i.e., DnaA would already be ATP-bound and localized at the origin). We hypothesize that a cellular signal, such as an increase in nutritional availability, entry into a developmental programme, or a cell-cycle event, stimulates the conversion of Soj from monomer to dimer (perhaps locally around oriC), thereby swiftly promoting the initiation of DNA replication.
Chromosomally encoded par genes are found throughout all branches of the bacterial kingdom (Livny et al, 2007). Nonetheless, many species do not harbour par genes and we hypothesize that regulation of DnaA helix formation, representing the active initiation complex, is achieved through different mechanisms in various bacteria. In E. coli, which does not have par genes, the rate of ATP binding appears to limit DnaA oligomerization potential (Kurokawa et al, 1999). In C. crescentus DnaA is degraded at the end of every cell cycle (Gorbatyuk and Marczynski, 2005), suggesting that helix formation could be limited by protein synthesis. Interestingly, although C. crescentus contains par genes, it appears that in this organism ParA is involved in segregation of the chromosome origin region (Ptacin et al, 2010; Scholefield et al, 2010). Whether this variation in ParA activity reflects the fact that DnaA oligomerization in C. crescentus is limited at a different step in the assembly pathway compared with B. subtilis is not known, but it will be informative to determine the rate-limiting step in DnaA oligomerization for a range of bacteria and correlate this with the activity of their respective ParA proteins.
The B. subtilis Par system: roles in DNA replication and chromosome segregation
par operons were originally identified on low-copy number plasmids where they ensure faithful segregation of plasmids into daughter cells (Gerdes et al, 2010). In these systems, the ParB protein binds to a centromere-like site on the plasmid (parS), followed by segregation of this nucleoprotein complex by the ATPase ParA. The chromosomal orthologues of Par proteins are found throughout the bacterial kingdom and have been shown to influence chromosome organization and segregation in a number of diverse species (for review see Gerdes et al, 2010). These results led to the hypothesis that Par proteins act in an analogous manner to their plasmid orthologues by forming a segregation machine that actively separates replicated chromosomes prior to cell division.
Despite the appeal of this model, rigorous examination of Soj/ParA activity during vegetative growth in B. subtilis has so far provided little support for a role in chromosome segregation or origin localization (Lee and Grossman, 2006). However, it has been shown that Spo0J/ParB is required for proper DNA segregation in B. subtilis (Ireton et al, 1994), and recent work indicates that Spo0J affects chromosome organization by recruiting Condensin (the SMC complex) to the origin region of the chromosome (Gruber and Errington, 2009; Sullivan et al, 2009). Therefore, the B. subtilis Par system is involved in accurate chromosome segregation, although apparently not in an analogous manner to related plasmid systems. Moreover, it appears that the activity of the B. subtilis Par system is more complex than initially imagined, with the Spo0J:parS complex acting as a regulator of, and perhaps coordinating the activities of, factors involved in DNA replication (Soj) and DNA organization/segregation (SMC) leading to possible coordination between the two systems.
AAA+ inter-protein interactions as a regulatory target in ORC
In eukaryotic organisms, DNA replication is initiated by the origin recognition complex (ORC; Orc1–6) in combination with Cdc6. Orc1, Orc4, Orc5, and Cdc6 contain AAA+ domains, while Orc2 and Orc3 are predicted to have AAA+-like folds. Orc1 and Orc5 have been shown to bind ATP while Orc1 and Cdc6 have intrinsic ATPase activity (Kawakami and Katayama, 2010). Strikingly, ORC has been shown to undergo an ATP-dependent conformational change, and the structure of the DnaA helix can be docked into a low-resolution structure of ORC (Clarey et al, 2006). These observations suggest that the ATP-dependent conformational changes in ORC result from a similar reorientation of the AAA+ domains observed in DnaA. Consistent with this model, the arginine finger residue in Orc4 and the ATPase activity of Cdc6 are both critical for reiterative helicase loading (Schepers and Diffley, 2001; Bowers et al, 2004). We suggest that detailed examination of DnaA conformational changes required for the assembly of an active initiation complex will likely underpin and inform the mechanistic understanding of related initiator complexes from higher organisms. Furthermore, our finding that DnaA helix formation is targeted for regulation by Soj opens up the possibility that the conformational changes observed for ORC will also be subject to regulation.
Materials and methods
Strains and plasmids
Strains and plasmids used in this study are described in the Supplementary data and listed in Supplementary Tables SI and SII and the relevant oligonucleotides in Supplementary Table SIII. E. coli strain DH5α (Invitrogen) was used for the construction of all plasmids, and strain BL21 (DE3) pLysS (Stratagene) was used to express all proteins. The plasmid pET21-d (Invitrogen) was used as the expression vector for all proteins.
Media and chemicals
Nutrient agar (NA; Oxoid) was used for routine selection and maintenance of both B. subtilis and E. coli strains. For experiments in B. subtilis, cells were grown in either Luria-Bertani (LB) medium or casein hydrolysate medium. Supplements were added as required: 20 μg/ml tryptophan, 5 μg/ml chloramphenicol, 2 μg/ml kanamycin, 50 μg/ml spectinomycin. For plasmid and protein expression in E. coli, cells were grown in LB medium or Nutrient Broth (Oxoid) and supplemented with 30 μg/ml (for single copy plasmids) or 75 μg/ml ampicillin and 10 μg/ml chloramphenicol. Unless otherwise stated, all chemicals and reagents were obtained from Sigma-Aldrich.
Marker frequency analysis
MFA was essentially done as previously described (Murray and Errington, 2008). For details see Supplementary data.
Purification of in vivo protein–protein complexes
This was done as previously described, for details see Supplementary data (Murray and Errington, 2008).
Primary amine crosslinking assay
Soj proteins (36 μM) were diluted into oligomer formation buffer (25 mM HEPES pH 7.6, 200 mM NaCl, 100 mM potassium glutamate, and 10 mM MgCl2) supplemented with 2 mM ATP and 3 nM pBSoriC4 at room temperature. DnaA (3 μM) was then immediately added and the reaction was incubated at 37°C for 15 min. The primary amine-specific crosslinker BS3 (Thermo Scientific) was then added (0.1 mM final) and the reaction was left to proceed for 3 min at 37°C. The reaction was quenched by the addition of TRIS (100 mM final pH 8) for 10 min at 37°C. Samples were separated by SDS–PAGE and bands were visualized by western blot analysis using α-DnaA polyclonal antibodies.
In vitro helix formation assay
Soj proteins (12, 24, and 36 μM) were diluted into oligomer formation buffer (see above) supplemented with 2 mM ATP and 3 nM pBSoriC4. DnaA (3 μM) was then added and the reaction was incubated at 37°C for 15 min. BMOE cysteine-specific crosslinker (Thermo Scientific) was added to a final concentration of 2 mM and the reaction was left to proceed for 3 min at 37°C. The reaction was quenched by the addition of cysteine (50 mM final) for 5 min at 37°C. Samples were separated by SDS–PAGE and bands were visualized by western blot analysis using α-DnaA polyclonal antibodies.
In vivo helix formation assay
Crosslinking media (Spizizen minimal media supplemented with 0.01 mg/ml Fe–NH4–citrate, 0.5% glucose, 6 mM MgSO4, and 0.02 mg/ml of all the natural amino acids except cysteine) was inoculated directly from −80°C with the required strain and left to grow overnight at 37°C. Cells were diluted 1:100 into fresh crosslinking media in a flask allowing for high aeration and grown at 30°C until the A600 reached ∼0.1 at which point xylose (1% final) was added or removed by washing to induce or to repress soj alleles, respectively. Cell growth was allowed to continue until the A600 reached ∼0.6. 10 ml of culture was collected by centrifugation at 14K r.p.m. for 1 min and resuspended in 2 ml in vivo crosslinking buffer (50 mM HEPES pH 7 and 10% Sucrose). For MFA, another 250 μl of cells was added directly to 25 μl of 10% sodium azide and then treated and analysed by qPCR. Cells were collected as above and resuspended in 100 μl in vivo crosslinking buffer and flash frozen in liquid nitrogen (flash freezing and the subsequent addition of DMSO did not affect cell viability). Frozen cells were thawed and BMOE was added to a final concentration of 2 mM. The reaction was left to proceed for 30 min at 37°C with shaking at 800 r.p.m. before being quenched by the addition of cysteine (100 mM final) for 5 min at 37°C. Cells were lysed by the addition of a 0.5 × reaction volume of SDS–PAGE sample loading buffer and DTT followed by heating to 90°C for 15 min and then briefly sonicated at 4°C. Cell debris was removed by centrifugation at 14K r.p.m. for 15 min at 4°C. Finally, samples were concentrated using a 10-kDa MWCO centrifugal unit (Millipore) and analysed by SDS–PAGE. Bands were visualized by western blot analysis using α-DnaA polyclonal antibodies.
Supplementary Material
Acknowledgments
We thank Dr Leendert Hameon, Dr Masayuki Su'etsugu, and Professor Kenn Gerdes for critical reading of this manuscript. This work was supported by a BBSRC studentship to GS, by Grant number 43/G18654 from the BBSRC to JE, and by a Royal Society University Research Fellowship to HM.
Author contributions: Experimental design, data acquisition, and data analysis were performed by GS and HM. The manuscript was co-written by GS, JE, and HM.
Footnotes
The authors declare that they have no conflict of interest.
References
- Abe Y, Jo T, Matsuda Y, Matsunaga C, Katayama T, Ueda T (2007) Structure and function of DnaA N-terminal domains: specific sites and mechanisms in inter-DnaA interaction and in DnaB helicase loading on oriC. J Biol Chem 282: 17816–17827 [DOI] [PubMed] [Google Scholar]
- Bowers JL, Randell JC, Chen S, Bell SP (2004) ATP hydrolysis by ORC catalyzes reiterative Mcm2-7 assembly at a defined origin of replication. Mol Cell 16: 967–978 [DOI] [PubMed] [Google Scholar]
- Carr KM, Kaguni JM (2001) Stoichiometry of DnaA and DnaB protein in initiation at the Escherichia coli chromosomal origin. J Biol Chem 276: 44919–44925 [DOI] [PubMed] [Google Scholar]
- Cassler MR, Grimwade JE, Leonard AC (1995) Cell cycle-specific changes in nucleoprotein complexes at a chromosomal replication origin. EMBO J 14: 5833–5841 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen LL, Rosa JJ, Turner S, Pepinsky RB (1991) Production of multimeric forms of CD4 through a sugar-based crosslinking strategy. J Biol Chem 266: 18237–18243 [PubMed] [Google Scholar]
- Clarey MG, Erzberger JP, Grob P, Leschziner AE, Berger JM, Nogales E, Botchan M (2006) Nucleotide-dependent conformational changes in the DnaA-like core of the origin recognition complex. Nat Struct Mol Biol 13: 684–690 [DOI] [PubMed] [Google Scholar]
- Duderstadt KE, Chuang K, Berger JM (2011) DNA stretching by bacterial initiators promotes replication origin opening. Nature 478: 209–213 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Duderstadt KE, Mott ML, Crisona NJ, Chuang K, Yang H, Berger JM (2010) Origin remodeling and opening in bacteria relies on distinct assembly states of the DnaA initiator. J Biol Chem 285: 28229–28239 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Erzberger JP, Mott ML, Berger JM (2006) Structural basis for ATP-dependent DnaA assembly and replication-origin remodeling. Nat Struct Mol Biol 13: 676–683 [DOI] [PubMed] [Google Scholar]
- Erzberger JP, Pirruccello MM, Berger JM (2002) The structure of bacterial DnaA: implications for general mechanisms underlying DNA replication initiation. EMBO J 21: 4763–4773 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Felczak MM, Simmons LA, Kaguni JM (2005) An essential tryptophan of Escherichia coli DnaA protein functions in oligomerization at the E. coli replication origin. J Biol Chem 280: 24627–24633 [DOI] [PubMed] [Google Scholar]
- Fujikawa N, Kurumizaka H, Nureki O, Terada T, Shirouzu M, Katayama T, Yokoyama S (2003) Structural basis of replication origin recognition by the DnaA protein. Nucleic Acids Res 31: 2077–2086 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Funnell BE, Baker TA, Kornberg A (1987) In vitro assembly of a prepriming complex at the origin of the Escherichia coli chromosome. J Biol Chem 262: 10327–10334 [PubMed] [Google Scholar]
- Gerdes K, Howard M, Szardenings F (2010) Pushing and pulling in prokaryotic DNA segregation. Cell 141: 927–942 [DOI] [PubMed] [Google Scholar]
- Gorbatyuk B, Marczynski GT (2005) Regulated degradation of chromosome replication proteins DnaA and CtrA in Caulobacter crescentus. Mol Microbiol 55: 1233–1245 [DOI] [PubMed] [Google Scholar]
- Gruber S, Errington J (2009) Recruitment of condensin to replication origin regions by ParB/SpoOJ promotes chromosome segregation in B. subtilis. Cell 137: 685–696 [DOI] [PubMed] [Google Scholar]
- Hassan AK, Moriya S, Ogura M, Tanaka T, Kawamura F, Ogasawara N (1997) Suppression of initiation defects of chromosome replication in Bacillus subtilis dnaA and oriC-deleted mutants by integration of a plasmid replicon into the chromosomes. J Bacteriol 179: 2494–2502 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hester CM, Lutkenhaus J (2007) Soj (ParA) DNA binding is mediated by conserved arginines and is essential for plasmid segregation. Proc Natl Acad Sci USA 104: 20326–20331 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ireton K, Gunther NWt, Grossman AD (1994) spo0J is required for normal chromosome segregation as well as the initiation of sporulation in Bacillus subtilis. J Bacteriol 176: 5320–5329 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Katayama T, Ozaki S, Keyamura K, Fujimitsu K (2010) Regulation of the replication cycle: conserved and diverse regulatory systems for DnaA and oriC. Nat Rev Microbiol 8: 163–170 [DOI] [PubMed] [Google Scholar]
- Kawakami H, Katayama T (2010) DnaA, ORC, and Cdc6: similarity beyond the domains of life and diversity. Biochem Cell Biol 88: 49–62 [DOI] [PubMed] [Google Scholar]
- Kurokawa K, Nishida S, Emoto A, Sekimizu K, Katayama T (1999) Replication cycle-coordinated change of the adenine nucleotide-bound forms of DnaA protein in Escherichia coli. EMBO J 18: 6642–6652 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee PS, Grossman AD (2006) The chromosome partitioning proteins Soj (ParA) and Spo0J (ParB) contribute to accurate chromosome partitioning, separation of replicated sister origins, and regulation of replication initiation in Bacillus subtilis. Mol Microbiol 60: 853–869 [DOI] [PubMed] [Google Scholar]
- Leonard AC, Grimwade JE (2005) Building a bacterial orisome: emergence of new regulatory features for replication origin unwinding. Mol Microbiol 55: 978–985 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leonard AC, Grimwade JE (2010) Regulating DnaA complex assembly: it is time to fill the gaps. Curr Opin Microbiol 13: 766–772 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leonard TA, Butler PJ, Lowe J (2005) Bacterial chromosome segregation: structure and DNA binding of the Soj dimer—a conserved biological switch. EMBO J 24: 270–282 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Livny J, Yamaichi Y, Waldor MK (2007) Distribution of centromere-like parS sites in bacteria: insights from comparative genomics. J Bacteriol 189: 8693–8703 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miller DT, Grimwade JE, Betteridge T, Rozgaja T, Torgue JJ, Leonard AC (2009) Bacterial origin recognition complexes direct assembly of higher-order DnaA oligomeric structures. Proc Natl Acad Sci USA 106: 18479–18484 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Molt KL, Sutera VA Jr, Moore KK, Lovett ST (2009) A role for nonessential domain II of initiator protein, DnaA, in replication control. Genetics 183: 39–49 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mott ML, Berger JM (2007) DNA replication initiation: mechanisms and regulation in bacteria. Nat Rev Microbiol 5: 343–354 [DOI] [PubMed] [Google Scholar]
- Murray H, Errington J (2008) Dynamic control of the DNA replication initiation protein DnaA by Soj/ParA. Cell 135: 74–84 [DOI] [PubMed] [Google Scholar]
- Ozaki S, Katayama T (2009) DnaA structure, function, and dynamics in the initiation at the chromosomal origin. Plasmid 62: 71–82 [DOI] [PubMed] [Google Scholar]
- Ozaki S, Katayama T (2011) Highly organized DnaA-oriC complexes recruit the single-stranded DNA for replication initiation. Nucleic Acids Res (advance online publication 3 November 2011; doi:; DOI: 10.1093/nar/gkr832) [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ozaki S, Kawakami H, Nakamura K, Fujikawa N, Kagawa W, Park SY, Yokoyama S, Kurumizaka H, Katayama T (2008) A common mechanism for the ATP-DnaA-dependent formation of open complexes at the replication origin. J Biol Chem 283: 8351–8362 [DOI] [PubMed] [Google Scholar]
- Ptacin JL, Lee SF, Garner EC, Toro E, Eckart M, Comolli LR, Moerner WE, Shapiro L (2010) A spindle-like apparatus guides bacterial chromosome segregation. Nat Cell Biol 12: 791–798 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rozgaja TA, Grimwade JE, Iqbal M, Czerwonka C, Vora M, Leonard AC (2011) Two oppositely-oriented arrays of low affinity recognition sites in oriC guide progressive binding of DnaA during E. coli pre-RC assembly. Mol Microbiol 82: 475–488 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schepers A, Diffley JF (2001) Mutational analysis of conserved sequence motifs in the budding yeast Cdc6 protein. J Mol Biol 308: 597–608 [DOI] [PubMed] [Google Scholar]
- Scholefield G, Veening JW, Murray H (2010) DnaA and ORC: more than DNA replication initiators. Trends Cell Biol 21: 188–194 [DOI] [PubMed] [Google Scholar]
- Scholefield G, Whiting R, Errington J, Murray H (2011) Spo0J regulates the oligomeric state of Soj to trigger its switch from an activator to an inhibitor of DNA replication initiation. Mol Microbiol 79: 1089–1100 [DOI] [PubMed] [Google Scholar]
- Simmons LA, Felczak M, Kaguni JM (2003) DnaA Protein of Escherichia coli: oligomerization at the E. coli chromosomal origin is required for initiation and involves specific N-terminal amino acids. Mol Microbiol 49: 849–858 [DOI] [PubMed] [Google Scholar]
- Speck C, Messer W (2001) Mechanism of origin unwinding: sequential binding of DnaA to double- and single-stranded DNA. EMBO J 20: 1469–1476 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sullivan NL, Marquis KA, Rudner DZ (2009) Recruitment of SMC by ParB-parS organizes the origin region and promotes efficient chromosome segregation. Cell 137: 697–707 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sutton MD, Carr KM, Vicente M, Kaguni JM (1998) Escherichia coli DnaA protein. The N-terminal domain and loading of DnaB helicase at the E. coli chromosomal origin. J Biol Chem 273: 34255–34262 [DOI] [PubMed] [Google Scholar]
- Tucker PA, Sallai L (2007) The AAA+ superfamily—a myriad of motions. Curr Opin Struct Biol 17: 641–652 [DOI] [PubMed] [Google Scholar]
- Weigel C, Schmidt A, Seitz H, Tungler D, Welzeck M, Messer W (1999) The N-terminus promotes oligomerization of the Escherichia coli initiator protein DnaA. Mol Microbiol 34: 53–66 [DOI] [PubMed] [Google Scholar]
- Wigley DB (2009) ORC proteins: marking the start. Curr Opin Struct Biol 19: 72–78 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.