Abstract
Simple Sequence Repeats (SSRs) or microsatellites constitute a significant portion of genomes however; their significance in organellar genomes has not been completely understood. The availability of organelle genome sequences allows us to understand the organization of SSRs in their genic and intergenic regions. In the present work, SSRs were identified and categorized in 14 mitochondrial and 22 chloroplast genomes of algal species belonging to Chlorophyta. Based on the study, it was observed that number of SSRs in non-coding region were more as compared to coding region and frequency of mononucleotides repeats were highest followed by dinucleotides in both mitochondrial and chloroplast genomes. It was also observed that maximum number of SSRs was found in genes encoding for beta subunit of RNA polymerase in chloroplast genomes and NADH dehydrogenase in mitochondrial genomes. This is the first and original report on whole genomes sequence analysis of organellar genomes of green algae.
Keywords: SSRs, microsatellites, Chlorophyta, chloroplast, mitochondria, green algae
Background
Microsatellites also known as simple sequence repeats (SSRs) or simple sequence length polymorphisms (SSLPs) are small arrays of tandem repeats of one to six nucleotides that are interspersed throughout the genome [1]. They are ubiquitous and highly abundant in prokaryotes and eukaryotes, present even in the smallest bacterial genomes. Microsatellites can be found anywhere in the genome, both in coding and non-coding regions, may arise through an error in the process responsible for copying of the genome during cell division and are unavoidable products of genome replication [2]. Microsatellites can be classified as perfect, imperfect and compound repeats. SSRs were initially considered to be evolutionary neutral but now they are known to play an important role in genome evolution and hot spot of recombination due to their high mutability [3]. They are thought to be involved in gene expression, regulation and function as transcriptional activating elements. SSRs are inherently unstable and are inherited in a mendelian manner and so they can be used for checking genetic relationships [4]. SSRs are highly variable that is number of repeat units in the array is different in different members of a species [5]. SSRs act as powerful genetic markers as they are locus-specific, co-dominant, PCR-based and highly polymorphic [6]. Microsatellites are highly applied in the field of forensics for DNA fingerprinting, paternity studies, diagnosis and identification of disease and population studies [2, 7, 3, 8].
The growing numbers of completed genome sequences in eukaryotic organisms from fungi to humans have greatly assisted understanding of SSRs at the genome- wide level. One obvious observation from such studies have been that the distribution of SSRs in the genome is not random in several respects; differential distribution in terms of abundance of SSRs in between intronic and intergenic regions 5' and 3' UTRs, and different chromosomes; and different species have different frequencies of SSR types and repeat units [9].
Most of the previous studies on microsatellites distribution are based on sequence databases in which coding or gene-rich regions are overrepresented and are generally nuclear- genome based [10] but in current sequenced genome revolution, the complete organellar genomes permit the determination of frequencies of SSRs at the whole genome level. A significant feature of organellar genomes is that they are uniparentally inherited and not perturbed by recombination [11] thus the observed variation may be relevant to understand their maternal mode of transmission. In present study, chloroplast and mitochondrial genomes of organisms belonging to Chlorophyta are analysed for SSRs identification and characterization to detect the degree of polymorphism in them.
Methodology
Organelle genome sequence source:
The mitochondrial and chloroplast genome sequence data, belonging to Chlorophyta, were retrieved from NCBI’s Genome data bank www.ncbi.nlm.nih.gov. The plant species used in study are listed in Table 1 (see supplementary material) (chloroplast) and Table 2 (see supplementary material) (mitochondria) with their accession numbers. Selection of organisms was based on the availability of completely sequenced chloroplast or mitochondrial genomes.
Identification of SSRs:
SSRs were identified using Perl script MISA http://pgrc.ipkgatersleben. de/misa/misa.html, which detects perfect SSRs only. The frequency of SSRs according to size and type of constituting SSRs were determined. MISA considered identifying motifs of one to six nucleotides in size. The minimum repeat unit was defined as ten for mononucleotides, six for dinucleotide and five for all the higher order motifs including tri-, tetra-, penta-, and hexanucleotides. MISA also detects compound microsatellites. The maximal number of interrupting base pairs in a compound microsatellite was set to 100. The occurrence of repeats in genic and intergenic regions and functional categorization of sequences having microsatellites was identified based on the sequence annotation information available in GenBank database.
Discussion
The results obtained from both organelles had not shown any relation between them, they were different in terms of frequency and type of microsatellite present. Chloroplast and mitochondrial genomes sequences are different from nuclear genome in terms of frequency and SSR patterns present in them.
Frequency of SSRs in chloroplast and mitochondrial genomes:
The number of SSRs was varying in different organelle genomes of Chlorophyta. The frequency of SSRs is highest in members of family Trebouxiophyceae and Chlorophyceae in chloroplast genomes and, in Chlorophyceae and Pendinophyceae in case of mitochondrial genomes. In comparison to chloroplast genomes the frequencies of SSRs in mitochondrial genomes were much lower. The chloroplast genomes also contain compound microsatellites in larger number as compared to mitochondrial genomes. The chloroplast genomes of members of Trebouxiophyceae and Chlorophyceae had represented the major compound SSRs containing genomes; in other family members either the number of compound SSR was much less or zero. In mitochondrial genomes no such family based characteristics were observed. The numbers of compound SSRs was consistently low in mitochondrial genomes. In chloroplast genome sequence of Ostreococcus tauri, Nephroselmis olivacea and mitochondrial genome sequence of Chlamydomonas reinhardtii, no SSRs were detected. In common algal species existing in both chloroplast and mitochondrial datasets, there was no relationship between the frequencies of simple and compound SSRs. Frequencies of SSRs were less in the Chlorophyta organelle genomes in comparison with organelle genomes of members of Streptophyta [12, 13].
Distribution of SSRs in genic and intergenic regions:
In both chloroplast and mitochondrial genomes the major proportion of SSRs were detected in intergenic regions as compared to genic and intragenic regions (Figure 1 & Figure 2). In chloroplast genome sequences maximum microsatellites in coding regions were represented by Chlorella vulgaris 1.48% of gross genic region. In other chloroplast genome sequences the microsatellite content in genic regions ranged from 0.01% to 0.57% of total coding content of genomes. In mitochondrial genomes the number of SSRs in genic regions was less which ranged from 0.02% to 0.68% and in almost 50% of available sequenced mitochondrial genomes no microsatellites in genic regions were observed. Interestingly the overall content of microsatellites in genic region of mitochondrial genomes was more than the chloroplast genomes, despite the fact that the former has overall less SSR content.
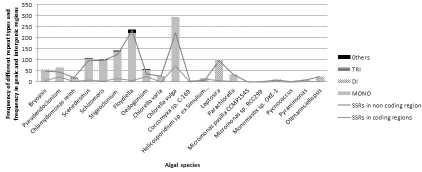
Figure 1.
Frequency of different type of microsatellite repeats and abundance of SSRs in genic and intergenic regions of chloroplast genomes
Figure 2.
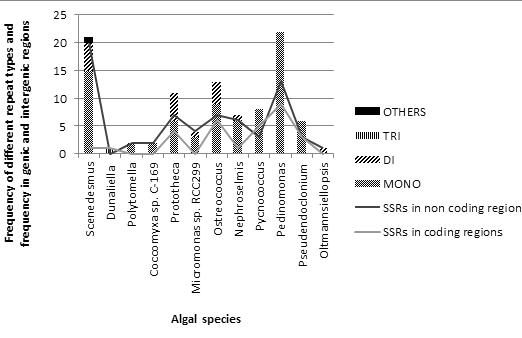
Frequency of different type of microsatellite repeats and abundance of SSRs in genic and intergenic regions of mitochondrial genomes.
The number of SSRs detected in intergenic regions of chloroplast and mitochondrial genomes is higher than SSRs in genic regions, which represented a similar pattern with SSR distribution in organelle genomes of Streptophytes [12, 13]. The differences in frequency of microsatellites in genic and intergenic regions suggest that the polymorphism associated with coding regions is lower than non-coding region. The density of microsatellites in mitochondrial genomes of Streptophyta ranged from 1 SSR/2.06 to SSR/75.27 kb [13] whereas in Chlorophyta this density ranged from 1 SSR/1.25 to 1 SSR/56.76 kb Table 2 (see supplementary material) Polymorphisms associated with a specific locus are due to the variation in length of the microsatellite, which in turn depends on the number of repetitions of the basic motif [14]. The lower substitution rate of chloroplast DNA compared to the nuclear genome has been documented [ 15] and the mutational processes at simple repeat loci in the chloroplast genome also occur less frequently than those in the nuclear genome [11]. The microsatellite patterns observed on various loci in chloroplast and mitochondrial genomes of Chlorophyta differed from each other making it impossible to measure associated relatedness and polymorphism present within alleles.
SSR Patterns:
Most prominent repeat patterns observed in organelle genomes of members of Chlorophyta were mono-nucleotide repeats, poly A/T. The length distribution of SSRs indicated that the frequency of repeats decreases with repeat length (Figure 1 & Figure 2).
Mono-Di-Trinucleotide and other repeats:
In both chloroplast and mitochondrial genomes the most prominent pattern of poly A/T repeats observed was A/T10 and others were A/T11-20, in chloroplast genomes and A/T11-14, in mitochondrial genomes. Poly C/G was either absent in most chloroplast and mitochondrial genomes sequences or in very less frequency in few genomes. Dinucleotide repeat patterns were less frequent as compared to mononucleotide repeats in both organelle genome sequences. In both genome sequences the maximum dinucleotide repeats observed was poly AT/TA and most frequent was AT/TA6. In chloroplast genomes other observations were AT/TA7-12 while in mitochondria, they were AT/TA8, AT/TA11-13. Another rare pattern, AC/GT6, was also detected in mitochondrial genome of Ostreococcus tauri. The major di-nucleotide repeat pattern observed in Chlorophyta organellar genomes had shown the similarity with organelle and nuclear genomes of higher taxa [12, 13, 16]; but in nuclear genome of Chlamydomonas it is AC/GT [16].
Trinucleotide repeats of AAT/TTA5 type were observed in chloroplast genomes in family members of Chlorophyceae and mitochondrial genomes of Dunaliella salina. In very minute amounts other repeat pattern tetra-, penta-, hexa- were also observed in chloroplast genomes of Chlorophycean members but mitochondrial genomes were devoid of more than trinucleotide repeats, except a single panta-nucleotide repeat observed in Scenedesmus obliquus.
Organelle genomes of streptophytes also revealed a similar pattern representing abundance of mononucleotide A/T in both the organellar genomes in comparison to C/G repeats [12, 13, 17] whereas SSRs in nuclear ESTs of other plant species and some cereal species tri-nucleotide repeats were the most abundant class followed by di-nucleotide repeats [8, 18].
Functional categorization of genes having microsatellites:
In all chloroplast genomes, maximum numbers of SSRs were detected in the genes encoding for beta subunit of RNA polymerase, followed by genes encoding for cell division proteins, photosystem II proteins, hypothetical proteins, photosystem I proteins, group II intron, endonuclease, DNA polymerase and transport proteins. In mitochondrial genomes, maximum number of genes containing SSRs encode for NADH dehydrogenase followed by genes encoding for ribosomal proteins, ATP synthase genes, hypothetical, cox2 and transporter proteins. Among the chloroplast and mitochondrial genomes of major cereals maximum number of SSRs was found in rpo and ndh gene clusters [12-.
Conclusion
To studying the distribution pattern of SSRs in organellar genomes of green algae, the mitochondrial and chloroplast genomes of Chlorophyta have been analysed and it is concluded that distribution pattern varies significantly in mitochondrial and chloroplast genomes. The distribution of SSR pattern diversed with the genomic regions, characterized by species and organelle examined. The number of SSRs in chloroplast genomes is higher in comparison to mitochondrial genomes and maximum numbers of SSRs are found in intergenic regions than genic regions in both chloroplast and mitochondrial genomes. Mononucleotides are found to be the most abundant repeat type in both organelles and the repeat motifs are not evenly distributed. The overall representation of SSRs in mitochondrial and chloroplast genomes of Chlorophyta demonstrated that distribution pattern of SSRs in organellar genomes is not uniform and two organelles showed different patterns and arrangement of microsatellites. The study is important in term of revealing the simple repeat patterns in organellar genomes of lower plant species and to measure their abundance and polymorphism. This can be used to suggest the evolution of chloroplast and mitochondrial genomes independent of nuclear genomes. Significant differences in SSR patterns among members of same family also suggest that organellar genomes can be affected by evolutionary factors with different rate and randomness in contrast to nuclear genomes.
Supplementary material
Acknowledgments
The authors acknowledge access to the Dept. of Biotechnology, Govt. of India, Centre for Bioinformatics facility at Department of Biosciences and Biotechnology, Banasthali University, India, through out during this research work.
Footnotes
Citation:Kuntal et al, Bioinformation 8(6): 255-259 (2012)
References
- 1.M Dhar, et al. Plant Sci. 1988;55:43. [Google Scholar]
- 2.S Subramanian, et al. Genome Biol. 2003;4:R13. doi: 10.1186/gb-2003-4-2-r13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.G Toth, et al. Genome Res. 2000;10:967. [Google Scholar]
- 4.P Jarne, PJ Lagoda. Trends Ecol Evol. 1996;11:424. doi: 10.1016/0169-5347(96)10049-5. [DOI] [PubMed] [Google Scholar]
- 5.M Legendre, et al. Genome Res. 2007;17:1787. doi: 10.1101/gr.6554007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.F Grellet, et al. J Mol Bio. 1986;187:495. [Google Scholar]
- 7.SK Shakyawar, et al. Bioinformation. 2009;4:158. [Google Scholar]
- 8.RK Varshney, et al. Cell Mol Biol Lett. 2002;7:537. [PubMed] [Google Scholar]
- 9.Lawson, Zhang Genome Biol. 2006;7:R14. doi: 10.1186/gb-2006-7-2-r14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.ML Zapiola, et al. Mol Ecol Resour. 2010;10:738. doi: 10.1111/j.1755-0998.2009.02828.x. [DOI] [PubMed] [Google Scholar]
- 11.J Provan, et al. Genetics. 1999;153:943. doi: 10.1093/genetics/153.2.943. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.P Rajendrakumar, et al. In Silico Biol. 2008;8:87. [PubMed] [Google Scholar]
- 13.H Kuntal, V Sharma. OMICS. 2011;15:783. doi: 10.1089/omi.2011.0074. [DOI] [PubMed] [Google Scholar]
- 14.MA Pagnotta, et al. Diversity. 2009;1:19. [Google Scholar]
- 15.KH Wolfe, et al. Proc Natl Acad Sci USA. 1987;84:9054. [Google Scholar]
- 16.M von Stackelberg, et al. BMC Plant Biol. 2006;6:9. doi: 10.1186/1471-2229-6-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.DM Melotto-Passarin, et al. Genet Mol Res. 2011;10:2024. [Google Scholar]
- 18.B Jayashree, et al. In Silico Biol. 2006;6:607. [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.