Abstract
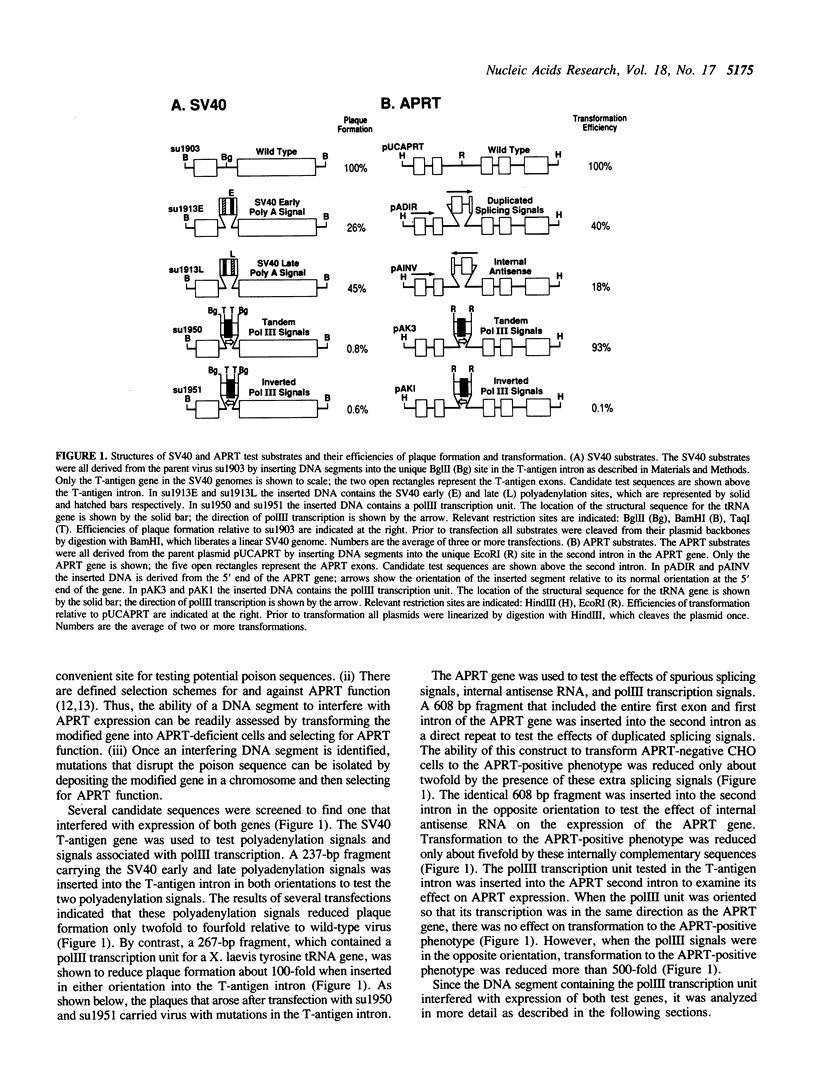
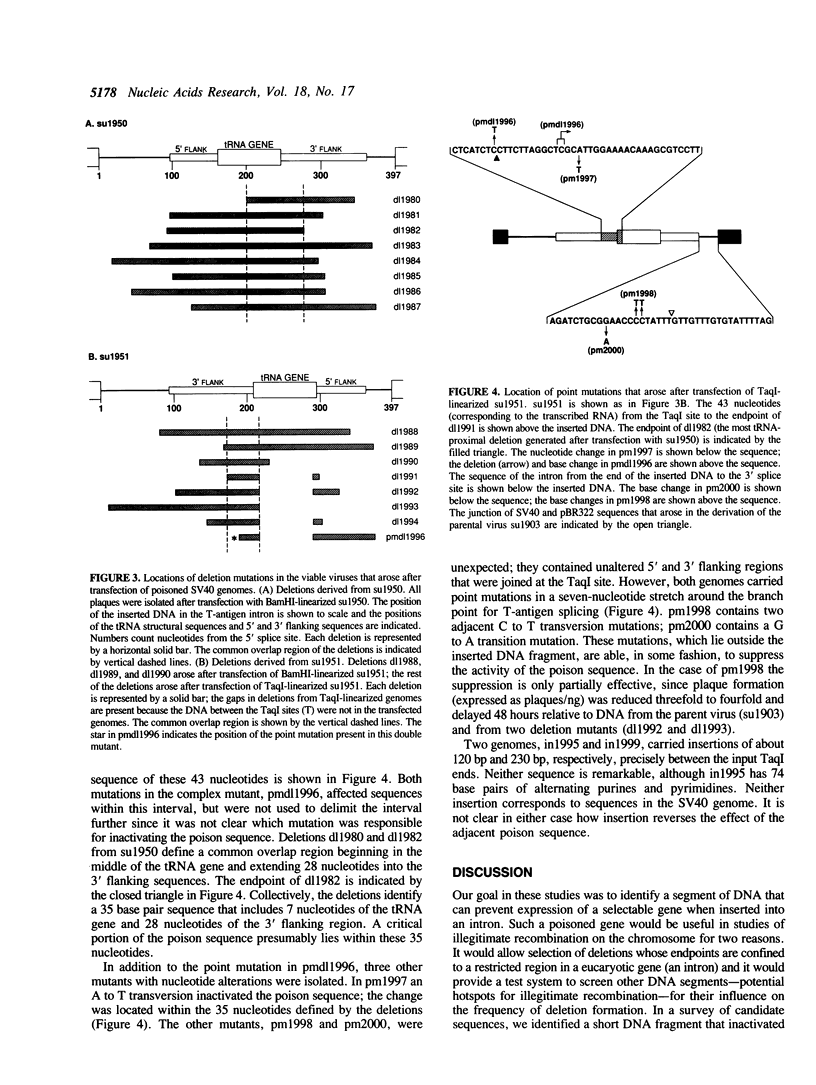
In order to identify a poison sequence that might be useful in studying illegitimate recombination of mammalian cell chromosomes, several DNA segments were tested for their ability to interfere with gene expression when placed in an intron. A tRNA gene and its flanking sequences (267 bp) were shown to inhibit SV40 plaque formation 100-fold, when inserted into the intron in the T-antigen gene. Similarly, when the same DNA segment was placed in the second intron of the adenosine phosphoribosyl transferase (APRT) gene from CHO cells, it inhibited transformation of APRT-CHO cells 500-fold. These two tests indicated that the 267-bp DNA segment contained a poison sequence. The poison sequence did not affect replication since the replication of poisoned SV40 genomes was complemented by viable SV40 genomes and poisoned APRT genes were stably integrated into cell chromosomes. Cleavage of the poison sequence in the SV40 T-antigen intron by restriction enzymes indicated that the tRNA structural sequences and the 5' flanking sequences were not required for inhibition of SV40 plaque formation. Sequence analysis of viable mutant SV40, which arose after transfection of poisoned genomes, localized the poison sequence to a 35 bp segment immediately 3' of the tRNA structural sequences.
Full text
PDF





Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Adair G. M., Carver J. H., Wandres D. L. Mutagenicity testing in mammalian cells. I. Derivation of a Chinese hamster ovary cell line heterozygous for the adenine phosphoribosyltransferase and thymidine kinase loci. Mutat Res. 1980 Sep;72(2):187–205. doi: 10.1016/0027-5107(80)90035-4. [DOI] [PubMed] [Google Scholar]
- Adair G. M., Nairn R. S., Wilson J. H., Seidman M. M., Brotherman K. A., MacKinnon C., Scheerer J. B. Targeted homologous recombination at the endogenous adenine phosphoribosyltransferase locus in Chinese hamster cells. Proc Natl Acad Sci U S A. 1989 Jun;86(12):4574–4578. doi: 10.1073/pnas.86.12.4574. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Adair G. M., Stallings R. L., Nairn R. S., Siciliano M. J. High-frequency structural gene deletion as the basis for functional hemizygosity of the adenine phosphoribosyltransferase locus in Chinese hamster ovary cells. Proc Natl Acad Sci U S A. 1983 Oct;80(19):5961–5964. doi: 10.1073/pnas.80.19.5961. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Adami G., Nevins J. R. Splice site selection dominates over poly(A) site choice in RNA production from complex adenovirus transcription units. EMBO J. 1988 Jul;7(7):2107–2116. doi: 10.1002/j.1460-2075.1988.tb03050.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gorman C. M., Merlino G. T., Willingham M. C., Pastan I., Howard B. H. The Rous sarcoma virus long terminal repeat is a strong promoter when introduced into a variety of eukaryotic cells by DNA-mediated transfection. Proc Natl Acad Sci U S A. 1982 Nov;79(22):6777–6781. doi: 10.1073/pnas.79.22.6777. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kootstra A., Lew L. K., Nairn R. S., MacLeod M. C. Preferential modification of GC boxes by benzo[a]pyrene-7,8-diol-9,10-epoxide. Mol Carcinog. 1989;1(4):239–244. doi: 10.1002/mc.2940010406. [DOI] [PubMed] [Google Scholar]
- Laski F. A., Belagaje R., RajBhandary U. L., Sharp P. A. An amber suppressor tRNA gene derived by site-specific mutagenesis: cloning and function in mammalian cells. Proc Natl Acad Sci U S A. 1982 Oct;79(19):5813–5817. doi: 10.1073/pnas.79.19.5813. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Levitt N., Briggs D., Gil A., Proudfoot N. J. Definition of an efficient synthetic poly(A) site. Genes Dev. 1989 Jul;3(7):1019–1025. doi: 10.1101/gad.3.7.1019. [DOI] [PubMed] [Google Scholar]
- Lowy I., Pellicer A., Jackson J. F., Sim G. K., Silverstein S., Axel R. Isolation of transforming DNA: cloning the hamster aprt gene. Cell. 1980 Dec;22(3):817–823. doi: 10.1016/0092-8674(80)90558-9. [DOI] [PubMed] [Google Scholar]
- Lusky M., Botchan M. Inhibition of SV40 replication in simian cells by specific pBR322 DNA sequences. Nature. 1981 Sep 3;293(5827):79–81. doi: 10.1038/293079a0. [DOI] [PubMed] [Google Scholar]
- Michaeli T., Prives C. pBR322 DNA inhibits simian virus 40 gene expression in Xenopus laevis oocytes. Nucleic Acids Res. 1987 Feb 25;15(4):1579–1594. doi: 10.1093/nar/15.4.1579. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Michel B., Ehrlich S. D. Illegitimate recombination occurs between the replication origin of the plasmid pC194 and a progressing replication fork. EMBO J. 1986 Dec 20;5(13):3691–3696. doi: 10.1002/j.1460-2075.1986.tb04701.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mulligan R. C., Berg P. Expression of a bacterial gene in mammalian cells. Science. 1980 Sep 19;209(4463):1422–1427. doi: 10.1126/science.6251549. [DOI] [PubMed] [Google Scholar]
- Nairn R. S., Humphrey R. M., Adair G. M. Transformation of UV-hypersensitive Chinese hamster ovary cell mutants with UV-irradiated plasmids. Int J Radiat Biol Relat Stud Phys Chem Med. 1988 Feb;53(2):249–260. doi: 10.1080/09553008814550601. [DOI] [PubMed] [Google Scholar]
- Nalbantoglu J., Phear G. A., Meuth M. Nucleotide sequence of hamster adenine phosphoribosyl transferase (aprt) gene. Nucleic Acids Res. 1986 Feb 25;14(4):1914–1914. doi: 10.1093/nar/14.4.1914. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peden K. W., Pipas J. M., Pearson-White S., Nathans D. Isolation of mutants of an animal virus in bacteria. Science. 1980 Sep 19;209(4463):1392–1396. doi: 10.1126/science.6251547. [DOI] [PubMed] [Google Scholar]
- Robberson B. L., Cote G. J., Berget S. M. Exon definition may facilitate splice site selection in RNAs with multiple exons. Mol Cell Biol. 1990 Jan;10(1):84–94. doi: 10.1128/mcb.10.1.84. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roth D. B., Chang X. B., Wilson J. H. Comparison of filler DNA at immune, nonimmune, and oncogenic rearrangements suggests multiple mechanisms of formation. Mol Cell Biol. 1989 Jul;9(7):3049–3057. doi: 10.1128/mcb.9.7.3049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roth D. B., Porter T. N., Wilson J. H. Mechanisms of nonhomologous recombination in mammalian cells. Mol Cell Biol. 1985 Oct;5(10):2599–2607. doi: 10.1128/mcb.5.10.2599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roth D. B., Wilson J. H. Nonhomologous recombination in mammalian cells: role for short sequence homologies in the joining reaction. Mol Cell Biol. 1986 Dec;6(12):4295–4304. doi: 10.1128/mcb.6.12.4295. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roth D. B., Wilson J. H. Relative rates of homologous and nonhomologous recombination in transfected DNA. Proc Natl Acad Sci U S A. 1985 May;82(10):3355–3359. doi: 10.1073/pnas.82.10.3355. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tegtmeyer P. Simian virus 40 deoxyribonucleic acid synthesis: the viral replicon. J Virol. 1972 Oct;10(4):591–598. doi: 10.1128/jvi.10.4.591-598.1972. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wake C. T., Gudewicz T., Porter T., White A., Wilson J. H. How damaged is the biologically active subpopulation of transfected DNA? Mol Cell Biol. 1984 Mar;4(3):387–398. doi: 10.1128/mcb.4.3.387. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wake C. T., Vernaleone F., Wilson J. H. Topological requirements for homologous recombination among DNA molecules transfected into mammalian cells. Mol Cell Biol. 1985 Aug;5(8):2080–2089. doi: 10.1128/mcb.5.8.2080. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wilson J. H., Berget P. B., Pipas J. M. Somatic cells efficiently join unrelated DNA segments end-to-end. Mol Cell Biol. 1982 Oct;2(10):1258–1269. doi: 10.1128/mcb.2.10.1258. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wilson J. H., DePamphilis M., Berg P. Simian virus 40-permissive cell interactions: selection and characterization of spontaneously arising monkey cells that are resistant to simian virus 40 infection. J Virol. 1976 Nov;20(2):391–399. doi: 10.1128/jvi.20.2.391-399.1976. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wilson J. H. Genetic analysis of host range mutant viruses suggests an uncoating defect in simian virus 40-resistant monkey cells. Proc Natl Acad Sci U S A. 1977 Aug;74(8):3503–3507. doi: 10.1073/pnas.74.8.3503. [DOI] [PMC free article] [PubMed] [Google Scholar]



