Abstract
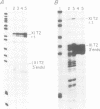
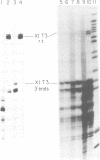
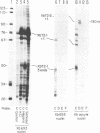
In the ribosomal genes of X. laevis, the sequence GACTTGCNC is found about 60bp upstream of the gene promoter (T3) and is necessary and sufficient to cause termination of RNA polymerase I transcription. At the 3' end of the 40S precursor coding region (T2) a sequence differing by one nucleotide, GACTTGCNG, directs RNA 3' end formation but allows polymerase to transcribe on into the intergenic spacer (Labhart and Reeder, 1989, Genes and Dev. 4: 269-276). Sites corresponding to T2 and T3 are also found in a related species, X. borealis. Inspection of the T2 sequence in X. borealis reveals that it contains two copies of the terminator sequence, GACTTGCNC, located 15 and 96 bp downstream of the 3' end of the 40S precursor coding region. Here we present functional tests of those two T2 elements that show that, as predicted from the sequence, they both show termination activity and are functionally indistinguishable from the T3 site in X. laevis. These results suggest that X. laevis T2 is an example of a naturally occurring point mutation, and the inability to terminate transcription at T2 is an exception to the general pattern of ribosomal gene transcription in higher eukaryotes.
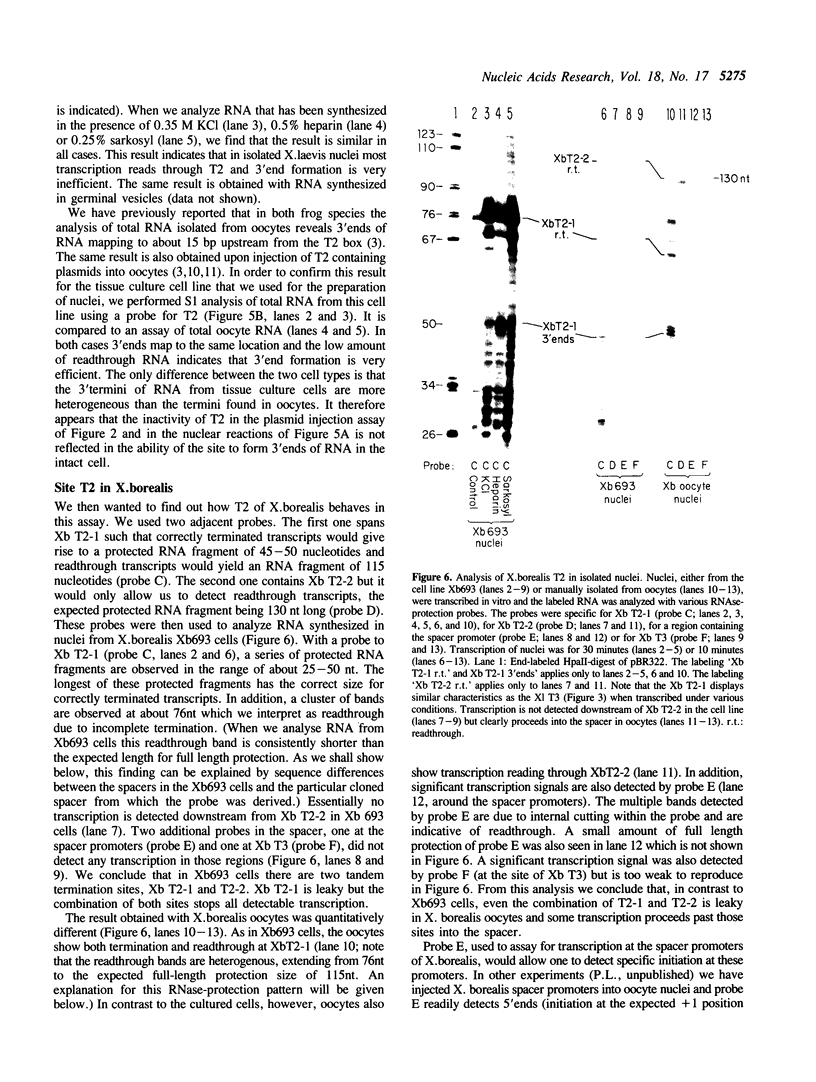
Full text
PDF




Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Bartsch I., Schoneberg C., Grummt I. Evolutionary changes of sequences and factors that direct transcription termination of human and mouse ribsomal genes. Mol Cell Biol. 1987 Jul;7(7):2521–2529. doi: 10.1128/mcb.7.7.2521. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chikaraishi D. M., Buchanan L., Danna K. J., Harrington C. A. Genomic organization of rat rDNA. Nucleic Acids Res. 1983 Sep 24;11(18):6437–6452. doi: 10.1093/nar/11.18.6437. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Crampton J. M., Woodland H. R. A cell-free assay system for the analysis of changes in RNA synthesis during the development of Xenopus laevis. Dev Biol. 1979 Jun;70(2):453–466. doi: 10.1016/0012-1606(79)90038-1. [DOI] [PubMed] [Google Scholar]
- Grummt I., Maier U., Ohrlein A., Hassouna N., Bachellerie J. P. Transcription of mouse rDNA terminates downstream of the 3' end of 28S RNA and involves interaction of factors with repeated sequences in the 3' spacer. Cell. 1985 Dec;43(3 Pt 2):801–810. doi: 10.1016/0092-8674(85)90253-3. [DOI] [PubMed] [Google Scholar]
- Harrington C. A., Chikaraishi D. M. Transcription of spacer sequences flanking the rat 45S ribosomal DNA gene. Mol Cell Biol. 1987 Jan;7(1):314–325. doi: 10.1128/mcb.7.1.314. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Henderson S., Sollner-Webb B. A transcriptional terminator is a novel element of the promoter of the mouse ribosomal RNA gene. Cell. 1986 Dec 26;47(6):891–900. doi: 10.1016/0092-8674(86)90804-4. [DOI] [PubMed] [Google Scholar]
- Kempers-Veenstra A. E., Oliemans J., Offenberg H., Dekker A. F., Piper P. W., Planta R. J., Klootwijk J. 3'-End formation of transcripts from the yeast rRNA operon. EMBO J. 1986 Oct;5(10):2703–2710. doi: 10.1002/j.1460-2075.1986.tb04554.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- La Volpe A., Simeone A., D'Esposito M., Scotto L., Fidanza V., de Falco A., Boncinelli E. Molecular analysis of the heterogeneity region of the human ribosomal spacer. J Mol Biol. 1985 May 25;183(2):213–223. doi: 10.1016/0022-2836(85)90214-1. [DOI] [PubMed] [Google Scholar]
- Labhart P., Reeder R. H. A 12-base-pair sequence is an essential element of the ribosomal gene terminator in Xenopus laevis. Mol Cell Biol. 1987 May;7(5):1900–1905. doi: 10.1128/mcb.7.5.1900. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Labhart P., Reeder R. H. A point mutation uncouples RNA 3'-end formation and termination during ribosomal gene transcription in Xenopus laevis. Genes Dev. 1990 Feb;4(2):269–276. doi: 10.1101/gad.4.2.269. [DOI] [PubMed] [Google Scholar]
- Labhart P., Reeder R. H. Characterization of three sites of RNA 3' end formation in the Xenopus ribosomal gene spacer. Cell. 1986 May 9;45(3):431–443. doi: 10.1016/0092-8674(86)90329-6. [DOI] [PubMed] [Google Scholar]
- Labhart P., Reeder R. H. DNA sequences for typical ribosomal gene spacers from Xenopus laevis and Xenopus borealis. Nucleic Acids Res. 1987 Apr 24;15(8):3623–3624. doi: 10.1093/nar/15.8.3623. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Labhart P., Reeder R. H. High initiation rates at the ribosomal gene promoter do not depend upon spacer transcription. Proc Natl Acad Sci U S A. 1989 May;86(9):3155–3158. doi: 10.1073/pnas.86.9.3155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Labhart P., Reeder R. H. Ribosomal precursor 3' end formation requires a conserved element upstream of the promoter. Cell. 1987 Jul 3;50(1):51–57. doi: 10.1016/0092-8674(87)90661-1. [DOI] [PubMed] [Google Scholar]
- McStay B., Reeder R. H. A termination site for Xenopus RNA polymerase I also acts as an element of an adjacent promoter. Cell. 1986 Dec 26;47(6):913–920. doi: 10.1016/0092-8674(86)90806-8. [DOI] [PubMed] [Google Scholar]
- Morgan G. T., Roan J. G., Bakken A. H., Reeder R. H. Variations in transcriptional activity of rDNA spacer promoters. Nucleic Acids Res. 1984 Aug 10;12(15):6043–6052. doi: 10.1093/nar/12.15.6043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moss T. A transcriptional function for the repetitive ribosomal spacer in Xenopus laevis. Nature. 1983 Mar 17;302(5905):223–228. doi: 10.1038/302223a0. [DOI] [PubMed] [Google Scholar]
- Reeder R. H. Regulatory elements of the generic ribosomal gene. Curr Opin Cell Biol. 1989 Jun;1(3):466–474. doi: 10.1016/0955-0674(89)90007-0. [DOI] [PubMed] [Google Scholar]
- Tautz D., Dover G. A. Transcription of the tandem array of ribosomal DNA in Drosophila melanogaster does not terminate at any fixed point. EMBO J. 1986 Jun;5(6):1267–1273. doi: 10.1002/j.1460-2075.1986.tb04356.x. [DOI] [PMC free article] [PubMed] [Google Scholar]