Abstract
Xeroderma pigmentosum (XP) is a devastating disease associated with dramatic skin cancer proneness. XP cells are deficient in nucleotide excision repair (NER) of bulky DNA adducts including ultraviolet (UV)-induced mutagenic lesions. Approaches of corrective gene transfer in NER-deficient keratinocyte stem cells hold great hope for the long-term treatment of XP patients. To face this challenge, we developed a retrovirus-based strategy to safely transduce the wild-type XPC gene into clonogenic human primary XP-C keratinocytes. De novo expression of XPC was maintained in both mass population and derived independent candidate stem cells (holoclones) after more than 130 population doublings (PD) in culture upon serial propagation (>1040 cells). Analyses of retrovirus integration sequences in isolated keratinocyte stem cells suggested the absence of adverse effects such as oncogenic activation or clonal expansion. Furthermore, corrected XP-C keratinocytes exhibited full NER capacity as well as normal features of epidermal differentiation in both organotypic skin cultures and in a preclinical murine model of human skin regeneration in vivo. The achievement of a long-term genetic correction of XP-C epidermal stem cells constitutes the first preclinical model of ex vivo gene therapy for XP-C patients.
Introduction
The mammalian epidermis is a squamous stratified epithelium endowed with a capacity of permanent renewal throughout life and fast regeneration upon accidental injury. Interfollicular epidermal stem cells are located in the innermost layer (basal layer) of the epithelium. Although they divide infrequently in vivo, epidermal stem cells can achieve more than 150 population doublings (PD) and generate a progeny of >1040 cells when cultivated under appropriate conditions in vitro.1 Clonal analyses have shown that, in vitro, the keratinocytes endowed with the highest proliferative potential generate large colonies whose progeny is composed of >95% clonogenic cells. These keratinocytes, known as holoclones, are thought to correspond to epidermal stem cells.2 Remarkably, cultured epidermal stem cells retain the ability to regenerate a fully differentiated epidermis when grafted back to an autologous donor as demonstrated by the successful treatment of thousands of severely burnt patients since the late 1970s.3,4 On this basis, it has been proposed that skin resurfacing using genetically corrected epidermal stem cells could greatly contribute to the clinical treatment of some devastating monogenic skin diseases that still lack appropriate treatment. Recent advances reported by Mavilio and colleagues demonstrated the benefits of ex vivo corrective gene transfer in combination with skin grafting for patients suffering from junctional epidermolysis bullosa.5
Xeroderma pigmentosum (XP) is one of those rare, life-threatening disorders. XP patients are highly sensitive to sunlight exposure and have a tremendous risk (2000×) of developing skin tumors in sun-exposed areas, mostly basal and squamous cell carcinomas, arising from epidermal keratinocytes, and malignant melanomas.6 XP cells are deficient in nucleotide excision repair (NER), a versatile DNA repair mechanism involved in the removal of bulky DNA adducts including ultraviolet (UV)-induced lesions such as cyclobutane pyrimidine dimers (CPD) and 6,4 pyrimidine-pyrimidone (6-4 PP). NER relies on the recognition of helix-distorting lesions followed by the assembly of a multiprotein machinery leading to (i) DNA unwinding around the lesion catalyzed by XPB and XPD helicases; (ii) excision of the DNA strand bearing the lesion thanks to the 5′ and 3′ endonuclease activities of XPF and XPG, respectively; (iii) replicative DNA synthesis and ligation.7 The NER process operates through two subpathways thought to differ only in the initial step of DNA damage recognition. In actively transcribed genes, stalling of RNA polymerase II at the DNA distortion initiates the assembly of the repair complex (transcription-coupled repair). In contrast, the global genome repair subpathway is triggered by the recognition of bulky lesions in nontranscribed DNA by the XPC-HR23B-Centrin2 complex.8 Seven XP groups of genetic complementation (XP-A to XP-G) corresponding to gene-specific alterations of the NER pathway have been described. Persistence of UV-induced DNA damage in NER-deficient XP cells results in elevated mutagenesis, eventually leading to the development of skin tumors in sun-exposed areas. Recent experimental evidence has suggested that UV-induced skin carcinomas may result from the accumulation of DNA damage in murine epidermal stem cells9,10 but direct evidence in human cells deserves further investigations. Nevertheless, one can anticipate that NER deficiency in XP stem cells and/or progenitors is expected to play an essential role in skin cancer development. Protecting stem cells from the accumulation of DNA lesions thus appears as the cornerstone of any perennial anticancer approach for XP patients.
In the absence of any curative treatment for XP patients, management of the disease mainly involves strict avoidance of sun exposure and surgical resection of newly developed skin tumors. In most severe cases, excision of large portions of skin can be followed by reconstructive surgery using photo-protected skin autografts.11,12 However, engrafted cells remain DNA repair-deficient and thus susceptible to UV-induced neoplastic transformation.13 Grafting genetically corrected skin in XP patients would certainly reduce the incidence of cancerous lesions as long as (i) skin grafts contain a sufficient proportion of stem cells to allow lifelong renewal of the regenerated epidermis, (ii) all stem cells are functionally corrected, i.e., are protected against mutagenesis, (iii) safety assessment excludes adverse effects such as oncogene activation linked to retroviral integration. XP group C (XP-C) is the best candidate for an ex vivo cutaneous gene therapy protocol since clinical traits of XP-C patients are mainly restricted to photo-exposed skin, generally without the neurological disorders that can be observed in other XP groups.14 In our previous attempt, however, we failed to obtain a sustained expression of XPC complementary DNA in XP-C primary keratinocytes, i.e., for more than 30 PD (ref. 15 and Arnaudeau, Chevallier and Magnaldo, unpublished results). To circumvent this limiting issue, we developed a safe, retroviral-based protocol based on immunoaffinity sorting of multiplying/transduced primary XP-C keratinocytes. For the first time, our data demonstrate that (i) retrovirus-mediated wild-type XPC gene transfer in XP-C primary keratinocytes fully restores DNA repair capacity and cell survival properties after UV irradiation; (ii) corrected XP-C keratinocytes exhibiting the long-term growth potential of stem cells can be isolated in culture after retroviral transduction and non-antibiotic cell selection; (iii) transgene expression in stem cells persists sufficiently to ensure long-term protection against UV challenge after 130 PD; (iv) transduced XP-C keratinocytes can regenerate, both in vitro and in vivo, a differentiated epidermis with a normal capacity to repair UV-induced DNA lesions.
Results
Correction of the genetic defect responsible for the XP disease
Our first aim here was to assess the presence of stably corrected stem cells after retroviral transduction of primary keratinocytes populations isolated from healthy and non-photo exposed skin from XP-C patients. To allow purification of transduced cells, we constructed a Moloney murine leukemia virus-derived retroviral vector containing the bicistronic cassette CD24-IRES-XPC driven by the cytomegalovirus promoter (Supplementary Figure S1). CD24 is a small cell surface marker of postmitotic epidermal keratinocytes.16 We previously reported that ectopic expression of CD24 in clonogenic keratinocytes allows their immnunoaffinity-based selection with subsequent enrichment in stem cells.17 Yet, as for other genodermatoses such as recessive junctional epidermolyis bullosa,5 the presence of a sufficient number of stem keratinocytes in patients' cell cultures and their safe transduction and selection remained to be demonstrated.
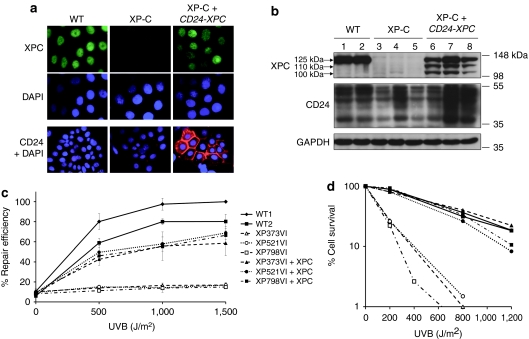
Primary keratinocytes from three independent XP-C donors (XP373VI, XP521VI, and XP798VI) were infected with high-titer (1.2 × 109 infectious viral genome/ml) retroviral supernatants and purified by magnetic-activated cell sorting using a monoclonal CD24 antibody. No evidence of altered morphology or cytotoxicity was observed after transduction and cell sorting (data not shown). Southern blot analysis demonstrated the presence of a 4.7 kb fragment corresponding to the full-length proviral sequence in the genome of transduced cells (Supplementary Figure S1). Expression of CD24 and XPC proteins in control (WT), parental (XP-C), and transduced XP-C cells (XP-C+CD24-XPC) was assessed by indirect immunofluorescence labeling and western blotting (Figure 1a,b). In WT and XP-C keratinocytes, CD24 was detected only at the periphery of stratified/differentiated cells (large cells); in contrast, exogenous CD24 was expressed at the membrane of transduced, clonogenic keratinocytes (small cells) from where it is normally absent16 (Figure 1a). Due to germinal mutations resulting in the production of a premature termination codon (Supplementary Table S1) and the use of an antibody raised against the carboxyterminal end of the protein, the XPC protein was not detected in XP-C keratinocytes. In contrast, all nuclei were positive in WT control cells as well as in transduced XP-C keratinocytes (Figure 1a). In transduced cells, levels of nuclear XPC were heterogeneous from one colony to another, suggesting that the sorted population was polyclonal. Western blotting analysis showed that the full-length XPC protein (XPC-FL, 125 kDa) was expressed in WT and transduced XP-C keratinocytes, but could not be detected in parental XP-C keratinocytes (Figure 1b). Surprisingly, two additional bands migrating as ~110 and ~100 kDa proteins, respectively, were also detected in transduced XP-C keratinocytes, suggesting retrovirus-driven expression of shorter forms of the XPC protein (see discussion).
Figure 1.
Restoration of XPC expression, UV-cell survival and nucleotide excision repair in transduced XP-C keratinocytes. (a) Immunofluorescence analysis of XPC (green) and CD24 (red) expression in WT (WT1) and XP-C (XP521VI) keratinocytes before (XP-C) and after (XP-C+CD24-XPC) retroviral transduction and CD24 selection. DAPI was used to stain nuclei. (b) Western blot analysis of WT (1: WT1; 2: WT2), XP-C (3: XP373VI; 4: XP521VI; 5: XP798VI), and transduced XP-C keratinocytes (6: XP373VI+CD24-XPC; 7: XP521VI+CD24-XPC; 8: XP798VI+CD24-XPC). Anti-XPC and anti-CD24 antibodies were used as probes; anti-GAPDH antibody was used as a loading control. (c) Determination of nucleotide excision repair (NER) efficiency after UVB irradiation by unscheduled DNA synthesis (UDS) at the indicated doses. Percent repair efficiency is determined by the ratio of the average number of grains in a strain to the average number of grains in WT#1 cells at 1,500 J/m2 (100%). Data are represented as mean ± SEM of at least two independent experiments. (d) UV survival curves of WT, XP-C, and transduced XP-C strains after UVB irradiation at the indicated doses. GAPDH, glyceraldehyde 3-phosphate dehydrogenase; NER, nucleotide excision repair; UDS, unscheduled DNA synthesis.
To assess the ability of transduced XP-C keratinoytes to perform NER, unscheduled DNA synthesis (UDS) was measured after UVB irradiation, as described15,18 (Figure 1c). In XP-C keratinocytes, the residual DNA repair capacity was low (10–15% compared to WT keratinocytes), and almost no increase was observed after UVB irradiation. Conversely, transduced XP-C keratinocytes recovered dose-dependent levels of UDS in the range of two independent NER-proficient strains of primary keratinocytes (about 70 and 90% UDS recovery after the highest UVB dose compared to WT1 and WT2 strains, respectively).
The ability of transduced, NER-reverted keratinocytes to counteract lethal effects of UV irradiation was then assessed by clonal analysis. In the absence of UV irradiation, colony-forming efficiencies (CFE) of WT, XP-C, and transduced XP-C keratinocytes ranged over 10–20%. After UVB irradiation, XP-C keratinocytes showed markedly reduced survival levels when compared to WT keratinocytes (Figure 1d). In contrast, transduced XP-C keratinocytes exhibited UVB cell survival comparable to that of WT cells (Figure 1d). Altogether, these data demonstrate efficient reversion of the DNA repair defect and UV sensitivity of XP-C keratinocytes following transduction with the CD24-IRES-XPC retroviral vector.
Long-term genetic correction of XP-C patient's keratinocyte stem cells ex vivo
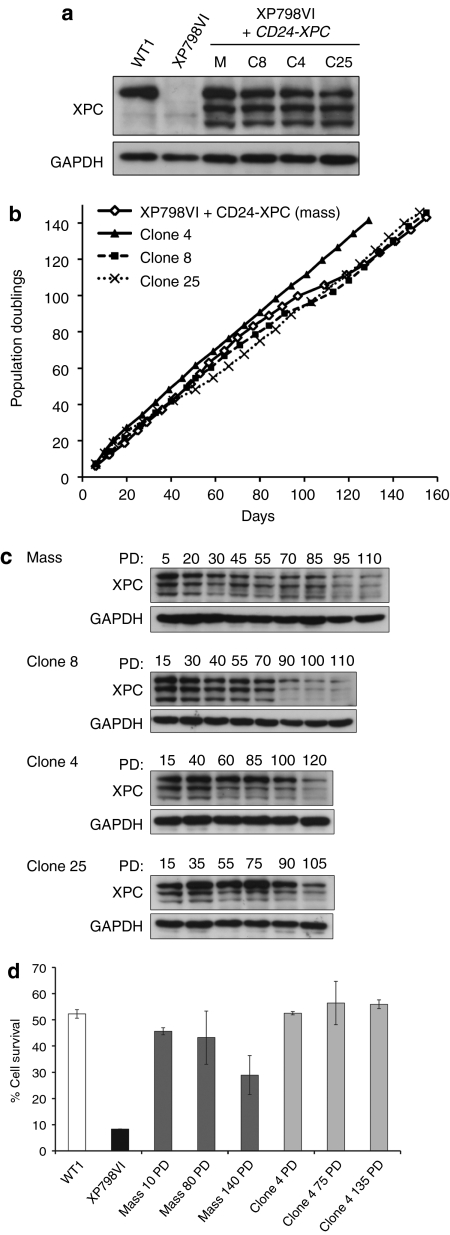
Successful gene therapy in rapidly renewing tissues requires efficient stem cell targeting, an essential prerequisite for long-term transgene expression.19 In vitro clonogenic studies have demonstrated that holoclones, which corresponds to epidermal stem cells in vitro, generate large colonies with a smooth perimeter (LSP) when seeded at clonal density on feeder cells.2 The presence of genetically corrected holoclones among transduced XP-C keratinocytes was thus assessed by clonal analysis and serial propagation. We isolated 32 candidate holoclones from the XP798VI+CD24-XPC mass population; all of them expressed the XPC protein, although at variable levels (Supplementary Figure S2). The long-term growth potential of three clones (clone 4, 8, and 25) with XPC-FL protein levels in the range of control cells (Figure 2a) was analyzed. The three clones as well as the corrected mass population were serially propagated for more than 140 days and achieved more than 140 PD (Figure 2b). These data unambiguously demonstrate the presence of true stem cells within the transduced population. Whether the reverted population and the derived holoclones expressed XPC-FL in the long-term was assessed by western blot analysis. In the reverted mass population subjected to serial propagation, the total amount of XPC was clearly reduced from 85 PD, although it was still detected after 110 PD (Figure 2c). In clone 8, XPC expression was roughly stable up to 70 PD before decreasing and being stably maintained at a low but detectable level until 110 PD (end of western blot experiment). In clone 4 and in clone 25, XPC expression was only decreased after 100 and 90 PD, respectively (Figure 2c).
Figure 2.
Presence of corrected stem cells in transduced XP-C keratinocytes cell populations. (a) Western blot analysis of XPC expression in holoclones (C8, C4, and C25) isolated from CD24-XPC-transduced XP798VI keratinocytes (M, corrected mass population). WT1, positive control; XP798VI, parental XP-C keratinocytes. Anti-GAPDH antibody was used as a loading control. (b) Cumulative cell doublings of CD24-XPC-transduced XP798VI keratinocytes (XP798VI+CD24-XPC (mass), open diamonds) and holoclones C4 (black triangles), C8 (black squares), and C25 (black cross). (c) Western blot analysis of XPC expression in CD24-XPC transduced XP798VI keratinocytes (mass) and its derived holoclones C8, C4, and C25 after the indicated population doublings (PD 1–120). (d) Survival of CD24-XPC-transduced XP798VI keratinocytes (mass) and its derived holoclone (clone 4) after exposure to a single UV-solar simulated radiation (UV-SSR, 4,000 J/m2 UVB + 41,300 J/m2 UVA) during long-term serial propagation. Data are represented as mean ± SEM of two independent experiments. GAPDH, glyceraldehyde 3-phosphate dehydrogenase; PD, population doublings.
To determine whether the decrease of XPC expression measured in the reverted mass population could be due to a loss of transduced cells, we isolated eight clones from the mass population after 120 PD. The full-length CD24-IRES-XPC cassette (4.7 kb) was detected in genomic DNA from all clones (Supplementary Figure S2), but expression of the XPC protein was uniformly weak in those eight clones (Supplementary Figure S2). Together, these data suggested that upon serial propagation clonal restriction could have occurred in favor of clones expressing low levels of XPC.
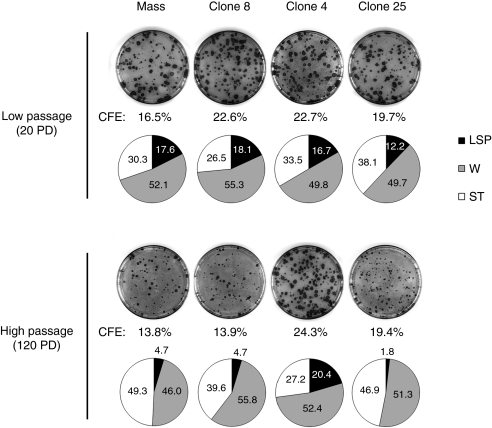
To demonstrate long-term functional protection against UV radiation, cell survival of transduced keratinocytes that were propagated over 10, 80, and 140 PD was assessed by clonal analysis after a single UV-solar simulated radiation (SSR) exposure (4,000 J/m2 UVB + 41,300 J/m2 UVA) (Figure 2d). At this dose, survival of WT and XP-C keratinocytes was about 50 and 10%, respectively. In the reverted mass population, decreased XPC expression in cells serially propagated over 140 PD was accompanied by a significantly higher (about 40%) cell mortality; resistance of transduced keratinocytes at 140 PD was nevertheless threefold higher than that of parental XP-C cells. In contrast UV-cell survival of clone 4 at 135 PD remained similar to that of control (WT) keratinocytes. Since holoclone 4 can be expanded over 140 PD and can still generate LSP colonies after 120 PD (Figure 3), it can be considered as a bona fide stem cell with stable genetic and phenotypic correction.
Figure 3.
Clonal analyses in corrected keratinocytes at lowand high population doublings. Colony-forming efficiency and colony-type distribution of corrected XP798VI mass population (mass) and reverted holoclones (clone 8, 4, and 25) at low passages (<20 PD) and high passages (>120 PD). Colonies were characterized by their size and morphological features: LSP, large colonies with smooth perimeter; W, wrinkled colonies; ST, small terminal colonies (for additional details, see “Materials and Methods”). Note the transition from a high (about 12–18%) to a reduced (1.8–4.7%) proportion of LSP colonies in mass population and in clone 8 and clone 25 after serial propagation (i.e., at 20 versus 120 PD). In contrast, clone 4 still generated a high proportion of LSPs colonies (about 20%) after 120 PD. “Mass” is for corrected XP798VI keratinocyte mass population; CFE, colony-forming efficiency; PD, population doublings. LSP, large (>4mm diameter) with smooth perimeter; W, intermediate (1 <∅ <4mm) wrinkled colonies; ST, small (∅ <1mm) terminally differentiated colonies (ST).
Safety of the genetic correction in individual clonogenic epidermal cells
To assess the safety of integration events in transduced keratinocytes, we performed a genome-wide analysis of the proviral integration sites in 10 clones isolated from the corrected XP798VI mass population after 20 PD. Libraries of vector-genome junctions were generated by linker-mediated (LM) nested PCR, and sequenced to saturation. This approach allowed unambiguous mapping of 33 independent integrations in genomic DNA and confirmed the polyclonality of the transduced mass population (Supplementary Table S2). As expected for γ-retroviruses, about 30% of the integrations occurred ≤30 kb upstream or downstream of 30 genes, and 15% were located at a distance ≤10 kb from transcriptional start sites. Twelve integrations (36%) were classified as intergenic and 11 (33%) were within the transcribed portion of referenced genes (Supplementary Tables S2 and S3). To investigate the putative emergence of specific clones during serial propagation, analysis of the proviral integration sites was also performed in nine clones isolated from the corrected XP798VI mass population after 120 PD. Fifteen independent integrations were found in these clones, among which five were also present in clones isolated after 20 PD (Supplementary Table S2). Since these integration sites were associated with different events at each time point, we concluded that no particular integration event was clonally selected.
Generation of genetically corrected skin ex vivo and in vivo
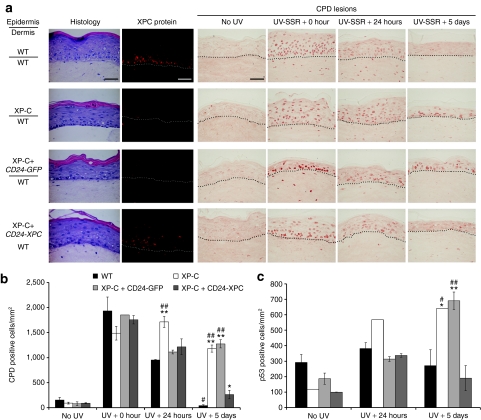
The ability of corrected XP-C keratinocytes to complete epidermal morphogenesis was then assessed in ex vivo organotypic skin cultures and after grafting onto athymic mice. In organotypic skin cultures, gene-corrected cells retained full stratification and differentiation potential (Figure 4a, Histology). As expected, the XPC protein was not detected in epidermis regenerated from both parental and CD24-GFP-transduced XP-C keratinocytes (Figure 4a, XPC protein). In WT epidermis, the XPC protein was preferentially detected in the basal layer, whereas in epidermis regenerated from CD24-XPC-transduced XP-C keratinocytes, XPC was detected at variable levels in all (basal and suprabasal) nuclei (Figure 4a, XPC protein). Organotypic skin cultures were exposed to a single dose of UV-SSR (1,600 J/m2 UVB + 17,000 J/m2 UVA) and DNA repair efficiency was measured by immunostaining of CPD lesions at various times after UV exposure (Figure 4a, CPD lesions and Figure 4b). Immediately after irradiation, CPD lesions were detected in all keratinocyte nuclei irrespective of the type of epidermis. Twenty-four hours after UVB irradiation, most nuclei remained positive in XP-C epidermis, whereas fewer CPD lesions were detected in WT, XP-C+CD24-GFP, and XP-C+CD24-XPC epidermis (Figure 4a,b). Five days after irradiation, about 75% CPD lesions persisted in all layers of XP-C and XP-C+CD24-GFP epidermis. In contrast, >95% of lesions were removed in WT epidermis and only 15% were still detectable in XP-C+CD24-XPC epidermis (Figure 4a,b).
Figure 4.
Restoration of proficient DNA repair in genetically corrected organotypic skin cultures. (a) Histological staining (HES, hematoxylin, eosin, and safran) shows that transduced XP-C (XP798VI) keratinocytes (XP-C+CD24-XPC) regenerate an epidermis with appropriate features of stratification and differentiation. XP-C keratinocytes transduced with pCMMP CD24-IRES-GFP retrovirus were used as a control (XP-C+CD24-GFP). XPC protein expression was revealed by immunofluorescence staining (XPC, red). The NER efficiency after a single UV-SSR exposure (1,600 J/m2 UVB + 17,000 J/m2 UVA) was assessed by indirect immunofluorescence staining of CPD DNA lesions at the indicated time points (CPD lesions, brown). The dash line delimits the dermal–epidermal junction. Bar, 50 µm. (b) Quantification of CPD repair in organotypic skin cultures. For each sample, CPD-positive cells were counted on four independent representative fields and their number was expressed with respect to the epidermis surface area, as estimated using ImageJ software. (c) After p53 immunolabeling, quantification of p53-positive cells was performed as described in b. Mean values obtained from each epidermis were compared to mean values in WT (*) or corrected XP-C epidermis (#) at the same time point. **/##P < 0.001; */#P < 0.01. All XP-C keratinocytes were from patient XP798VI. CPD, cyclobutane pyrimidine dimers; NER, nucleotide excision repair.
In accordance, the P53 protein, which expression is stabilized in the presence of DNA lesions, rapidly accumulated in the nuclei of basal keratinocytes of all samples 24 hours after irradiation (Figure 4c). Five days after irradiation, the number of P53-positive cells had returned to basal levels in WT and XP-C+CD24-XPC epidermis, whereas it remained elevated in NER-deficient epidermis (XP-C and XP-C+CD24-GFP).
Recently a skin-humanized mouse model of XP-C was achieved upon engraftment of bioengineered skin regenerated from XP-C patients cells onto immunodeficient mice.20 Following this approach we generated mice engrafted either with WT, XP-C+CD24-GFP or XP-C+CD24-XPC keratinocytes (Figure 5c), Histology. As previously observed in XP-C skin-humanized mice,21 animals engrafted with corrected XP-C cells presented normal skin architecture (Figure 5c, Histology and Supplementary Figure S3).
Figure 5.
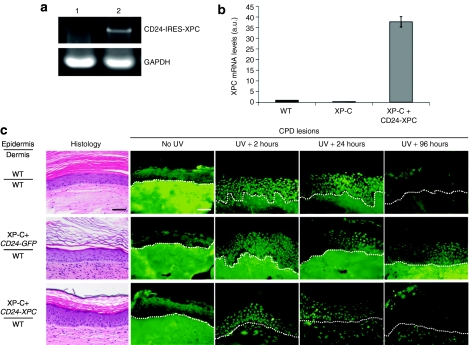
Restoration of normal DNA repair capacity in vivo. (a) RT-PCR analysis of the full-length CD24-IRES-XPC transcript (4.7 kb) in XP-C+CD24-GFP (lane 1) and XP-C+CD24-XPC (lane 2) regenerated skins in vivo, 12 weeks after grafting. (b) Real-time quantitative-PCR analysis of XPC mRNA levels in WT, XP-C+CD24-GFP and XP-C+CD24-XPC regenerated skins in vivo, 12 weeks after grafting. GAPDH and PPIA mRNA levels were quantified in each sample for normalization. (c) Histological appearance of WT, XP-C+CD24-GFP, and XP-C+CD24-XPC epidermis was revealed by HES staining 8 weeks after grafting. The NER efficiency was assessed after labeling of CPD lesions by indirect immunofluorescence at the indicated times to assess the ability to repair CPD DNA lesions after a single UVB exposure (3,360 J/m2). The dash line indicates the dermal–epidermal junction. Bar, 50 µm. Histological aspect of the grafts at each time point following UV exposure is shown in Supplementary Figure 3. CPD, cyclobutane pyrimidine dimers; GAPDH, glyceraldehyde 3-phosphate dehydrogenase; HES, hematoxylin, eosin, and safran; mRNA, messenger RNA; NER, nucleotide excision repair; RT-PCR, reverse transcriptase-PCR.
Immunostaining of XPC using our polyclonal mouse antibody on the grafts section was not reliable (see Material and Methods). Nevertheless, the full-length CD24-IRES-XPC transcript (4.7 kb) could be detected in the corrected skins 12 weeks after grafting (Figure 5a). Real-time quantitative-PCR analyses revealed persistence of high levels of XPC messenger RNA in corrected (CD24-IRES-XPC) grafts compared to control (WT) and mocktransduced (CD24-IRES-GFP) grafts (Figure 5b).
Human skin grafts were then submitted to a single UVB exposure at a dose equivalent to twice the biological efficient dose, based on previously described criteria.22 Two hours after UV irradiation, CPD lesions were detectable in all nuclei of the regenerated skins and persisted in an amount virtually identical 24 hours later (Figure 5c, CPD lesions). However, owing to some background staining, reliable quantification of CPD lesions could not be performed as for organotypic skin cultures (Figure 4); nevertheless, we could definitely determine that 4 days after UV exposure (96 hours), CPD lesions were completely eliminated from WT and XP-C+CD24-XPC regenerated skins whereas numerous CPD-positive nuclei could still be detected in XP-C+CD24-GFP epidermis (Figure 5c, CPD lesions). These data indicated that genetically corrected XP-C keratinocytes recovered full NER capcity in the three-dimensional context of skin in vivo.
Discussion
We report herein the first long-term genetic correction of epidermal stem cells from NER-deficient/cancer-prone XP-C patients. Until now, most approaches of retroviral/lentiviral mediated-corrective gene transfer in XP cells have been limited to fibroblasts.23,24,25,26 However, since most skin tumors arise from epidermal keratinocytes, cutaneous gene therapy for XP patients must rely on the stable genetic correction of XP keratinocyte stem cells. Since prospects of skin grafting require the complete elimination of cancer-prone cells, and thus the selection of genetically corrected keratinocytes, here we used a strategy allowing the safe sorting of transduced keratinocyte stem cells.17 Using this method, primary keratinocytes from three independent XP-C patients were transduced with a retroviral vector containing the CD24-IRES-XPC cassette. The choice of a retroviral vector was dictated by a much higher efficiency of transduction of human primary epidermal keratinocytes compared to lentiviral vectors (data not shown). In primary keratinocytes, other studies reported either similar efficiency of lentiviruses and γ-retroviruses,27 or a threefold higher efficiency of retroviruses over lentiviruses.28 γ-retroviruses are also superior over lentiviruses for transducing the collagen VII complementary DNA in human and dog cells from individual suffering from recessive dystrophic epidermolysis bullosa.29,30
In the three strains studied here, as in about 95% XP-C patients, the XPC protein could not be detected, presumably due to nonsense-mediated mRNA decay of transcripts containing in-frame premature stop codons.31 Unexpectedly, expression of the full-length (125 kDa) XPC protein in transduced XP-C keratinocytes was accompanied by the expression of two shorter proteins of ~100 and 110 kDa. The XPC coding sequence bears two in-frame AUG codons (positions +457 and +685, respectively, downstream the bona fide initiation codon (+106)). Translation initiation from these codons could theoretically generate proteins of 823 (XPC (118–940), ~110 kDa] and 747 (XPC (194–940), ~100 kDa] amino acids, respectively. In 1992, Legerski et al. reported the cloning of a human XPC complementary DNA lacking the 117 first amino acids (XPC (118–940)] expression of this protein in immortalized XP-C cells restored normal UDS levels and UV survival properties.32 This protein presumably corresponds to the 110 kDa protein observed in our transduced cells. Whether XPC (194–940), is functional and retains the capacity to interact with other DNA repair proteins such as OGG1 and XPA7,33,34 is not known yet. However, the absence of cytotoxicity, the full recovery of UDS and cell survival after UVB irradiation in transduced XP-C keratinocytes suggest that this shorter XPC form neither acts as a dominant negative regulator of NER nor noticeably modifies cell behavior. For safety reasons, improved retroviral constructs containing an internal promoter instead of the IRES sequence (as described in ref. 29) are being developed and should avoid the production of these XPC isoforms.
In contrast to hematopietic stem cells that can be enriched a priori by immunologic sorting from bone marrow samples, there is no reliable means (except the selection of holoclones) to isolate interfollicular keratinocyte stem cells in culture before genetic manipulation. A major objective here was thus to assess safe and successful genetic correction of XP-C keratinocyte stem cells. Our results indicate that (i) mass populations of genetically corrected XP-C keratinocytes contain clones with the morphological characteristics of holoclones and (ii) candidate holoclones could be individually propagated ex vivo for more than 140 PD, which definitively demonstrates the presence of stem cells in the transduced population. A second essential question concerned the sustainability of corrective gene expression in transduced XP-C keratinocyte stem cells. Our previous attempts showed that retroviral-mediated expression of the XPC protein in XP-C keratinocytes was limited to 6–7 weeks (<30 PD),15 presumably due to antibiotic selection of transduced cells.19 Our CD24-selection procedure yielded a much better sustainability of XPC expression, yet the latter was decreased in the very long term (>85 PD) in the mass population after serial propagation. In individual clones derived from this mass population, the amount of XPC protein(s) was roughly stable up to 70 (clone 8), 100 (clone 4), and 90 (clone 25) PD, respectively; then, expression of all three forms of XPC was reduced, although it was still clearly detectable after 100 PD (Figure 2). Since the average proviral copy number was diminished by about 35% between 20 and 120 PD (Supplementary Figure S2), a progressive selection of clonogenic cells bearing less copies of the proviral genome could account for the decrease of XPC levels in the transduced mass culture. XPC is a DNA binding protein thought to permanently scan the chromatin for the presence of bulky DNA adducts.8 Inappropriately elevated levels of XPC could thus interfere with the replication and/or transcription processes and result in slower growth of cells expressing high levels of XPC. Finally, we cannot rule out that silencing of the transgene, through methylation of the provirus regulatory sequences,35 or post-translational modification of XPC protein stability, could also contribute to the decrease of XPC expression, in particular in isolated holoclones. Importantly, after 130 PD, residual amount of the XPC protein was still sufficient to confer protection against UV to the transduced mass population, as well as to holoclones. Considering that human epidermis contains ~1010 cells and is entirely renewed every month (i.e., about 1,000 renewals in 80 years), less than 45 PD from one stem cell should be largely sufficient to produce the cell progeny required for complete epidermal regeneration throughout life (about 1013 cells). Since we were able to isolate stably transduced stem cells in which functional expression of XPC was maintained over 100 PD, pools of transduced holoclones carefully chosen after mapping of retroviral insertion sites36 should be largely sufficient for lifelong replacement of a dilapidated skin area by a fully sun-resistant counterpart.
Beyond protection against UV irradiation, an important question was to assess their capacity to generate a normal epidermis in organotypic skin cultures and after grafting on immunodeficient mice. Due to its transcriptional control by the hybrid long terminal repeat -cytomegalovirus promoter, XPC was expressed in all living layers of the epidermis regenerated from transduced XP-C keratinocytes, whereas it was found mostly in the basal layer of control (WT) reconstructed epidermis, as in human skin.37 Organotypic skin cultures and skin grafts regenerated from transduced keratinocytes exhibited normal features of proliferation, stratification, and differentiation (data not shown), suggesting that pan-epidermal XPC expression had no detectable impact on epidermal homeostasis.
Although, irrespective of the host species of antibodies, we failed to reliably detect the XPC protein in situ on sections from skin-humanized mice (see Materials and Methods), long-term presence and expression of XPC in corrected skin could be verified by real-time quantitative-PCR 12 weeks after grafting. Importantly, in both organotypic skin cultures and skin regenerated in vivo from reverted XP-C keratinocytes (8 weeks after grafting), complete elimination of CPD DNA lesions could be observed by 96 hours following a single UVB irradiation. Thus, in a 3D context, the expression of XPC in both basal and suprabasal epidermal layers remained compatible with normal rates of DNA repair by NER in corrected XP-C epidermis. Together with efficient NER, the absence of prolonged nuclear accumulation of P53 in corrected XP-C epidermis suggests recovery of a normal cellular response to UV-induced DNA damage.
In further experiments it will be essential to determine whether corrected XP-C epidermal cells are also protected in the long-term from UV-induced tumorigenesis. Using recombinant adenoviruses, Marchetto et al. have shown that in vivo delivery of human XPA complementary DNA to Xpa-mutant mice prevented UVB-induced squamous cell carcinoma development.26 However, as the use of adenoviral vectors in clinical trials is hampered by the strong immune response they elicit in the host,38 lentiviruses and Moloney murine leukemia virus-derived retroviruses currently remain the most suitable vectors for optimal transduction of primary keratinocytes.5,30,39 Here, safety assessment of retroviral integration sites did not reveal clonal expansion of cells containing specific integration events after serial propagation in vitro. Although a clonal restriction might still occur in vivo, Kolodka et al. suggested that the number of PD required to maintain the grafted epidermis for 12 weeks is probably not sufficient to generate this restriction in vivo.40 In accordance, Mavilio and colleagues reported the absence of clonal selection 4 months after grafting corrected keratinocytes in a patient suffering from junctional epidermolysis bullosa.5 This first successful clinical trial of ex vivo cutaneous gene therapy was based on a conventional MoMLV retroviral vector.5 As described in the latter study,5 the only accurate way to identify transduced stem cells is to isolate and propagate clonogenic cells in vitro. Such an approach may appear as a limitation in the frame of clinical applications but the present study shows that appropriate and highly controlled culture conditions give excellent results. Then, in the case of XP patients, a first attempt would be to graft genetically corrected epidermal sheets at the place of surgically resected tumors. Prior such clinical application, we aim at further minimizing the potential adverse effects of insertional mutagenesis and oncogene activation41 using self-inactivating vectors in which the transgene would be under the control of the endogenous XPC promoter.29
Materials and Methods
Skin biopsies and cell culture. All XP-C patients presented a marked photosensitivity and were unrelated (Supplementary Table S1). XP-C skin biopsies from non-photo exposed sites (buttock) were obtained with the patients' or parents' written informed consent in accordance with the declaration of Helsinki.
Keratinocytes were seeded at a density of 4,000 cells/cm2 in complete F12 Adenine DMEM (cFAD) medium, on a feeder layer of lethally X-ray–irradiated 3T3-J2 fibroblasts as described.42 Cells were cultured at 37 °C in a 10% CO2 atmosphere.
Retroviral production and transduction. Infectious retroviral particles pseudotyped with the vesicular stomatitis virus glycoprotein were produced following triple transfection of helper plasmids and the proviral genome (Supplementary Figure S1) in 293T cells as described.43 RV particles were resuspended in serum-free medium (Invitrogen, Cergy-Pontoise, France). Keratinocyte were seeded at a density of 8,000 cells/cm2 and were transduced with concentrated RV supernatants (1.2 × 109 vg/ml) in serum-free medium for 16 hours as described.17 (Sigma-Aldrich, St Louis, MO). They were then refed with cFAD medium until reaching 80–90% confluence.
Magnetic-activated cell sorting CD24 cell sorting. Keratinocytes were dissociated and incubated for 20 minutes at 4 °C with CD24 monoclonal antibody (ABL9; Immunotech, Luminy, France) in cold phosphate-buffered saline containing 0.5% bovine serum albumin, 2 mmol/l EDTA. After two washes, they were incubated for 15 minutes at 4 °C with goat anti-mouse IgG-coupled microbeads (Miltenyi Biotec, Bergisch Gladbach, Germany), and then sorted on magnetic-activated cell sorting as described.17
Irradiation sources and procedures. UVB irradiation was performed using a Philips TL20W/12 fluorescent tube as described.44 UV-SSR using a 1000 W Xenon lamp equipped with a dichroic mirror (Oriel, Les Ulis, France) filtered by UG5 (2 mm) and WG 320 (1.5 mm) filters (Schott, Clichy, France) was performed as described.45 Organotypic skin cultures were exposed without medium to 18,600 J/m2 UV-SSR (1,600 J/m2 UVB + 17,000 J/m2 UVA).
Western blot analysis. Proteins in 8 mol/l urea buffer were separated on 10% SDS–polyacrylamide gels, transferred onto a polyvinyl difluoride membrane (GE Healthcare, Saclay, France) and probed using mouse monoclonal XPC antibody (dilution 1/1,000; Abcam, Cambridge, UK), CD24 monoclonal antibody (ALB9; Immunotech; dilution 1/400) or GAPDH monoclonal antibody (clone 9484; Abcam; dilution 1/2,000). Blots were revealed using electrochemiluminescence reagents (GE Healthcare).
Southern blot. Genomic DNA (20 µg) was digested overnight with AgeI/EcoRV restriction enzymes (Roche, Meylan, France) to release the full-length CD24-IRES-XPC cassette (4.7 kb) (Supplementary Figure S1). Digested DNA was separated on a 0.7% agarose gel, transferred to a nylon membrane (Hybond; GE Healthcare) and hybridized with a [32P]-labeled XPC-specific probe obtained by PCR amplification (XPC: 5′AAAACACGATGATAATATGGCC/5′GCAGAGTAAATAGCAAATCTCC).
UDS analysis. DNA repair after UVB irradiation was followed by [3H]thymidine incorporation as described.18,44 The mean number of grains per nucleus was obtained by counting at least 60 non-S phase nuclei for each UV dose. All UDS experiments were performed at least twice.
Real-time quantitative-PCR. Biopsies from regenerated skins (~0.2 cm2) were excised 12 weeks after grafting. Samples were crushed using Lysing Matrix D beads and FastPrep 24 device (MP Biomedicals, Illkirch, France). Total RNA extraction was then performed using RNeasy Mini Kit (Qiagen, Hilden, Germany). Reverse transcription was performed using Advantage RT for PCR kit (Clontech, Saint-Germain-en-Laye, France) according to the manufacturer's protocol. The full-length CD24-IRES-XPC cassette was amplified using 5′TAATCCGGATCCTCTAGAGC/5′CCCTCAAAGTAGACGG-CATC. Real-timequantitative-PCR was performed according to the manufacturer instructions using Hs01104212_m1 primer set for XPC, Hs99999905_m1 for GAPDH, and Hs999999904_m1 for PPIA (Peptidylpropyl isomerase A/cyclophilin A).
Cell survival analysis. Keratinocytes were grown in 60-mm dishes. After UV exposure, cells were dissociated and seeded at clonal density (17 cells/cm2) onto lethally γ-irradiated 3T3-J2 feeders. After 12 days, keratinocyte colonies were fixed in 3.7% formaldehyde in phosphate-buffered saline and colored by 1% rhodamine B (Sigma-Aldrich). The relative cell survival was calculated as follow: (number of colonies obtained after UV irradiation/number of colonies obtained from non-irradiated cells) × 100.
Clonal analysis and long-term serial propagation of individual clones. Clonal types were determined as described2,44: (i) large colonies (∅>4mm) with a LSP; (ii) small (∅ <1mm) terminally differentiated colonies; (iii) intermediate (1 <∅ <4mm) wrinkled colonies (W). LSP colonies were individually trypsinized; one-fourth of each clone was transferred into a 100-mm culture dish to perform secondary clonal analysis and three-fourths of each clone was replated for serial propagation and further analysis. Corrected XP-C mass population and selected clones were serially passaged during 22 weeks (1 passage per week). Proteins were prepared every 2 weeks. PD was calculated as follow: PD = (log N/N0)/log2, where N0 represents the number of colony-forming cells plated and N the total number of cells obtained at passage.
Organotypic skin cultures. Detailed procedure was as described.46 Keratinocytes overlying the dermal equivalent (lattice) were then incubated for 7 days at 37 °C, 5% CO2, immersed in the culture medium, before being raised at the air–liquid interface for 8 days. Each condition was performed in duplicate. After SSR exposure, samples were immediately harvested or maintained for 24 hours or 5 days at the air–liquid interface, and then processed for analyses.
In vivo regeneration of genetically engineered human epidermis. A fibrin matrix populated with live fibroblasts was used as the dermal component of the bioengineered skin following the procedure previously described.21 WT, gene-corrected or uncorrected XP-C human keratinocytes (0,5–1 × 106 cells/well) were then seeded on the fibrin matrix to form the epidermal layer of the bioengineered skin. When confluent, bioengineered skins were manually detached from tissue culture wells and grafted onto immunodeficient mice. All animal studies have been approved by Centro de Investigaciones Energéticas Medioambientales y Tecnológicas's (CIEMAT) institutional review board and all experimental procedures were conducted according to European and Spanish laws and regulations. Animals were housed in pathogen-free conditions, in individually ventilated type II cages, with 25 air changes per hour and 10 KGy γ-irradiated soft wood pellets as bedding.
Immunostaining analysis. Immunolabeling of XPC and CD24 proteins were performed on cells or on air-dried 5-µm cryosections using mouse polyclonal XPC antibody47 (dilution 1/400) and CD24 monoclonal antibody (ALB9; Immunotech; 1/50). Note that the murine polyclonal XPC antibody generated an unacceptable background in human skin grafted in mice. We thus failed to reliably detect the XPC protein in human skin sections after grafting. Detection of thymine dimers on cryosections was carried out as previously described, using mouse monoclonal H3 antibody (1/300; Sigma-Aldrich).22 For detection of p53 and thymine dimers on formalin-fixed paraffin-embedded sections, mouse monoclonal p53 (1/50; Diaclone, Besançon, France) and KTM53 (1/1,000; Kamiya Biomedicals, Seattle, WA) antibodies were used. Staining was revealed using a biotin/streptavidin detection kit (Vector Laboratories, Burlingame, CA).
Analysis of retroviral integration sites. Integration sites were cloned by LM-PCR, as described.5 Genomic DNA was digested with MseI and PstI, and ligated to an MseI double-strand linker. LM-PCR was performed with nested primers specific for the LTR and the linker. PCR products were shotgun cloned by the TOPO TA cloning kit (Invitrogen) into libraries of integration junctions, which were sequenced to saturation. Sequences were mapped onto the human genome by the BLAT genome browser (UCSC Human Genome Project Working Draft, May 2004; http://www.genome.ucsc.edu).
Statistical analysis. Results are expressed as means ± SD. Mean values were compared using two-tailed Student's t-test. P values < 0.01 were considered statistically significant.
SUPPLEMENTARY MATERIAL Figure S1. Southern analysis of provirus integration into the host genome. Figure S2. XPC expression in independent keratinocyte clones at low and high population doublings. Figure S3. Histological features of in vivo regenerated skin after UV exposure. Table S1. Patients and cells characteristics. Table S2. Complete list of proviral integrations in corrected clones isolated after 20 and 120 population doublings. Table S3. Distribution of proviral integration sites.
Acknowledgments
We are indebted to Jacques Leclaire for his continuous support and encouragements. We thank Dr Sandra Del Bino for helpful advices and Juliette Sok for her excellent and enthusiastic experimental help with RNA extraction from skin samples. We thank Gaëlle Gendronneau for her kind help with qPCR analysis. We thank Christian Baudoin for his help with Southern Blot analysis. We thank the Production and Control department of Genethon which is supported by the Association Française contre les Myopathies (13158). L'Oréal Recherche is acknowledged for exclusive financial support of the in vitro experiments performed in this study. We are indebted to the Fondation de l'Avenir (ET9-551) for his continuous support. E.W. thanks the Association Nationale de la Recherche et de la Technologie (ANRT) and gratefully acknowledges grants from the Fondation René Touraine and the Ligue Nationale Contre le Cancer. AS is thankful to the “Association des Enfants de la Lune” (Tercis, France). F.L. was supported in part by grants PI081054 from ISCIII and PBIO-0306-2006 from Comunidad de Madrid (CAM). M.D.R. was supported by grant SAF2010-16976 from MICINN. The authors declared no conflict of interest.
Supplementary Material
Southern analysis of provirus integration into the host genome.
XPC expression in independent keratinocyte clones at low and high population doublings.
Histological features of in vivo regenerated skin after UV exposure.
Patients and cells characteristics.
Complete list of proviral integrations in corrected clones isolated after 20 and 120 population doublings.
Distribution of proviral integration sites.
REFERENCES
- Rheinwald JG., and, Green H. Epidermal growth factor and the multiplication of cultured human epidermal keratinocytes. Nature. 1977;265:421–424. doi: 10.1038/265421a0. [DOI] [PubMed] [Google Scholar]
- Barrandon Y., and, Green H. Three clonal types of keratinocyte with different capacities for multiplication. Proc Natl Acad Sci USA. 1987;84:2302–2306. doi: 10.1073/pnas.84.8.2302. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gallico GG., 3rd, , O'Connor NE, Compton CC, Kehinde O., and, Green H. Permanent coverage of large burn wounds with autologous cultured human epithelium. N Engl J Med. 1984;311:448–451. doi: 10.1056/NEJM198408163110706. [DOI] [PubMed] [Google Scholar]
- Pellegrini G, Ranno R, Stracuzzi G, Bondanza S, Guerra L, Zambruno G.et al. (1999The control of epidermal stem cells (holoclones) in the treatment of massive full-thickness burns with autologous keratinocytes cultured on fibrin Transplantation 68868–879. [DOI] [PubMed] [Google Scholar]
- Mavilio F, Pellegrini G, Ferrari S, Di Nunzio F, Di Iorio E, Recchia A.et al. (2006Correction of junctional epidermolysis bullosa by transplantation of genetically modified epidermal stem cells Nat Med 121397–1402. [DOI] [PubMed] [Google Scholar]
- Kraemer KH, Lee MM., and, Scotto J. Xeroderma pigmentosum. Cutaneous, ocular, and neurologic abnormalities in 830 published cases. Arch Dermatol. 1987;123:241–250. doi: 10.1001/archderm.123.2.241. [DOI] [PubMed] [Google Scholar]
- Cleaver JE, Lam ET., and, Revet I. Disorders of nucleotide excision repair: the genetic and molecular basis of heterogeneity. Nat Rev Genet. 2009;10:756–768. doi: 10.1038/nrg2663. [DOI] [PubMed] [Google Scholar]
- Sugasawa K, Akagi J, Nishi R, Iwai S., and, Hanaoka F. Two-step recognition of DNA damage for mammalian nucleotide excision repair: Directional binding of the XPC complex and DNA strand scanning. Mol Cell. 2009;36:642–653. doi: 10.1016/j.molcel.2009.09.035. [DOI] [PubMed] [Google Scholar]
- Morris RJ, Coulter K, Tryson K., and, Steinberg SR. Evidence that cutaneous carcinogen-initiated epithelial cells from mice are quiescent rather than actively cycling. Cancer Res. 1997;57:3436–3443. [PubMed] [Google Scholar]
- Nijhof JG, van Pelt C, Mulder AA, Mitchell DL, Mullenders LH., and, de Gruijl FR. Epidermal stem and progenitor cells in murine epidermis accumulate UV damage despite NER proficiency. Carcinogenesis. 2007;28:792–800. doi: 10.1093/carcin/bgl213. [DOI] [PubMed] [Google Scholar]
- Akizuki T., and, Harii K. Microsurgical facial reconstruction of a xeroderma pigmentosum patient after skin tumor resection: case report. J Reconstr Microsurg. 1990;6:129–134. doi: 10.1055/s-2007-1006812. [DOI] [PubMed] [Google Scholar]
- Atabay K, Celebi C, Cenetoglu S, Baran NK., and, Kiymaz Z. Facial resurfacing in xeroderma pigmentosum with monoblock full-thickness skin graft. Plast Reconstr Surg. 1991;87:1121–1125. doi: 10.1097/00006534-199106000-00018. [DOI] [PubMed] [Google Scholar]
- Ergün SS, Cek DI., and, Demirkesen C. Is facial resurfacing with monobloc full-thickness skin graft a remedy in xeroderma pigmentosum. Plast Reconstr Surg. 2002;110:1290–1293. doi: 10.1097/01.PRS.0000025230.84677.C7. [DOI] [PubMed] [Google Scholar]
- Khan SG, Oh KS, Emmert S, Imoto K, Tamura D, Digiovanna JJ.et al. (2009XPC initiation codon mutation in xeroderma pigmentosum patients with and without neurological symptoms DNA Repair (Amst) 8114–125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arnaudeau-Bégard C, Brellier F, Chevallier-Lagente O, Hoeijmakers J, Bernerd F, Sarasin A.et al. (2003Genetic correction of DNA repair-deficient/cancer-prone xeroderma pigmentosum group C keratinocytes Hum Gene Ther 14983–996. [DOI] [PubMed] [Google Scholar]
- Magnaldo T., and, Barrandon Y. CD24 (heat stable antigen, nectadrin), a novel keratinocyte differentiation marker, is preferentially expressed in areas of the hair follicle containing the colony-forming cells. J Cell Sci. 1996;109 ( Pt 13:3035–3045. doi: 10.1242/jcs.109.13.3035. [DOI] [PubMed] [Google Scholar]
- Bergoglio V, Larcher F, Chevallier-Lagente O, Bernheim A, Danos O, Sarasin A.et al. (2007Safe selection of genetically manipulated human primary keratinocytes with very high growth potential using CD24 Mol Ther 152186–2193. [DOI] [PubMed] [Google Scholar]
- Sarasin A, Blanchet-Bardon C, Renault G, Lehmann A, Arlett C., and, Dumez Y. Prenatal diagnosis in a subset of trichothiodystrophy patients defective in DNA repair. Br J Dermatol. 1992;127:485–491. doi: 10.1111/j.1365-2133.1992.tb14845.x. [DOI] [PubMed] [Google Scholar]
- Mathor MB, Ferrari G, Dellambra E, Cilli M, Mavilio F, Cancedda R.et al. (1996Clonal analysis of stably transduced human epidermal stem cells in culture Proc Natl Acad Sci USA 9310371–10376. [DOI] [PMC free article] [PubMed] [Google Scholar]
- García M, Llames S, García E, Meana A, Cuadrado N, Recasens M.et al. (2010In vivo assessment of acute UVB responses in normal and Xeroderma Pigmentosum (XP-C) skin-humanized mouse models Am J Pathol 177865–872. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Del Rio M, Larcher F, Serrano F, Meana A, Muñoz M, Garcia M.et al. (2002A preclinical model for the analysis of genetically modified human skin in vivo Hum Gene Ther 13959–968. [DOI] [PubMed] [Google Scholar]
- Bernerd F., and, Asselineau D. Successive alteration and recovery of epidermal differentiation and morphogenesis after specific UVB-damages in skin reconstructed in vitro. Dev Biol. 1997;183:123–138. doi: 10.1006/dbio.1996.8465. [DOI] [PubMed] [Google Scholar]
- Carreau M, Quilliet X, Eveno E, Salvetti A, Danos O, Heard JM.et al. (1995Functional retroviral vector for gene therapy of xeroderma pigmentosum group D patients Hum Gene Ther 61307–1315. [DOI] [PubMed] [Google Scholar]
- Quilliet X, Chevallier-Lagente O, Eveno E, Stojkovic T, Destée A, Sarasin A.et al. (1996Long-term complementation of DNA repair deficient human primary fibroblasts by retroviral transduction of the XPD gene Mutat Res 364161–169. [DOI] [PubMed] [Google Scholar]
- Zeng L, Quilliet X, Chevallier-Lagente O, Eveno E, Sarasin A., and, Mezzina M. Retrovirus-mediated gene transfer corrects DNA repair defect of xeroderma pigmentosum cells of complementation groups A, B and C. Gene Ther. 1997;4:1077–1084. doi: 10.1038/sj.gt.3300495. [DOI] [PubMed] [Google Scholar]
- Marchetto MC, Muotri AR, Burns DK, Friedberg EC., and, Menck CF. Gene transduction in skin cells: preventing cancer in xeroderma pigmentosum mice. Proc Natl Acad Sci USA. 2004;101:17759–17764. doi: 10.1073/pnas.0406304101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuhn U, Terunuma A, Pfutzner W, Foster RA., and, Vogel JC. In vivo assessment of gene delivery to keratinocytes by lentiviral vectors. J Virol. 2002;76:1496–1504. doi: 10.1128/JVI.76.3.1496-1504.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ghazizadeh S, Katz AB, Harrington R., and, Taichman LB. Lentivirus-mediated gene transfer to human epidermis. J Investig Dermatol Symp Proc. 2004;9:269–275. doi: 10.1111/j.1087-0024.2004.09302.x. [DOI] [PubMed] [Google Scholar]
- Titeux M, Pendaries V, Zanta-Boussif MA, Décha A, Pironon N, Tonasso L.et al. (2010SIN retroviral vectors expressing COL7A1 under human promoters for ex vivo gene therapy of recessive dystrophic epidermolysis bullosa Mol Ther 181509–1518. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gache Y, Pin D, Gagnoux-Palacios L, Carozzo C., and, Meneguzzi G. Correction of dog dystrophic epidermolysis bullosa by transplantation of genetically modified epidermal autografts. J Invest Dermatol. 2011;131:2069–2078. doi: 10.1038/jid.2011.172. [DOI] [PubMed] [Google Scholar]
- Khan SG, Oh KS, Shahlavi T, Ueda T, Busch DB, Inui H.et al. (2006Reduced XPC DNA repair gene mRNA levels in clinically normal parents of xeroderma pigmentosum patients Carcinogenesis 2784–94. [DOI] [PubMed] [Google Scholar]
- Legerski R., and, Peterson C. Expression cloning of a human DNA repair gene involved in xeroderma pigmentosum group C. Nature. 1992;359:70–73. doi: 10.1038/359070a0. [DOI] [PubMed] [Google Scholar]
- Bunick CG, Miller MR, Fuller BE, Fanning E., and, Chazin WJ. Biochemical and structural domain analysis of xeroderma pigmentosum complementation group C protein. Biochemistry. 2006;45:14965–14979. doi: 10.1021/bi061370o. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bernardes de Jesus BM, Bjoras M, Coin F., and, Egly JM. Dissection of the molecular defects caused by pathogenic mutations in the DNA repair factor XPC. Mol Cell Biol. 2008;28:7225–7235. doi: 10.1128/MCB.00781-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Challita PM., and, Kohn DB. Lack of expression from a retroviral vector after transduction of murine hematopoietic stem cells is associated with methylation in vivo. Proc Natl Acad Sci USA. 1994;91:2567–2571. doi: 10.1073/pnas.91.7.2567. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Larcher F, Dellambra E, Rico L, Bondanza S, Murillas R, Cattoglio C.et al. (2007Long-term engraftment of single genetically modified human epidermal holoclones enables safety pre-assessment of cutaneous gene therapy Mol Ther 151670–1676. [DOI] [PubMed] [Google Scholar]
- de Feraudy S, Boubakour-Azzouz I, Fraitag S, Berneburg M, Chan L, Chew K.et al. (2010Diagnosing xeroderma pigmentosum group C by immunohistochemistry Am J Dermatopathol 32109–117. [DOI] [PubMed] [Google Scholar]
- Yang Y, Nunes FA, Berencsi K, Furth EE, Gönczöl E., and, Wilson JM. Cellular immunity to viral antigens limits E1-deleted adenoviruses for gene therapy. Proc Natl Acad Sci USA. 1994;91:4407–4411. doi: 10.1073/pnas.91.10.4407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gagnoux-Palacios L, Hervouet C, Spirito F, Roques S, Mezzina M, Danos O.et al. (2005Assessment of optimal transduction of primary human skin keratinocytes by viral vectors J Gene Med 71178–1186. [DOI] [PubMed] [Google Scholar]
- Kolodka TM, Garlick JA., and, Taichman LB. Evidence for keratinocyte stem cells in vitro: long term engraftment and persistence of transgene expression from retrovirus-transduced keratinocytes. Proc Natl Acad Sci USA. 1998;95:4356–4361. doi: 10.1073/pnas.95.8.4356. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hacein-Bey-Abina S, Von Kalle C, Schmidt M, McCormack MP, Wulffraat N, Leboulch P.et al. (2003LMO2-associated clonal T cell proliferation in two patients after gene therapy for SCID-X1 Science 302415–419. [DOI] [PubMed] [Google Scholar]
- Rheinwald JG., and, Green H. Serial cultivation of strains of human epidermal keratinocytes: the formation of keratinizing colonies from single cells. Cell. 1975;6:331–343. doi: 10.1016/s0092-8674(75)80001-8. [DOI] [PubMed] [Google Scholar]
- Merten OW. State-of-the-art of the production of retroviral vectors. J Gene Med. 2004;6 ( suppl. 1:S105–S124. doi: 10.1002/jgm.499. [DOI] [PubMed] [Google Scholar]
- Otto AI, Riou L, Marionnet C, Mori T, Sarasin A., and, Magnaldo T. Differential behaviors toward ultraviolet A and B radiation of fibroblasts and keratinocytes from normal and DNA-repair-deficient patients. Cancer Res. 1999;59:1212–1218. [PubMed] [Google Scholar]
- Fagot D, Asselineau D., and, Bernerd F. Matrix metalloproteinase-1 production observed after solar-simulated radiation exposure is assumed by dermal fibroblasts but involves a paracrine activation through epidermal keratinocytes. Photochem Photobiol. 2004;79:499–505. doi: 10.1562/yg-03-11-r1.1. [DOI] [PubMed] [Google Scholar]
- Bernerd F, Asselineau D, Vioux C, Chevallier-Lagente O, Bouadjar B, Sarasin A.et al. (2001Clues to epidermal cancer proneness revealed by reconstruction of DNA repair-deficient xeroderma pigmentosum skin in vitro Proc Natl Acad Sci USA 987817–7822. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Charbonnier JB, Renaud E, Miron S, Le Du MH, Blouquit Y, Duchambon P.et al. (2007Structural, thermodynamic, and cellular characterization of human centrin 2 interaction with xeroderma pigmentosum group C protein J Mol Biol 3731032–1046. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Southern analysis of provirus integration into the host genome.
XPC expression in independent keratinocyte clones at low and high population doublings.
Histological features of in vivo regenerated skin after UV exposure.
Patients and cells characteristics.
Complete list of proviral integrations in corrected clones isolated after 20 and 120 population doublings.
Distribution of proviral integration sites.