Abstract
Recent genetic studies attribute the negative correlation between population genetic diversity and distance from Africa to a serial founder effects (SFE) evolutionary process. A recent linguistic study concluded that a similar decay in phoneme inventories in human languages was also the product of the SFE process. However, the SFE process makes additional predictions for patterns of neutral genetic diversity, both within and between groups, that have not yet been tested on phonemic data. In this study, we describe these predictions and test them on linguistic and genetic samples. The linguistic sample consists of 725 widespread languages, which together contain 908 distinct phonemes. The genetic sample consists of 614 autosomal microsatellite loci in 100 widespread populations. All aspects of the genetic pattern are consistent with the predictions of SFE. In contrast, most of the predictions of SFE are violated for the phonemic data. We show that phoneme inventories provide information about recent contacts between languages. However, because phonemes change rapidly, they cannot provide information about more ancient evolutionary processes.
Keywords: serial founder effects, coevolution, historical linguistics
1. Introduction
The study of linguistic and biological coevolution began over 150 years ago when Darwin proposed that ‘If we possessed a perfect pedigree of mankind, a genealogical arrangement of the races of man would afford the best classification of the various languages now spoken throughout the world’ (p. 422 in [1]). In 1988, Cavalli-Sforza and colleagues [2] tested this phylogenetic model of coevolution by comparing trees constructed from global genetic and linguistic samples. Linguists broadly rejected their conclusion that the trees were congruent because of flaws in the method used to infer linguistic relationships, and the lack of critical thinking about the rate and reticulate nature of language evolution [3–5].
Since then, both geneticists and linguists have continued to refine our understanding of the evolutionary processes that have moulded genetic and linguistic variation, often employing similar evolutionary approaches and statistical methods [6]. Within genetics, there is a growing consensus that the global pattern of neutral genetic diversity is largely the product of a special case of the phylogenetic model [7], termed serial founder effects (SFE). The SFE process views human evolution as a series of successive population splits, movements into unoccupied territory and relative isolation. Evidence presented for the process so far consists largely of a stepwise decrease in population genetic diversity with increasing distance from Africa [8–10].
Atkinson [11] recently documented a similar, albeit weaker, pattern of decay for phoneme (distinctive sound) inventories in a global sample of languages and concluded that the decay was also the product of the SFE process. If true, he has not only provided evidence for a single origin for human languages, but also an important new source of information about human prehistory at deep temporal and geographical scales. The study, however, examines only one prediction of the SFE process: that of decreasing diversity with increasing geographical distance from the founder. The process makes additional empirical predictions, not only for the pattern of within-group diversity, but also for the between-group pattern. The goal of this study is to outline and test these predictions (see [12] for alternative viewpoints).
(a). Model and predictions
Our genetic sample consists of 614 unlinked, autosomal microsatellite loci assayed in 2251 individuals in 100 populations. Microsatellite loci consist of short tandem repeat DNA sequences. Mutation at the loci consists of stepwise increases or decreases in the number of repeats [13]. The stepwise process produces multiple variants, or alleles, at each locus. Founder effects eliminate some alleles and alter the frequencies of others, the net effect being a sharp reduction in the level of genetic variation in daughter populations relative to parents. Over time, daughter populations grow, mutation regenerates variation and the genetic signature of founder effects is erased, though the rate of recovery is slow [14].
Our phoneme sample consists of 908 phonemes scored as present or absent in 725 languages. Phonemes do not vary extensively within languages and are therefore not strictly analogous to alleles [15]. Nonetheless, to the extent that the same processes that mould genetic variation also mould phonemic variation, the SFE process makes several testable predictions for the pattern of genetic and phonemic variation. First, because humans are a young species, groups in all regions will still share alleles and phonemes [16,17]. However, because we originated in Africa, and because founder effects reduce variation, Africans will possess more unique alleles and phonemes than other regions [16,18]. Second, following each founder event, the new daughter group will carry only a subset of the variation of its parental group [8]. Because daughter groups continually expand into unoccupied territory, this loss will manifest as a negative correlation between within-group variation and geographical distance from the African origin [8]. Third, the pattern of among-group variation will be tree-like, and the trees will be rooted in Africa [16]. A corollary of this third prediction is that the degree of difference among groups will reflect the pattern of branching in the tree [16,19,20]. Under this prediction, any correlation that arises between patterns of among-group variation and geographical distance will be purely a by-product of the splitting and movement process, and not exchange between neighbouring groups. These among-group predictions are as yet untested on phonemic data.
2. Results
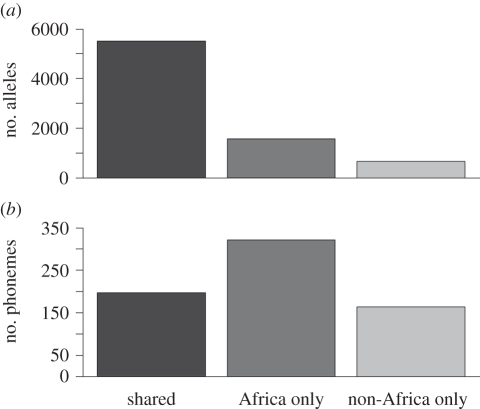
Figure 1 compares alleles and phonemes in Africans and non-Africans. As predicted, most alleles are shared by both groups, but Africans have more unique alleles than non-Africans. African languages also contain more unique phonemes, but relatively few phonemes are shared between the two groups. Low sharing probably reflects high rates of phonemic change.
Figure 1.
African versus non-African (a) alleles and (b) phonemes.
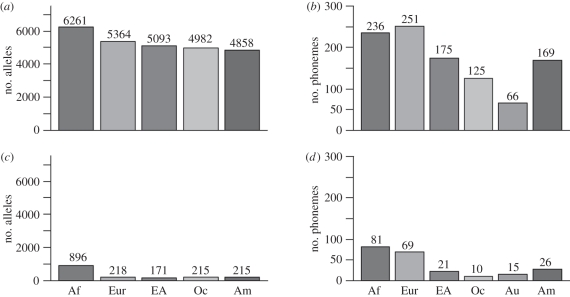
Figure 2 shows the number of alleles and phonemes for each region. The total alleles plot in figure 2a shows the SFE-predicted pattern of variation: highest in Africa and decreasing steadily through Eurasia, Oceania and the Americas. In contrast, the total number of phonemes is highest in Eurasia (figure 2b), and it is higher in the Americas than in Oceania. Since the genetic sample contains only one Australian population, we were unable to compare allelic and phonemic patterns there.
Figure 2.
Total and private alleles and phonemes. (a) Total alleles; (b) total phonemes; (c) private alleles; (d) private phonemes. The single Australian sample is omitted from the genetic analysis. Af, Africa; Eur, Eurasia; EA, East Asia; Oc, Oceania; Am, Americas.
The number of alleles that are unique or ‘private’ to each region is given in figure 2c. There are many fewer private versus total alleles, reflecting the high degree of allelic sharing described above. Africa has substantially more private alleles than any other region, reflecting the African origin of our species. Though the number of private alleles is relatively low in all other regions, east Asia has a relative deficit. Figure 2d gives the sample size-adjusted number of private phonemes in each region. Though Africa also has more private phonemes than any other region, outside of Africa, Oceania has a relative deficit (compared with private alleles), and the Americas have a relative excess.
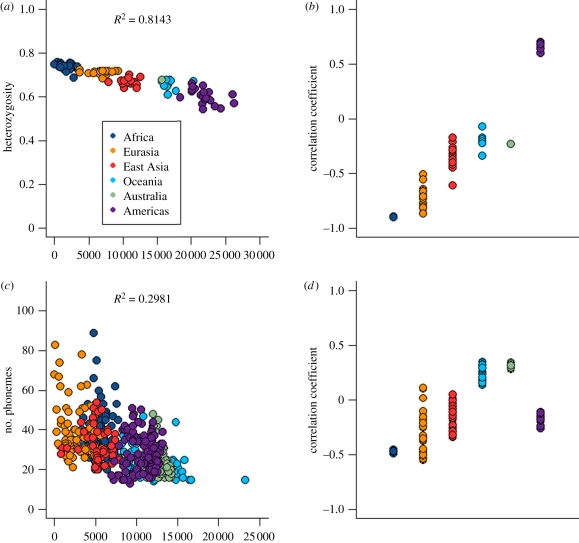
Figure 3a plots the microsatellite heterozygosity within each of the 100 populations versus geographical distance from Africa. Heterozygosity is highest in African populations and decreases steadily through to the Americas. Consistent with this geographical pattern, the correlations between heterozygosity and geographical distance were lowest (most negative) when African populations were chosen as the location of origin, and highest when Native American populations were chosen as the location of origin (figure 3b).
Figure 3.
Within-group diversity versus geographical distance from origin. (a) Microsatellite heterozygosity versus geographical distance, best-fit origin. (b) Correlation coefficients for microsatellite heterozygosity versus geographical distance when using each population location as the origin. (c) Number of phonemes versus geographical distance, best-fit origin. (d) Correlation coefficients for number of phonemes versus geographical distance when using each language location as the origin. Geographical distances are through waypoints on land.
Figure 3c shows the relationship between phoneme numbers and geographical distance. In contrast to the genetic pattern, the number of phonemes is highest on average in Eurasia; a Eurasian origin produced the lowest correlation, and an Oceanic location produced the highest correlation (figure 3d).
We also examined correlations between phoneme counts and geographical distance from hypothesized origins for the five largest language families in our sample (see electronic supplementary material, table S3). The correlation reached statistical significance only for Indo-European (IE), but the correlation was positive instead of negative (r = 0.322, p = 0.025). These results suggest that, even if founder effects might have reduced phoneme levels during the initial radiation of languages from Africa, they did not reduce phoneme levels during more recent expansions.
Figure 4a shows the rooted neighbour-joining (NJ) tree for the genetic data. The test of treeness (electronic supplementary material, figure S1) indicates that the tree is a good representation of the pattern of genetic variation, and that geographical distance explains little of the pattern of among-population genetic distance independent of the tree. All trees rooted on an African branch of the tree fit better than all trees rooted on a non-African branch of the tree. The best-fitting of all possible rooted trees separates the African San from the remaining 99 populations, and the level of variation within populations decreases steadily away from this node (signified by ever-increasing terminal branch lengths).
Figure 4.
(a) Rooted NJ tree for the genetic data. (b) Between-population heterozygosity versus geographical distance. (c) Midpoint-rooted 50% majority-rule tree for the phonemic data. (d) Between-language phonemic difference versus geographical distance.
Figure 4b shows why the best-fitting tree separates the San from the remaining populations. The plot shows heterozygosity versus geographical distance between each pair of populations. The tiered pattern of variation in the plot corresponds to the pattern of branching in the NJ tree. The top, grey-coloured points show the heterozygosity between the African San and the other 99 populations (branch 1 in the NJ tree in figure 4a). The level of heterozygosity is roughly uniform whether populations are located nearby in Africa, or thousands of kilometres away. This uniformity reflects (i) a single African origin for all humans, (ii) a split between the population that would become the San and the founder of the remaining 99 populations, and (iii) relative subsequent isolation between these two groups.
The next tier of dark blue points shows the heterozygosity between the remaining 19 African populations and the 80 non-African populations. The level of heterozygosity is again uniform over thousands of kilometres. This uniformity reflects common ancestry for all non-African populations associated with an ancient out-of-Africa founder event (branch 2). The remaining tiers are the product of subsequent splits and founder effects associated with the peopling of major geographical regions (branches 3–5). The pattern is not perfectly tree-like owing to the admixture between long-separated groups in Oceania and the Americas, and gene flow between neighbouring groups (electronic supplementary material). Still, the correspondence between the tiers of heterozygosity and the branches in the NJ tree attests to the dramatic impact of ancient founder effects on global patterns of neutral genetic diversity.
Several methods were used to construct a phoneme tree. Each produced a different topology; none were similar in topology to the microsatellite NJ tree and diagnostic output from each method strongly implies that phonemic variation is not tree-like. Figure 4c shows a midpoint-rooted phoneme tree produced using a Bayesian approach. Though there is some regional clustering, it contains considerably less geographical structure than the microsatellite NJ tree in figure 4a. Though the midpoint root does separate an African language from the remaining languages, African languages are dispersed throughout the tree.
Consistent with a lack of treeness, the among-region pattern of phonemic variation is not tiered (figure 4d), but there is some evidence of a correlation between phonemic difference and the geographical distance in the plot. The correlation could be a by-product of SFE (i.e. it could reflect the tendency of phylogenetically related languages to be located near to one another). Alternatively, it could reflect sustained phonemic exchange (borrowing) between geographical neighbours, in which case it is inconsistent with SFE.
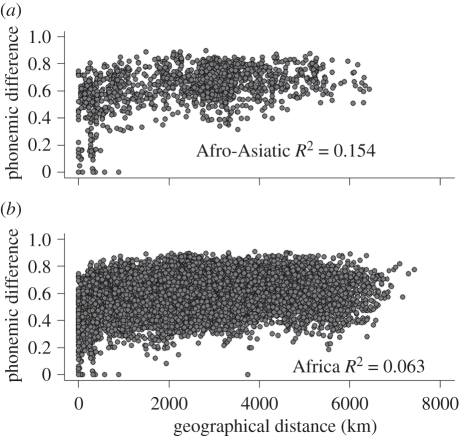
To distinguish among these alternatives, we examined (i) the correlation between phonemic difference and geographical distance within the five best-sampled language families in our sample and within each geographical region, and (ii) partial correlations between phonemic difference and geographical distance within each region controlling language family membership. The Afro-Asiatic family plot in figure 5a confirms that nearby languages share more phonemes than distant languages. Three of the other families show this same relationship (electronic supplementary material, figure S2). There is also a statistically significant (but weaker) correlation within the geographical regions (figure 5b; electronic supplementary material, figure S3). Partial correlation analyses indicate that this regional correlation is independent of language family membership in Africa, Asia and Oceania, indicating that at least some of the correlation is the product of local exchange, not common ancestry within families.
Figure 5.
Phonemic difference versus geographical distance. (a) Afro-Asiatic language family. (b) Africa. Figures S2 and S3 in the electronic supplementary material show the plots for other language families and geographical regions.
Persistent phonemic exchange between neighbouring languages, especially when combined with high rates of phonemic mutation, would quickly erode any evidence of an SFE process that might have once existed. The rapid levelling of phonemic difference in most plots (figure 5; electronic supplementary material, S2 and S3) is consistent with a high mutation rate, as is the relatively low proportion of phonemic sharing between African and non-African languages shown in figure 1. The IE language tree in figure S4 in the electronic supplementary material provides even more evidence for a high rate.
3. Discussion
Predictions of the SFE process were met for the genetic data. Some aspects of the global phonemic pattern also appear consistent with the predictions. There are more private phonemes in Africa than elsewhere (figure 2d), and phoneme levels correlate with distance from possible ancestral locations (figure 3c). There are also important differences between the genetic and phonemic patterns. First, African and non-African languages share relatively few phonemes, implying a rapid rate of phonemic change. Such a high rate would obscure evidence of ancient founder effects. Second, in contrast to Atkinson [11], we find that the correlation between phoneme levels and distance from putative origins is most negative when the origin is located in Eurasia, not Africa (figure 3d), implying that phonemic diversity has not been moulded at the global level by the same evolutionary processes that shaped neutral genetic diversity.
Also contrary to the predictions of SFE, the correlation between phoneme inventory size and geographical distance was most positive when the origin was located in Oceania rather than in the Americas. This is because there is a relative deficit of total and private phonemes in Oceania, and an excess in the Americas. As the phonemic and genetic samples contain substantial numbers of both Austronesian-speaking and non-Austronesian-speaking groups, the difference is unlikely to reflect biases in sample composition. Perhaps alleles were disproportionately preserved in admixed Oceanic groups (Austronesian and non-Austronesian) compared with phonemes. Conceivably, admixture with archaic hominids also contributed to the excess of alleles in Oceania compared with phonemes [21]. As Oceanic languages have fewer phonemes more generally, the lack of private phonemes may simply reflect the fact that small phoneme inventories disproportionally contain common consonants [22]. Whatever the cause, regional phonemic and genetic patterns are discordant here.
The non-tree-like pattern of phonemic variation is also inconsistent with the predictions of SFE. Because we were unable to construct a robust phoneme tree, we are unable to determine whether the observed correlations between phonemic difference and geographical distance are a by-product of the SFE process or the result of phonemic exchange between neighbouring languages. The fact that the correlations exist within regions independent of language family status, however, indicates that local exchange is responsible for at least some of the correlation. This is unsurprising, given that languages are known to borrow phonemes both through individual lexical items and through adstratal effects [23].
We also examined the correlation between phoneme number and distance from family origin within the largest language families in our dataset. Having diversified within the last 10 000 years, currently attested language families are young relative to the age of our species, and specialists have had success reconstructing the evolutionary process in many of them [3,24–28]. Only the IE correlation reached statistical significance, but the correlation was positive. Both the failure to find significance and the positive IE correlation might also reflect borrowing. Perhaps each newly formed language retained phonemes (through shared lexicon) from the parent language and picked up phonemes from resident languages through loans or substrate effects.
(a). Additional theoretical issues
Phonemes are the sound categories that signal a difference in meaning between two words. For example, /d/ and /t/ are distinct phonemes in English because they contrast in the words <bad> and <bat>. But both /d/ and /t/ have a range of sub-phonemic allophones that are conditioned by both location within the word and non-linguistic demographic factors such as social class [29]. For example, /d/ has voicing when it occurs between vowels, but it is partially or fully devoiced for most English speakers in word-final position. Variation in allophones is found in all languages and is a major driver of language change [30–32]. In contrast, the level of phonemic variation within a language is small [15]. Thus, if an SFE model does apply to language, it is more likely to affect allophonic variation. A daughter population would contain a subset of the allophonic diversity found in the parent, and the daughter would then be subject to processes of allophonic change, drift and selection that lead to sound change. Crucially, such changes are largely neutral with respect to phoneme inventory size [33,34]. Unfortunately, there currently exist no databases of allophonic variation that would allow this hypothesis to be tested. In contrast, borrowing effects would be expected to be revealed in phonemic inventories as neighbouring languages converge on similar inventories due to contact.
The SFE process implies a mechanism of rapid phoneme loss in small populations followed by slow recovery, even despite rapid increase in speaker numbers. We can envision no linguistic or social reason why there would be a bias towards loss, as there are no such biases in phonemic change generally [35]. There is no notion of effective size for languages that could keep phonemic diversity low, and our analysis suggests a rapid rate of phonemic change.
Analysing phoneme inventory size rather than composition ignores the fact that languages may have identical inventory sizes but distinct inventories with little overlap. For example, from Proto-IE to Proto-Balto-Slavic, the consonant inventory shrank from 25 to 19 consonants, though only 15 of those consonants were present in Proto-IE (electronic supplementary material, figure S4). Languages often show simultaneous gains and losses that obfuscate a direct relationship between inventory size and language change.
4. Conclusions
The genetic signature of founder effects persists in human populations in part because they accumulate variation slowly. In language, however, even if founder effects initially eliminated phonemes, rates of phonemic change are so high that the signal of loss would quickly disappear. While we reject the SFE process for phonemic data on both empirical and theoretical grounds, these data do provide information about recent contacts between languages.
5. Material and methods
(a). Data
The linguistic data comprise 908 phonemes scored as present or absent in 725 languages present across the world [36,37]. Scoring and analysing phonemes in this format assume that phoneme categories are directly comparable: for example, phoneme /t/ in one language is comparable with phoneme /t/ in another. We recognize that this is a simplification; however, our analyses primarily rely on the number of contrasts, rather than their realization.
The Eurasian grouping was guided by previous genetic studies that combined European, Middle Eastern and south central Asian groups [17,38]. In relevant analyses, we treated south central Asia as a separate group; reported results were unaffected by the grouping decision.
Details of the sample are provided in table S1 in the electronic supplementary material. Unlike Atkinson [11], who used a single gross estimate of inventory size [39], we used full consonant and vowel inventory figures. As our goal was to identify any existing phylogenetic signal in the data, we removed cases of obvious recent borrowing and creole languages in all regions. We also removed dialects of languages with identical phoneme inventories, as well as two IE languages from Africa, three IE languages from the Americas and one click language (!Xóõ) that treated all the click-consonant clusters as though they were separate phonemes, thereby artificially inflating the phoneme count to 162. Though we only report results from the edited sample, we reran key analyses on the full dataset; reported results were unaffected by the removal of languages.
The genetic data were compiled from published sources [38,40–42]. They consist of 614 autosomal microsatellite loci genotyped in 2251 individuals in 100 populations located in the same six regions (electronic supplementary material, table S2). We included approximately equal numbers of populations from all regions except Australia, where only one sample has been genotyped to date.
(b). Allele and phoneme analyses
Asia contained the fewest individuals in our genetic sample, at 282, while Africa contained the most, at 573. A finding of more total or private alleles in Africa could be the result of greater diversity in Africa, as predicted by SFE, or it could be an artefact of the larger African sample. We used the program ADZE [17] to control for this sample-size effect. ADZE uses a rarefaction approach to prune unequal sample sizes to a level equal to that in the smallest regional sample. We created a program to control for sample-size effects for the phonemic data, where our Asian sample had the fewest number of languages, at 48. The program randomly sampled 48 languages in each region and recorded the number of total and private phonemes in this sub-sample. We report the averages of this process over 100 000 trials.
To control for sample-size effects for the nested subset analyses, we divided the language sample into an African parental group and a non-African daughter group. We then created a program that randomly sampled each phoneme in the larger daughter group n times, where n was the number of languages in the smaller parental group. The program then summed the number of phonemes that were (i) unique to the parental group, (ii) unique to the sample-size-adjusted daughter group and (iii) shared by both groups. We report the averages of this process over 100 000 trials. The same program was used for the genetic data.
(c). Within-group diversity
We calculated the correlations between the number of phonemes in a language and geographical distance 725 times, each time using a different language as the point of origin. Partial correlations controlling the number of language speakers (see electronic supplementary material, table S1) did not affect the reported results (results not shown). Geographical distances were calculated both directly between populations and through waypoints on land using PASSaGE [43]. The same analyses were applied to unbiased heterozygosity for the microsatellite data [44]. We also computed correlations between the number of phonemes and distance from putative origin locations for five language families (electronic supplementary material, table S3).
(d). Between-group diversity
For the genetic data, we plotted the heterozygosity between all 4950 pairs of populations against the geographical distance between the pairs. For the phonemic data, we calculated the number of phonemes that differed between a pair of languages divided by the total number that could have differed. Geographical distances were calculated directly between population pairs.
We constructed an NJ tree [45] for the genetic data from minimum genetic distances [44] and used a generalized hierarchical modelling approach to test the treeness of the genetic data and to identify the root location [46]. The electronic supplementary material provides the details of the method.
We constructed the phoneme tree using the approach described by Huelsenbeck & Ronquist [47] and implemented in MrBayes. The tree is midpoint-rooted. Phonemes were coded as standard data, and we employed a general time-reversible evolutionary model with gamma-distributed rate variation across sites. We used the default option that runs two simultaneous, independent analyses starting from different random trees. The two runs failed to converge on the same tree. A neighbour net [48] of the phonemic data produced a delta score of 9.878, also implying that the data are non-treelike. We constructed a 1000-replicate, bootstrapped NJ tree. Bootstrap support was low for most branches in the tree, especially deeper internal branches, where it seldom exceeded 5 per cent.
Acknowledgements
We thank Barry Alpher, Pattie Epps and Patrick McConvell for thoughtful comments on the manuscript. This research was funded by National Science Foundation grant BCS-902114.
References
- 1.Darwin C. 1859. On the origin of species by means of natural selection. London, UK: John Murray [Google Scholar]
- 2.Cavalli-Sforza L. L., Piazza A., Menozzi P., Mountain J. 1988. Reconstruction of human evolution: bringing together genetic, archaeological, and linguistic data. Proc. Natl Acad. Sci. USA 85, 6002–6006 10.1073/pnas.85.16.6002 (doi:10.1073/pnas.85.16.6002) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Campbell L. 1997. American Indian languages: the historical linguistics of native America. New York, NY: Oxford University Press [Google Scholar]
- 4.O'Grady R. T., Goddard I., Bateman R. M., DiMichele W. A., Funk V. A., Kress W. J., Mooi R., Cannell P. F. 1989. Genes and tongues. Science 243, 1651. 10.1126/science.2928798 (doi:10.1126/science.2928798) [DOI] [PubMed] [Google Scholar]
- 5.McMahon R. 2004. Genes and languages. Commun. Genet. 7, 2–13 10.1159/000080298 (doi:10.1159/000080298) [DOI] [PubMed] [Google Scholar]
- 6.Atkinson D., Gray R. D. 2005. Curious parallels and curious connections—phylogenetic thinking in biology and historical linguistics. Syst. Biol. 54, 513–526 10.1080/10635150590950317 (doi:10.1080/10635150590950317) [DOI] [PubMed] [Google Scholar]
- 7.Cavalli-Sforza L. L., Edwards A. W. 1967. Phylogenetic analysis. Models and estimation procedures. Am. J. Hum. Genet. 19, 233–257 [PMC free article] [PubMed] [Google Scholar]
- 8.Ramachandran S., Deshpande O., Roseman C. C., Rosenberg N. A., Feldman M. W., Cavalli-Sforza L. L. 2005. Support from the relationship of genetic and geographic distance in human populations for a serial founder effect originating in Africa. Proc. Natl Acad. Sci. USA 102, 15 942–15 947 10.1073/pnas.0507611102 (doi:10.1073/pnas.0507611102) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Prugnolle F., Manica A., Balloux F. 2005. Geography predicts neutral genetic diversity of human populations. Curr. Biol. 15, R159–R160 10.1016/j.cub.2005.02.038 (doi:10.1016/j.cub.2005.02.038) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.DeGiorgio M., Jakobsson M., Rosenberg N. A. 2009. Out of Africa: modern human origins special feature: explaining worldwide patterns of human genetic variation using a coalescent-based serial founder model of migration outward from Africa. Proc. Natl Acad. Sci. USA 106, 16 057–16 062 10.1073/pnas.0903341106 (doi:10.1073/pnas.0903341106) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Atkinson Q. D. 2011. Phonemic diversity supports a serial founder effect model of language expansion from Africa. Science 332, 346–349 10.1126/science.1199295 (doi:10.1126/science.1199295) [DOI] [PubMed] [Google Scholar]
- 12.Plank F. (ed.) Linguist. Typol. 15, 147–534 [Google Scholar]
- 13.Weber J., Wong C. 1993. Mutation of short tandem repeats. Hum. Mol. Genet. 2, 1123–1128 10.1093/hmg/2.8.1123 (doi:10.1093/hmg/2.8.1123) [DOI] [PubMed] [Google Scholar]
- 14.Chakraborty R., Nei M. 1977. Bottleneck effects on average heterozygosity and genetic distance with the stepwise mutation model. Evolution 31, 347–356 10.2307/2407757 (doi:10.2307/2407757) [DOI] [PubMed] [Google Scholar]
- 15.Bowern C. 2011. Out of Africa? The logic of phoneme inventories and founder effects. Linguist. Typol. 15, 207–216 [Google Scholar]
- 16.Long J. C., Li J., Healy M. E. 2009. Human DNA sequences: more variation and less race. Am. J. Phys. Anthropol. 139, 23–34 10.1002/ajpa.21011 (doi:10.1002/ajpa.21011) [DOI] [PubMed] [Google Scholar]
- 17.Szpiech Z. A., Jakobsson M., Rosenberg N. A. 2008. ADZE: a rarefaction approach for counting alleles private to combinations of populations. Bioinformatics 24, 2498–2504 10.1093/bioinformatics/btn478 (doi:10.1093/bioinformatics/btn478) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Yu N., et al. 2002. Larger genetic differences within Africans than between Africans and Eurasians. Genetics 161, 269–274 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Cavalli-Sforza L. L., Piazza A. 1975. Analysis of evolution: evolutionary rates, independence and treeness. Theor. Popul. Biol. 8, 127–165 10.1016/0040-5809(75)90029-5 (doi:10.1016/0040-5809(75)90029-5) [DOI] [PubMed] [Google Scholar]
- 20.Hunley K., Healy M., Long J. 2009. The global pattern of gene identity variation reveals a history of long-range migrations, bottlenecks, and local mate exchange: implications for biological race. Am. J. Phys. Anthropol. 139, 35–46 10.1002/ajpa.20932 (doi:10.1002/ajpa.20932) [DOI] [PubMed] [Google Scholar]
- 21.Reich D., et al. 2010. Genetic history of an archaic hominin group from Denisova Cave in Siberia. Nature 468, 1053–1060 10.1038/nature09710 (doi:10.1038/nature09710) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Maddieson I. 1986. The size and structure of phonological inventories: analysis of UPSID. In Experimental phonology (eds Ohala J., Jaeger J.), pp. 105–123 Orlando, FL: Academic Press. [Google Scholar]
- 23.Thomason S. G., Kaufman T. 1988. Language contact, creolization, and genetic linguistics. Berkeley, CA: University of California Press [Google Scholar]
- 24.Gray R. D., Atkinson Q. D. 2003. Language-tree divergence times support the Anatolian theory of Indo-European origin. Nature 426, 435–439 10.1038/nature02029 (doi:10.1038/nature02029) [DOI] [PubMed] [Google Scholar]
- 25.Gray R. D., Drummond A. J., Greenhill S. J. 2009. Language phylogenies reveal expansion pulses and pauses in Pacific settlement. Science 323, 479–483 10.1126/science.1166858 (doi:10.1126/science.1166858) [DOI] [PubMed] [Google Scholar]
- 26.Alpher B. 2004. Pama-Nyungan: phonological reconstruction and status as a phylogenetic group. In Australian languages: classification and the comparative method (eds Bowern C., Koch H.), pp. 93–126 Amsterdam, The Netherlands/Philadelphia, PA: John Benjamins [Google Scholar]
- 27.Fortson B. 2004. Indo-European language and culture: an introduction. Oxford, UK: Blackwell [Google Scholar]
- 28.Holden C. J. 2002. Bantu language trees reflect the spread of farming across sub-Saharan Africa: a maximum-parsimony analysis. Proc. R. Soc. Lond. B 269, 793–799 10.1098/rspb.2002.1955 (doi:10.1098/rspb.2002.1955) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Labov W. 2001. Principles of linguistic change: social factors. Oxford, UK: Blackwell [Google Scholar]
- 30.Ohala J. 2009. Languages' sound inventories: the devil in the details. In Approaches to phonological complexity (eds Pellegrino F., Marsico E., Chitoran I., Coupé C.), pp. 47–58 Berlin, Germany: Walter de Gruyter [Google Scholar]
- 31.Garrett A., Johnson K. 2011. Phonetic bias in sound change. Berkeley, CA: University of California [Google Scholar]
- 32.Blevins J. 2004. Evolutionary phonology: the emergence of sound patterns. Cambridge, UK: Cambridge University Press [Google Scholar]
- 33.Gordon E., Campbell L., Hay J., Maclagan M., Sudbury A., Trudgill P. 2004. New Zealand English: its origins and evolution. Cambridge, UK: Cambridge University Press [Google Scholar]
- 34.Trudgill P., Gordon E., Lewis G., Maclagan M. 2000. Determinism in new dialect formation and the genesis of New Zealand English. J. Linguist. 36, 299–318 10.1017/S0022226700008161 (doi:10.1017/S0022226700008161) [DOI] [Google Scholar]
- 35.Hock H., Joseph B. 1996. Language history, language change, and language relationship. Berlin, Germany: Mouton de Gruyter [Google Scholar]
- 36.Mielke J. 2008. The emergence of distinctive features. Oxford, UK: Oxford University Press [Google Scholar]
- 37.Round E. 2011. What phonological diversity does and doesn't tell us: the consonants of Australian indigenous languages. CUNY Phonology Forum, CUNY Graduate Center, New York, 12–14 January 2011. See www.cunyphonologforum.net/endan.php [Google Scholar]
- 38.Rosenberg N. A., Mahajan S., Ramachandran S., Zhao C., Pritchard J. K., Feldman M. W. 2005. Clines, clusters, and the effect of study design on the inference of human population structure. PLoS Genet. 1, e70. 10.1371/journal.pgen.0010070 (doi:10.1371/journal.pgen.0010070) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Dryer M. S., Haspelmath M. 2011. The world atlas of language structures online. Munich, Germany: Max Planck Digital Library [Google Scholar]
- 40.Friedlaender J., et al. 2008. The genetic structure of Pacific Islanders. PLoS Genet. 4, e19. 10.1371/journal.pgen.0040019 (doi:10.1371/journal.pgen.0040019) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Tishkoff S. A., et al. 2009. The genetic structure and history of Africans and African Americans. Science 324, 1035–1044 10.1126/science.1172257 (doi:10.1126/science.1172257) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Wang S., et al. 2007. Genetic variation and population structure in Native Americans. PLoS Genet. 3, e185. 10.1371/journal.pgen.0030185 (doi:10.1371/journal.pgen.0030185) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Rosenberg M. S., Anderson C. D. 2011. PASSaGE: pattern analysis, spatial statistics and geographic exegesis. Version 2. Methods Ecol. Evol. 2, 229–232 10.1111/j.2041-210X.2010.00081.x (doi:10.1111/j.2041-210X.2010.00081.x) [DOI] [Google Scholar]
- 44.Nei M. 1987. Molecular evolutionary genetics. New York, NY: Columbia University Press [Google Scholar]
- 45.Saitou N., Nei M. 1987. The neighbor-joining method: a new method for reconstructing phylogenetic trees. Mol. Biol. Evol. 4, 406–425 [DOI] [PubMed] [Google Scholar]
- 46.Long J. C., Kittles R. A. 2003. Human genetic diversity and the nonexistence of biological races. Hum. Biol. 75, 449–471 10.1353/hub.2003.0058 (doi:10.1353/hub.2003.0058) [DOI] [PubMed] [Google Scholar]
- 47.Huelsenbeck J. P., Ronquist F. 2001. MRBAYES: Bayesian inference of phylogenetic trees. Bioinformatics 17, 754–755 10.1093/bioinformatics/17.8.754 (doi:10.1093/bioinformatics/17.8.754) [DOI] [PubMed] [Google Scholar]
- 48.Huson D. H., Bryant D. 2006. Application of phylogenetic networks in evolutionary studies. Mol. Biol. Evol. 23, 254–267 10.1093/molbev/msj030 (doi:10.1093/molbev/msj030) [DOI] [PubMed] [Google Scholar]