Abstract
Little is known about whether autophagic mechanisms are active in hematopoietic stem cells (HSCs) or how they are regulated. FIP200 (200-kDa FAK-family interacting protein) plays important roles in mammalian autophagy and other cellular functions, but its role in hematopoietic cells has not been examined. Here we show that conditional deletion of FIP200 in hematopoietic cells leads to perinatal lethality and severe anemia. FIP200 was cell-autonomously required for the maintenance and function of fetal HSCs. FIP200-deficient HSC were unable to reconstitute lethally irradiated recipients. FIP200 ablation did not result in increased HSC apoptosis, but it did increase the rate of HSC proliferation. Consistent with an essential role for FIP200 in autophagy, FIP200-null fetal HSCs exhibited both increased mitochondrial mass and reactive oxygen species. These data identify FIP200 as a key intrinsic regulator of fetal HSCs and implicate a potential role for autophagy in the maintenance of fetal hematopoiesis and HSCs.
Introduction
FIP200 (focal adhesion kinase family interacting protein of 200 kDa) was initially identified as a putative protein inhibitor of focal adhesion kinase and its related kinase Pyk2.1 Subsequent studies suggested that FIP200 regulates diverse cellular functions including cell size, survival, proliferation, spreading, and migration through its interaction with multiple other proteins.2 FIP200 is widely expressed in various human tissues and is an evolutionarily conserved protein present in human, mouse, rat, frog, fly, and worm,3 suggesting potentially important functions for metazoan FIP200 in vivo. Consistent with this and its diverse cellular activities in vitro, we showed recently that germ line deletion of FIP200 in mice resulted in embryonic lethality at mid/late gestation associated with heart failure and liver degeneration.4
Recent reports by several groups identified FIP200 as a component of the ULK-Atg13-FIP200 complex, leading to the assumption that it acts as a mammalian counterpart of yeast Atg17 protein despite limited sequence homology. This complex is essential for the induction of autophagy in mammalian cells.5–8 Although the primary function of autophagy is to supply amino acids during starvation, a basal level of constitutive autophagy is independent of nutrient stress. Constitutive autophagy also plays an important role in maintaining cellular homeostasis. Consistent with a potential role of FIP200 in autophagy as identified in these studies in vitro, we showed recently that neural-specific deletion of FIP200 resulted in abnormal accumulation of ubiquitinated protein aggregates and p62/sequestosome-1(SQSTM1), increased apoptosis and neurodegeneration.9–11 However, it was unclear whether FIP200 or basal autophagy might also be required to regulate hematopoietic stem cells (HSCs), as protein quality control might be unusually dependent upon autophagy in postmitotic cells such as neurons.12
Here we report experiments in which FIP200 was deleted in the hematopoietic cells of mice bearing a homozygous conditional FIP200 allele. These results reveal a cell-autonomous requirement for FIP200 in fetal HSCs. Deletion of FIP200 led to HSC depletion, loss of HSC reconstituting capacity, and a block in erythroid maturation. We also observed increased cell division by fetal HSCs and aberrant expansion of myeloid cells associated with an increase in mitochondrial mass and reactive oxygen species (ROS). These results implicate FIP200 in the regulation of fetal HSC homeostasis.
Methods
Mice and blood cell counts
The floxed FIP200 and Tie2-Cre mice were described previously.4,13 Mx1-Cre mice were obtained from The Jackson Laboratory. All mice were backcrossed for at least 6 generations onto a C57BL/6 background. Mice were housed and handled according to local, state, and federal regulations, and all experimental procedures were carried out with the approval of the Institutional Animal Care and Use Committee at the University of Michigan. Mice genotyping for FIP200 and Cre alleles were performed by polymerase chain reaction analysis of tail DNA, essentially as described previously.4 For analysis of blood counts, peripheral blood was collected in a heparinized microtube (SARSTEDT) and analyzed with a hematology analyzer (Advia 120 hematology system).
Protein extraction, sodium dodecyl sulfate– polyacrylamide gel electrophoresis, and Western blotting
Mouse fetal livers were collected from control or CKO mice at E14.5. The protein lysates were prepared by homogenization in CelLytic buffer (Sigma-Aldrich) supplemented with protease inhibitors (5 μg/mL leupeptin, 5 μg/mL aprotinin, and 1mM phenylmethylsulfonyl fluoride). The protein extraction and Western blotting procedures were performed as described previously.11 Antibodies against FIP200 were prepared as described previously.1 Anti-p62/SQSTM1 and anti-vinculin antibodies were purchased from Enzo Life Science and Sigma-Aldrich, respectively.
Histology and in situ detection of apoptosis
Mice were killed using CO2. E14.5 and E16.5 embryos were recovered and fixed in freshly made, prechilled (4°C) phosphate-buffered saline (PBS)–buffered formalin at 4°C. The liver tissues were sectioned and then embedded in paraffin, sectioned at 6 μm, and stained with hematoxylin and eosin (H&E) for histologic examination or left unstained for TUNEL assays. H&E-stained sections were examined under a BX41 light microscope (Olympus America), and images were captured with an Olympus digital camera (model DP70) using DP Controller software Version 1.2.1.108. For TUNEL assays, fetal liver sections were deparaffinized, incubated in methanol containing 0.3% H2O2 for 30 minutes, washed, and incubated with proteinase K (20 μg/mL) in PBS for 15 minutes at room temperature. Apoptotic cells were detected as described in the ApopTag Peroxidase In Situ Apoptosis Detection kit (Millipore). Sections were counterstained with methyl green.
Flow cytometry
Fetal livers were triturated with Hanks buffered salt solution without calcium or magnesium, supplemented with 2% heat-inactivated calf serum (HBSS+; Gibco) and filtered through a nylon mesh screen (45 mm; Sefar America) to obtain single cell suspensions. To examine different lineages, fetal liver cells were incubated with conjugated monoclonal antibodies of lineage markers including Ter119(Ter119)–fluorescein isothiocyanate (FITC), B220(6B2)-FITC, Mac1(M1/70)–phycoerythrin (PE), and Gr1(8C5)–allophycocyanin (APC). For the analysis of erythroid maturation, whole fetal liver cells were incubated with anti–Ter119-FITC and anti–CD71-PE (BD Biosciences). For the detection of fetal liver HSCs, whole fetal liver cells were incubated with FITC-conjugated antibody to CD41 (MWReg30), CD48 (HM48-1-PE), Ter119 (Ter119), PE-conjugated antibody to CD150 (26D12:DNAX), APC-conjugated Mac1 (M1/70), and biotin-conjugated Sca1 (Ly6A/E-biotin), followed by staining with streptavidin conjugated to APC-Cy7 (PharRed, PR; Becton Dickinson). Cells were resuspended in 2 μg/mL DAPI (4′,6-diamidino-2-phenylindole) to distinguish live from dead cells. Flow cytometry was performed on a FACSVantage SE-dual laser, 3-line flow cytometer or a FACSCanto (BD Biosciences).
In some experiments, BrdU (Sigma-Aldrich) was injected intraperitoneally into pregnant mice at 100 mg/kg 2 hours before killing the animals. BrdU staining was performed according to producer's manual.
In other experiments, after labeling with specific surface markers, cells were stained by MitoTracker (Invitrogen) at 20nM for 15 minutes at 37°C, or by 2′-7′-dichlorofluorscein diacetate (DCF-DA; Invitrogen) at 10μM for 15 minutes at 37°C, according to manufacturer's instructions.
To detect apoptotic cells, fetal liver cells were stained with lineage markers followed by staining with DAPI and annexin V (R&D Systems).
Blood cell staining
Dried blood smears were stained with Wright-Giemsa stain (WG16; Sigma-Aldrich) according to manufacturer's instructions. For neutral benzidine staining, dried smears were fixed for 4 minutes in methanol, incubated in a 1% o-dianisidine (D9143; Sigma-Aldrich) in methanol for 2 minutes, and then in 0.9% H2O2 in 50% ethanol for 1.5 minutes, rinsed with water, and then air-dried.
Long-term competitive repopulation assay
Adult recipient mice (CD45.1) were irradiated with an Orthovoltage X-ray source delivering approximately 300 rad/min in 2 equal doses of 570 rad, delivered at least 2 hours apart. Cells were injected into the retro-orbital venous sinus of anesthetized recipients. Each recipient mouse received 500 000 CD45.1 marrow cells for radioprotection. Beginning 4 weeks after transplantation and continuing for at least 16 weeks, blood was obtained from the tail veins of recipient mice, subjected to ammonium-chloride red cell lysis, and stained with directly conjugated antibodies to CD45.2 (104, FITC), B220 (6B2), Mac-1 (M1/70), CD3 (KT31.1), and Gr-1 (8C5) to monitor donor cell engraftment.
Polyinosine-polycytosine (pIpC) administration
pIpC (Amersham Pharmacia Biotech) was administered to mice as previously described.14 Briefly, pIpC was resuspended in Dulbecco PBS (D-PBS) at 50 μg/mL, and mice were injected with 0.4 μg/g body weight every other day for 10 days.
Results
FIP200 deletion leads to erythroblastic anemia and perinatal lethality
To test whether FIP200 might play a role in fetal hematopoiesis, we mated floxed FIP200 (FIP200F/F) mice4 with Tie2-Cre transgenic mice to generate FIP200F/F;Tie2-Cre mice (designated as CKO mice). Tie2-Cre mice express Cre recombinase in hematopoietic and endothelial cells during embryonic development.13 As shown in Table 1, CKO and littermate controls (FIP200F/+; Tie2-Cre, FIP200F/F, and FIP200F/+; the latter 2 genotypes were used as controls in all experiments) were observed at normal Mendelian ratios at E14.5 and E16.5. There was a slight decrease in the observed number of CKO embryos at E17.5 and E18.5, and virtually all CKO mice died within the first week of birth.
Table 1.
Genotypes of progeny from crosses between male FIP200F/+;Tie2-Cre and female FIP200F/F mice
| Genotype | E14.5 | E16.5 | E17.5 | E18.5 | P7 |
|---|---|---|---|---|---|
| FIP200F/+ | 39 | 13 | 13 | 39 | 32 |
| FIP200F/+;Cre | 37 | 9 | 17 | 33 | 26 |
| FIP200F/F | 38 | 10 | 10 | 38 | 31 |
| FIP200F/F;Cre (CKO) | 41 | 12 | 8 | 30 | 3 |
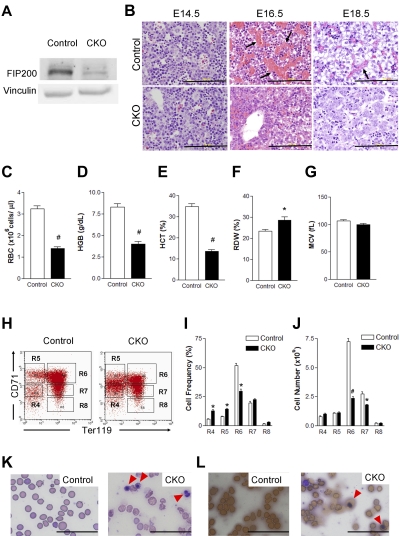
To investigate the effect of FIP200 deficiency on fetal hematopoiesis, we first evaluated deletion of FIP200 by Western blotting of lysates from E14.5 fetal livers of CKO and control embryos. As shown in Figure 1A, a significantly reduced level of FIP200 was found in the sample from CKO mice compared with control mice, indicating efficient deletion of FIP200 as expected. We then histologically examined fetal liver sections from CKO and control mice. At E14.5, no apparent differences were detected in CKO fetal liver sections compared with control sections (Figure 1B). In contrast, at E16.5 we observed robust erythropoiesis characterized by abundant sinuses filled with mature red blood cells (RBCs) in the fetal liver of control mice but not in CKO fetal liver sections, suggesting that FIP200 deletion significantly impaired fetal erythropoiesis (Figure 1B). At E18.5, we observed numerous mature RBCs within vascular structures of control fetal livers, but very few erythrocytes within vascular structures of CKO fetal livers (Figure 1B). These histologic features were consistent with the grossly paler appearance of CKO fetal livers at E16.5 and E18.5 (but not E14.5) compared with littermate control fetal livers (data not shown).
Figure 1.
Conditional deletion of FIP200 by Tie2-Cre causes severe anemia in developing embryos. (A) Lysates were prepared from E14.5 liver of control or CKO mice and analyzed by Western blotting using anti-FIP200 (top) or anti-vinculin (bottom) antibodies. (B) H&E staining of E14.5, E16.5, E18.5 fetal livers from control and CKO mice. Arrows indicate the enucleated RBCs. Scale bars, 200 μm. (C-G) RBC parameters of peripheral blood from E18.5 control and CKO mice: RBC numbers (C), hemoglobin (D), hematocrit (E), RBC distribution width (F), and mean corpuscular volume (G). n = 7-17, #P < .01, *P < .05. Data are mean ± SE. (H) Representative fluorescence-activated cell sorting (FACS) profile of erythroid maturation in E14.5 fetal liver of control and CKO embryos. The cells were double-labeled with anti-CD71 and anti-TER119 antibodies. Regions R4 to R8 are defined by characteristic staining pattern of cells, including CD71medTER119−, CD71highTER119−, CD71highTER119+, CD71medTER119+, and CD71lowTER119+, respectively. (I-J) The cell frequency (I) and number (J) of R4-R8 population. Note that there were increased immature population (R4 and R5) and decreased R6 in CKO. n = 5-8, *P < .05, #P < .01. Data are mean ± SE. (K) Wright-Giemsa staining of the blood smears from E18.5 control and CKO embryos. The arrowheads indicate the erythroblasts. (L) Benzidine staining of the blood smears as in panel K. Arrowheads indicate the positively stained erythroblasts. Scale bars, 100 μm.
Although Tie2-Cre is expressed in endothelial cells, we did not detect hemorrhaging or edema and immunochemical analysis also showed apparently normal density of vasculature in CKO embryos (data not shown). There were no significant gross or histologic defects in other major organs such as the lungs and heart in CKO embryos (supplemental Figure 1, available on the Blood Web site; see the Supplemental Materials link at the top of the online article). These results suggest that deficient fetal hematopoiesis is the primary defect in CKO embryos.
To further characterize the effects of FIP200 deletion on defective erythropoiesis, we first analyzed the peripheral blood of CKO and control embryos at E18.5. Consistent with the reduced erythropoiesis observed in fetal livers, CKO embryos displayed severely diminished numbers of RBCs (Figure 1C), decreased hemoglobin content (Figure 1D), a lower hematocrit (Figure 1E), and elevated RBC width distribution (Figure 1F) compared with the circulating blood of control embryos. All of these parameters indicated a severe impairment of definitive erythropoiesis upon genetic loss of FIP200, although we did not observe any change in mean corpuscular volume (Figure 1G). We then examined the fetal liver cells at E14.5 using the surface markers CD71 and Ter119 to determine possible erythroid maturation defects that may contribute to the defective erythropoiesis in CKO mice. We observed an increase in the frequency of immature erythroid cells (R4 population: CD71medTer119− and R5 population: CD71highTer119−) and a decrease in a maturing erythroid population (R6 population: CD71highTer119+) as well as a significant reduction in the absolute number of maturing erythroid cells (R6 and R7) in CKO embryos compared with control embryos (Figure 1H-J). These data suggested compromised erythroid maturation in CKO embryos. Analysis of the peripheral blood at E18.5 using the same cell surface markers CD71 and Ter119 showed a significant decrease in the frequency as well as the absolute number of CD71lowTer119+ mature RBCs (the R8 population) in CKO mice compared with control mice (supplemental Figure 2), providing further support for a profound anemia in these mice. The anemia appeared to be erythroblastic as evidenced by the significantly increased numbers of erythroblasts in the peripheral blood of E18.5 CKO mice by Wright-Giemsa staining (Figure 1K). Indeed, differential counting identified 72% of the nucleated cells to be erythroblasts and 25% to be neutrophils. The increase in erythroblasts was further confirmed by Benzidine staining (Figure 1L). Taken together, these results suggest a crucial role for FIP200 in fetal erythropoiesis and demonstrate that its loss in hematopoietic cells leads to perinatal lethality associated with severe erythroblastic anemia.
FIP200 deletion cell-autonomously leads to fetal HSC depletion
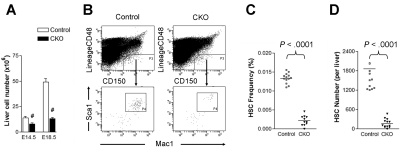
Although CKO fetal livers were histologically indistinguishable from control fetal livers at E14.5 (see Figure 1B), we noted a decrease in total fetal liver cell number in CKO embryos at this stage and a more dramatic reduction at E18.5 compared with control embryos (Figure 2A). These data raised the intriguing possibility that the deficient hematopoiesis observed in CKO mice might be elicited at the level of HSCs. To explore this possibility, we examined the frequency of CD150+CD48−Lin−Mac-1+Sca-1+ cells in the E14.5 fetal liver of CKO and control mice. These cells include all fetal liver HSC activity and are highly enriched for HSCs.15 As shown in Figure 2B-C, the frequency of immunophenotypic HSCs was 6-fold lower in CKO fetal livers compared with control samples. Coupled with the overall decrease in fetal liver cellularity in CKO embryos, the absolute number of immunophenotypic HSCs was diminished roughly 10-fold in the fetal livers of these embryos (Figure 2D). These results suggest that FIP200 deletion resulted in decreased HSC numbers in CKO embryos.
Figure 2.
FIP200 deletion depletes fetal HSCs. (A) Cell numbers in E14.5 and E18.5 fetal livers of control and CKO mice. n = 5-13, #P < .01. Data are mean ± SE (B) Representative FACS analysis of fetal liver cells from E14.5 control and CKO embryos. HSCs were gated as CD150+CD48−Lin−Mac1+Sca1+ cells. (C-D) The frequency (C) and number (D) of fetal HSCs in control and CKO embryos shown in panel B.
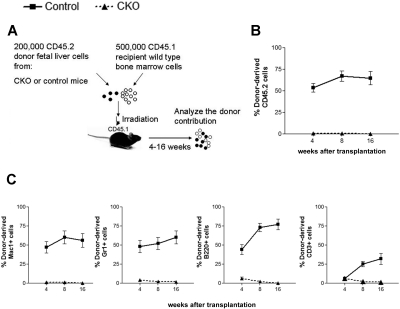
To address the hypothesis that FIP200 loss impaired fetal HSC function and to exclude the possibility that HSCs simply changed their immunophenotype upon FIP200 deletion, we functionally assessed the repopulating capacity of fetal liver cells from CKO mice in vivo. Competitive reconstitution experiments were performed, in which 200 000 E14.5 fetal liver cells from CD45.2+ CKO or CD45.2+ control donors were coinjected with 500 000 young adult bone marrow cells from congenic CD45.1+ mice into lethally irradiated CD45.1+ recipients (Figure 3A). We tracked donor cell reconstitution by analyzing the blood of recipients 4, 8, and 16 weeks after transplantation. Consistent with the relatively high proliferative potential of fetal liver cells,16 200 000 control fetal liver cells competed slightly better than 500 000 adult bone marrow cells in reconstituting the peripheral blood (Figure 3B), and contributed to the myeloid, B-cell, and T-cell lineages (Figure 3C and supplemental Figure 3) throughout the length of the experiment. In contrast, FIP200-null fetal liver cells failed to give long-term multilineage reconstitution of any recipients. These results confirmed the hypothesis that FIP200 loss results in a reduced number of HSCs in CKO embryos.
Figure 3.
FIP200 is essential for the maintenance of fetal HSCs. (A) Diagram of the competitive repopulation experimental design for data in panels B-C. Fetal liver cells (200 000) from CD45.2+ control or CKO mice were injected into lethally irradiated CD45.1+ wild-type recipients along with 500 000 CD45.1 bone marrow cells. Reconstitution of peripheral blood by donor cells was monitored for 16 weeks after the transplantation. (B) Contribution of fetal liver-derived CD45.2-expression (donor) cells to peripheral blood leukocytes in reconstituted mice. (C) Contribution of donor cells to peripheral multilineages, including myeloid lineage (Mac1+, Gr1+), B-cell lineage (B220+), and T-cell lineage (CD3+). Data represent the average donor chimerism levels from 3 independent experiments with a total of 12 recipients per genotype.
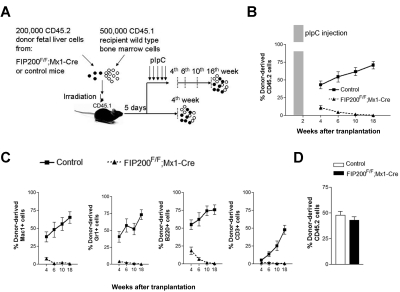
Although these transplantation results were consistent with HSC maintenance defects in vivo, it was possible that the failure of fetal liver cells from CKO mice to repopulate recipients was due to their inability to home to the bone marrow after transplantation and/or the 6-fold reduction in HSC number in the input fetal liver cells before transplantation. Moreover, other extrinsic mechanisms could account for the failure of CKO cells in transplantation. For example, FIP200 deletion in endothelial cells (as Tie2-Cre is also expressed in this tissue) could potentially cause irreversible damage to fetal HSCs by damaging an indispensable endothelial niche, as has been suggested for HSCs.17,18 Even though no apparent defects in vascular development were observed in CKO mice, we nevertheless wished to test these possibilities. We generated inducible FIP200 deletion mice (designated FIP200F/F;Mx1-Cre mice) by crossing the floxed FIP200 (FIP200F/F) mice with Mx1-Cre transgenic mice. We performed similar transplantation experiments as above by transplanting 200 000 floxed E14.5 fetal liver cells from CD45.2+ FIP200F/F;Mx1-Cre or CD45.2+ FIP200F/F embryos along with 500 000 young adult bone marrow cells from congenic CD45.1+ mice into lethally irradiated CD45.1+ recipients (Figure 4A). In one group of recipients, we administered 5 doses of pIpC every other day to induce deletion of FIP200 in the transplanted cells of the FIP200F/F;Mx1-Cre donors 5 days after transplantation (both experimental and control donor mice received pIpC treatment), which would allow sufficient time for homing.14 The peripheral blood was then analyzed 4 to 18 weeks after transplantation to monitor donor cell reconstitution. Control donor cells successfully reconstituted all recipient mice in all lineages throughout the length of the experiment (Figure 4B-C). In contrast, we noted a significantly reduced reconstitution by FIP200F/F;Mx1-Cre donor cells starting 4 weeks after transplantation. By 18 weeks, there was a complete loss of donor cell reconstitution from FIP200F/F;Mx1-Cre cells in all hematopoietic lineages (Figure 4B-C and supplemental Figure 4), suggesting that FIP200 is autonomously required for the repopulating ability of fetal HSCs. Interestingly, we noticed that B-cell reconstitution by FIP200F/F;Mx1-Cre cells decreased more gradually than myeloid cell reconstitution, which is consistent with the observation that myeloid cells are much shorter lived than lymphoid cells. In a second group of recipients, mice were not treated with pIpC, and their peripheral blood was analyzed at 4 weeks after transplantation. Donor cell reconstitution levels from FIP200F/F;Mx1-Cre cells (in the absence of Cre recombinase induction) were roughly equal to that of control cells at 4 weeks after transplantation (Figure 4D), suggesting that reconstitution defects observed in pIpC-treated FIP200F/F;Mx1-Cre donor cells were indeed due to induced deletion of FIP200.
Figure 4.
FIP200 is cell-autonomously required for the maintenance of fetal HSCs. (A) Diagram of the competitive repopulation experimental design for data in panels B-C. Fetal liver cells (200 000) from CD45.2+ FIP200F/F;Mx1-Cre or control mice were injected into lethally irradiated CD45.1+ wild-type recipients along with 500 000 CD45.1 bone marrow cells. Five pIpC injections were administered to one set of recipients every other day beginning 5 days after transplantation. Reconstitution of peripheral blood by donor cells was then monitored for additional 16 weeks (at the 4th, 6th, 10th, and 18th weeks after transplantation). No pIpC was administered to the other set of recipients. Reconstitution of peripheral blood by donor cells was examined at the 4th week after transplantation. The dashed vertical lines indicate the time points to monitor the donor contribution. (B) Contribution of fetal liver-derived CD45.2-expression (donor) cells to peripheral blood leukocytes in reconstituted mice in the group with pIpC treatment. Shaded bar in panel B indicates the pIpC administration. (C) Contribution of donor cells to peripheral blood lineages, including myeloid (Mac1+, Gr1+), B-cell (B220+), and T-cell (CD3+) lineages in the group with pIpC treatment. (D) Contribution of donor cells to peripheral blood leucocytes in reconstituted mice in the group without pIpC treatment. Data represent the average donor chimerism levels from 3 independent experiments with a total of 9 recipients per genotype.
Taken together, these results demonstrate that FIP200 is cell-autonomously required for the maintenance of fetal HSCs. This requirement is consistent with the observed dramatic reduction in HSC frequency and number in E14.5 CKO embryos before the onset of severe anemia and defects in in vivo reconstituting ability of FIP200-deficient HSCs. It is likely that the loss of fetal HSC activity contributed significantly to the perinatal lethality and anemia in CKO mice.
Increased HSC cycling and aberrant myeloid expansion after FIP200 deletion
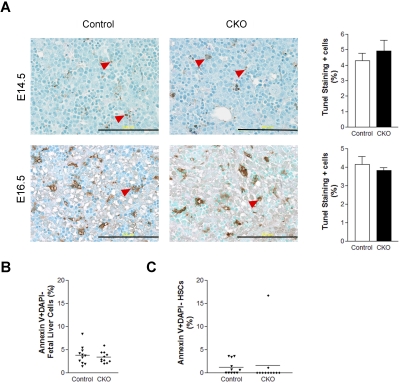
Our previous studies showed that FIP200 deletion led to increased apoptosis in several cell types in germ line and conditional deletion mutant mice.4,11 Thus, we wondered if increased apoptosis of HSCs upon FIP200 deletion led to the depletion of HSCs in CKO mice. To examine this possibility, we measured apoptosis of fetal liver cells in CKO and control embryos at E14.5 and E16.5 by performing terminal deoxynucleotidyl transferase dUTP nick end labeling (TUNEL) assays on fetal liver sections. Low levels (approximately 5%) of apoptotic cells were observed in the livers of both CKO and control embryos (Figure 5A). These results were further confirmed in E14.5 fetal liver cells by staining for annexin V followed by analysis using flow cytometry (Figure 5B). We also examined HSC cell death at E14.5 by annexin V staining and found that the levels of apoptosis in both CKO and control HSCs were, on average, comparably lower than in unfractionated fetal liver cells (Figure 5C). These results suggest that FIP200 deletion in hematopoietic cells did not appreciably increase apoptosis of either unfractionated fetal liver cells or HSCs and that HSC depletion in CKO mice was not caused by increased apoptosis.
Figure 5.
FIP200 deletion did not affect fetal liver cell apoptosis. (A) Fetal liver tissue sections from control and CKO mice at E14.5 and E16.5 were analyzed by TUNEL assays. Arrowheads indicate the positively stained apoptotic cells. Data on the right- side graphs are mean ± SE. n = 4-5. Scale bars, 200 μm. (B-C) Annexin V labeling of fetal liver cells (B) or fetal HSCs (C) from E14.5 control and CKO embryos. Data are mean ± SE.
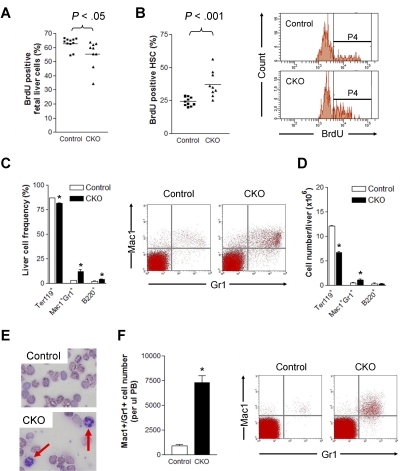
We also evaluated the effect of FIP200 deletion on the proliferation of HSCs by BrdU incorporation assays. BrdU was administered intraperitoneally into pregnant mothers carrying E14.5 embryos 2 hours before killing animals for analysis. After this short pulse, we noted a slightly lower level of BrdU incorporation in the unfractionated fetal liver cells of CKO embryos compared with control embryos (Figure 6A). Interestingly, a significant increase in BrdU incorporation was detected in FIP200-null HSCs (37.3% ± 9.9%) compared with control HSCs (24.3% ± 3.3%; Figure 6B). These results demonstrate that FIP200 plays a quantitative functional role in the cell cycle of fetal HSCs.
Figure 6.
FIP200 deletion led to increased HSC cell cycling and myeloid differentiation. (A-B) The percentage of BrdU positive fetal liver cells (A) and fetal HSCs (B) in E14.5 control and CKO mice after 2 hours BrdU pulse. Representative FACS profile of fetal HSCs is shown on the right in panel B. P4 represents the BrdU+ population. (C-D) Frequency (C) and number (D) of different lineage fetal liver cells of E14.5 control and CKO mice. Representative FACS profile of Mac1+Gr1+ population is shown on the right in panel C. n = 6-13, *P < .05. Data are mean ± SE. (E) Wright-Giemsa staining of the blood smears from E18.5 control and CKO embryos. The arrows indicate neutrophils. (F) Number of Mac1+Gr1+ cells/μL peripheral blood of E18.5 control and CKO embryos. Representative FACS profile is shown on the right. n = 3-15, *P < .05. Data are mean ± SE.
We next investigated potential aberrant differentiation and lineage progression of hematopoietic cells in CKO mice. Fetal liver cells were isolated from CKO and control embryos at E14.5 and analyzed by flow cytometry. While only slight changes were found for erythroid and B-cell lineages, a significant increase in the number of myeloid lineage cells (> 4-fold) was detected in CKO liver compared with control mice (Figure 6C). Moreover, despite the decrease in overall liver cellularity (see Figure 2A) and the corresponding decrease in the number of erythrocytes, an increased absolute number of myeloid cells was present per fetal liver in CKO embryos compared with that of control embryos (Figure 6D). The increase in frequency and absolute number of myeloid cells indicated enhanced myelopoiesis in the fetal liver of CKO mice. Consistent with this, Wright-Giemsa staining of peripheral blood from E18.5 embryos displayed an increase in the number of neutrophils in CKO embryos compared with control embryos (Figure 6E). Further analysis of the peripheral blood by flow cytometry indicated approximately 17-fold (from approximately 0.03% to 0.52%) and 8-fold increases in the frequency and numbers per microliter, respectively, of myeloid cells in the blood of CKO embryos compared with control embryos (Figure 6F). Taken together, these data suggest significantly increased myeloid lineage expansion in CKO mice.
Increased mitochondrial mass and ROS in FIP200-null hematopoietic cells
Several recent studies identified FIP200 as an essential component of the ULK1-Atg13-FIP200 complex involved in the generation of autophagosomes.6–8,19,20 Autophagy is thought to mediate the clearance of damaged and/or excess organelles including mitochondria, which are a major source of intracellular ROS.21,22 While normal HSCs contain a low level of ROS, an abnormal increase of ROS has been associated with increased cell-cycle progression and depletion of adult HSCs.23–25 Therefore, we investigated the possibility that autophagic defects in hematopoietic cells upon FIP200 deletion may cause an abnormal accumulation of mitochondria and increased ROS level.
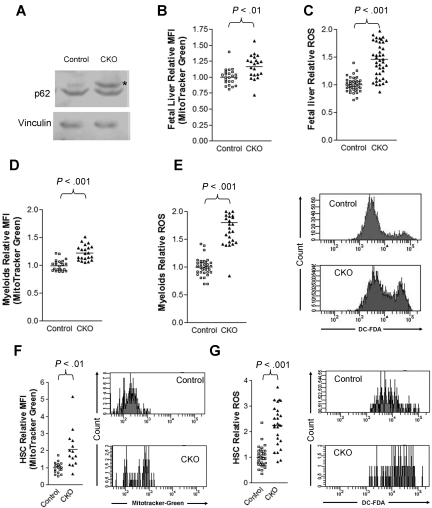
Consistent with a defect in autophagy, we observed an accumulation of p62, a selective substrate for autophagy, in CKO fetal liver cells compared with control cells (Figure 7A). Moreover, we observed an increase in mitochondrial mass in E14.5 CKO fetal liver cells compared with control cells by staining cells with the cell-permeant MitoTracker Green probe and analyzing by flow cytometry (Figure 7B). Lastly, we detected a 50% increase in ROS levels in CKO fetal liver cells by DCF-DA (Figure 7C), consistent with a model in which impaired autophagic clearance led to the accumulation of mitochondria and increased ROS levels. We also observed that mitochondrial mass (Figure 7D) and ROS levels (Figure 7E) increased in Mac-1+Gr-1+ cells in the E14.5 CKO fetal liver compared with controls. Likewise, we detected greater increases in both mitochondrial content (Figure 7F) and ROS levels (Figure 7G) in E14.5 CKO fetal liver HSCs compared with control fetal liver HSCs. Taken together, these results suggest that FIP200 deletion results in the increased mitochondrial mass and ROS levels in hematopoietic cells, which could contribute to the various hematopoietic defects in CKO embryos.
Figure 7.
FIP200 deletion led to increased mitochondrion mass and ROS in fetal hematopoietic cells. (A) Lysates from E14.5 liver of control or CKO mice were analyzed by Western blotting using anti-p62 (top) or anti-vinculin (bottom) antibodies. Asterisk on the right indicates the p62 band. (B-C) Relative mitochondrial mass (B) and ROS (C) of E14.5 fetal liver cells as measured by mean fluorescent intensity (MFI) of MitoTracker Green staining and DCF-DA staining, respectively. (D-E) Relative mitochondrial mass (D) and ROS (E) of myeloid cells from E14.5 fetal liver cells as measured by MFI of MitoTracker Green staining and DCF-DA staining, respectively. Representative FACS profiles of DCF-DA staining are shown on the right in panel E. (F-G) Relative mitochondrial mass (F) and ROS (G) of fetal HSCs from E14.5 embryos as measured by MFI of MitoTracker Green staining and DCF-DA staining, respectively. Representative FACS profiles of MitoTracker Green staining and DCF-DA staining are shown on the right.
Discussion
Compared with our understanding of the regulation of adult HSCs by both intrinsic factors and extrinsic niches,23,26–33 relatively little is known about the regulatory pathways for the maintenance of fetal HSCs. Here, we present data showing that deletion of FIP200 in hematopoietic cells resulted in the depletion of fetal HSCs in a cell-autonomous manner. Decreased liver cellularity was observed in CKO mice as early as E14.5 and progressed throughout development in the fetal liver. A massive decrease in fetal HSC frequency as well as number was found in CKO embryos. More importantly, fetal liver cells from CKO embryos completely lost the multilineage reconstituting ability when transplanted to wild-type recipient mice and so did those from FIP200F/F;Mx1-Cre mice after injection of pIpC to delete floxed FIP200 after transplantation.
The mechanisms by which inactivation of FIP200 led to fetal HSC depletion are still not clear at present. Although FIP200 deletion has been shown to increase apoptosis of cardiomyocytes, hepatocytes, and neurons in previous studies,4,11 comparable levels of apoptosis were found in fetal HSCs of CKO and control mice, excluding altered cell survival as a major mechanism of fetal HSC depletion upon FIP200 inactivation. Likewise, we did not find decreased proliferation of fetal HSCs in CKO mice compared with control cells, indicating that depletion of CKO HSCs was not caused by deficient proliferation of fetal HSCs. Surprisingly, we actually found a significant increase in the proliferation of FIP200-null fetal HSCs compared with control cells. It is well known that aberrantly increased cell cycling can lead to the depletion of adult HSCs, which are quiescent under normal conditions.28,30,34–38 Our data raise the interesting possibility that abnormally increased cell-cycle progression in fetal HSCs could also lead to their depletion, even though the fetal HSC pool expands rapidly under normal conditions in contrast to the quiescent nature of adult HSCs.
Consistent with our previous observations in neurons,11 deletion of FIP200 in hematopoietic cells led to increased accumulation of p62/SQSTM1 as a consequence of deficient autophagy. Moreover, we also observed increased mitochondrial mass in FIP200-null fetal HSCs as well as myeloid cells and total fetal liver cells by MitoTracker labeling, which is also consistent with the accumulation of damaged mitochondria in FIP200-null Purkinje cells as visualized directly by electron microscopy.11 Autophagy defects and associated accumulation and/or damage of mitochondria often results in an abnormal increase in the cellular level of ROS.39–42 Increased ROS has been associated with increased cell-cycle progression and depletion of adult HSCs.23,24 Therefore, our data are consistent with the idea that, similar to adult HSCs, the increased mitochondrial mass and ROS after deletion of FIP200 could contribute at least in part to the depletion of fetal HSCs in CKO mice.
We observed significantly increased myelopoiesis in CKO fetal livers, although the total cellularity already significantly decreased in these mice compared with control mice at E14.5. The abnormal myelopoiesis was also confirmed by the presence of nearly a 10-fold increase in the absolute number of myeloid cells in the peripheral blood of CKO mice (see Figure 6F). It is interesting to note that increased ROS caused by deletion of FoxO, a transcriptional activator of antioxidant scavenger protein expression, also led to myeloid lineage expansion in adult mice,23 similar to our observation of increased myeloid cells in fetal liver and embryonic peripheral blood of CKO mice. Furthermore, a recent report showed that in Drosophila, increasing the hematopoietic progenitor ROS beyond their basal level triggers precocious differentiation into all 3 mature blood cell types with functions reminiscent of the vertebrate myeloid lineage.43 Thus increased myelopoiesis may occur in response to elevated ROS levels, contributing to the depletion of HSCs, although it remains unknown whether FIP200 deficiency increases myeloid lineage commitment by fetal HSCs or the expansion of myeloid lineage progenitors.
Similar to several recent reports implicating a role of autophagy in erythropoiesis,41,44–46 we observed a significant decrease in erythroid maturation in CKO fetal livers, which likely also contributed to the severe anemia, beyond the fetal HSC defects in these mice. Although to a lesser extent than fetal HSCs, mitochondrial mass was increased in total fetal liver cells (which are mostly composed of erythrocyte lineage cells) from CKO mice. It is possible that deficient autophagy to clear mitochondria in FIP200-null erythroid cells was responsible for their compromised maturation as observed in Atg7−/− cells.45,47 Moreover, erythroid cells accumulate hemoglobin as they mature and are highly prone to oxidative damage. The lifespan of the erythrocyte depends on an adequate antioxidant response.48 It was shown that the increased ROS caused by FoxO3 deficiency had a ROS-mediated shortened lifespan associated with oxidative damage as well as decreased erythroid maturation.49 Interestingly, we also observed increased ROS in the R4 population of CKO embryos compared with control mice (supplemental Figure 5). Therefore, the increased ROS level in FIP200-null erythrocytes could also contribute to the maturation defect as well as anemia in CKO mice.
In conclusion, our study identified FIP200 as a critical cell-autonomous regulator of fetal HSCs. Given the recently described function of FIP200 in autophagy, these results provide the first suggestion for a potential role of autophagy in HSCs. While several previous studies established a role for autophagy in hematopoiesis, particularly in erythropoiesis,44–46,50 no specific defects in HSCs were described in those studies. It should be noted, however, FIP200 has other cellular functions through interaction with other proteins besides autophagy in ULK1-Atg13-FIP200 complex, which could also potentially contribute to the fetal HSCs defects in CKO mice. Indeed, neural deletion of FIP200 exhibited similar as well as distinctive phenotypes compared with mice with Atg5 or Atg7 inactivation in the same neurons.11 Future studies examining potential defects in HSCs upon deletion of other key components of autophagy such as Atg5 or Atg7 will help to clarify autophagy-dependent and -independent functions of FIP200 in the regulation of HSCs.
Supplementary Material
Acknowledgments
We are grateful to Dr Tracy Stokol of Cornell University for her help in the initial stage of the studies and Dr Lihong Shi of University of Michigan for her assistance in erythroblasts analysis by staining, and Drs Chong Chen and Yang Liu of University of Michigan for MitoTracker reagent and assistance in the experiments. We thank our laboratory colleagues for helpful comments of the manuscript.
This research was supported by National Institutes of Health grants HL073394 and GM052890 to J.-L.G.; F.L. was supported by a NIDCR training grant (T32DE007057).
Footnotes
The online version of this article contains a data supplement.
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 USC section 1734.
Authorship
Contribution: F.L., J.Y.L., and H.W. designed and performed research and analyzed data; O.T. and J.D.E. contributed analytical tools and analyzed data; S.J.M. and J.-L.G. designed research and analyzed data; and F.L. and J.-L.G. wrote the paper with contribution from other authors.
Conflict-of-interest disclosure: The authors declare no competing financial interests.
Correspondence: Jun-Lin Guan, Division of Molecular Medicine and Genetics, Department of Internal Medicine, and Department of Cell and Developmental Biology, University of Michigan Medical School, 109 Zina Pitcher, Rm 3027 BSRB, Ann Arbor, MI 48109; e-mail: jlguan@umich.edu.
References
- 1.Ueda H, Abbi S, Zheng C, Guan JL. Suppression of Pyk2 kinase and cellular activities by FIP200. J Cell Biol. 2000;149(2):423–430. doi: 10.1083/jcb.149.2.423. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Gan B, Guan JL. FIP200, a key signaling node to coordinately regulate various cellular processes. Cell Signal. 2008;20(5):787–794. doi: 10.1016/j.cellsig.2007.10.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Bamba N, Chano T, Taga T, Ohta S, Takeuchi Y, Okabe H. Expression and regulation of RB1CC1 in developing murine and human tissues. Int J Mol Med. 2004;14(4):583–587. [PubMed] [Google Scholar]
- 4.Gan B, Peng X, Nagy T, Alcaraz A, Gu H, Guan JL. Role of FIP200 in cardiac and liver development and its regulation of TNFα and TSC-mTOR signaling pathways. J Cell Biol. 2006;175(1):121–133. doi: 10.1083/jcb.200604129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Ganley IG, Lam du H, Wang J, Ding X, Chen S, Jiang X. ULK1.ATG13.FIP200 complex mediates mTOR signaling and is essential for autophagy. J Biol Chem. 2009;284(18):12297–12305. doi: 10.1074/jbc.M900573200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Hara T, Mizushima N. Role of ULK-FIP200 complex in mammalian autophagy: FIP200, a counterpart of yeast Atg17? Autophagy. 2009;5(1):85–87. doi: 10.4161/auto.5.1.7180. [DOI] [PubMed] [Google Scholar]
- 7.Hosokawa N, Hara T, Kaizuka T, et al. Nutrient-dependent mTORC1 association with the ULK1-Atg13-FIP200 complex required for autophagy. Mol Biol Cell. 2009;20(7):1981–1991. doi: 10.1091/mbc.E08-12-1248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Jung CH, Jun CB, Ro SH, et al. ULK-Atg13-FIP200 complexes mediate mTOR signaling to the autophagy machinery. Mol Biol Cell. 2009;20(7):1992–2003. doi: 10.1091/mbc.E08-12-1249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Hara T, Nakamura K, Matsui M, et al. Suppression of basal autophagy in neural cells causes neurodegenerative disease in mice. Nature. 2006;441(7095):885–889. doi: 10.1038/nature04724. [DOI] [PubMed] [Google Scholar]
- 10.Komatsu M, Waguri S, Chiba T, et al. Loss of autophagy in the central nervous system causes neurodegeneration in mice. Nature. 2006;441(7095):880–884. doi: 10.1038/nature04723. [DOI] [PubMed] [Google Scholar]
- 11.Liang CC, Wang C, Peng X, Gan B, Guan JL. Neural-specific deletion of FIP200 leads to cerebellar degeneration caused by increased neuronal death and axon degeneration. J Biol Chem. 2010;285(5):3499–3509. doi: 10.1074/jbc.M109.072389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Mizushima N, Levine B, Cuervo AM, Klionsky DJ. Autophagy fights disease through cellular self-digestion. Nature. 2008;451(7182):1069–1075. doi: 10.1038/nature06639. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Shen TL, Park AY, Alcaraz A, et al. Conditional knockout of focal adhesion kinase in endothelial cells reveals its role in angiogenesis and vascular development in late embryogenesis. J Cell Biol. 2005;169(6):941–952. doi: 10.1083/jcb.200411155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Kim I, Saunders TL, Morrison SJ. Sox17 dependence distinguishes the transcriptional regulation of fetal from adult hematopoietic stem cells. Cell. 2007;130(3):470–483. doi: 10.1016/j.cell.2007.06.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Kim I, He S, Yilmaz OH, Kiel MJ, Morrison SJ. Enhanced purification of fetal liver hematopoietic stem cells using SLAM family receptors. Blood. 2006;108(2):737–744. doi: 10.1182/blood-2005-10-4135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Morrison SJ, Hemmati HD, Wandycz AM, Weissman IL. The purification and characterization of fetal liver hematopoietic stem cells. Proc Natl Acad Sci U S A. 1995;92(22):10302–10306. doi: 10.1073/pnas.92.22.10302. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Kiel MJ, Yilmaz OH, Iwashita T, Yilmaz OH, Terhorst C, Morrison SJ. SLAM family receptors distinguish hematopoietic stem and progenitor cells and reveal endothelial niches for stem cells. Cell. 2005;121(7):1109–1121. doi: 10.1016/j.cell.2005.05.026. [DOI] [PubMed] [Google Scholar]
- 18.Butler JM, Nolan DJ, Vertes EL, et al. Endothelial cells are essential for the self-renewal and repopulation of Notch-dependent hematopoietic stem cells. Cell Stem Cell. 2010;6(3):251–264. doi: 10.1016/j.stem.2010.02.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Hara T, Takamura A, Kishi C, et al. FIP200, a ULK-interacting protein, is required for autophagosome formation in mammalian cells. J Cell Biol. 2008;181(3):497–510. doi: 10.1083/jcb.200712064. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Nishida Y, Arakawa S, Fujitani K, et al. Discovery of Atg5/Atg7-independent alternative macroautophagy. Nature. 2009;461(7264):654–658. doi: 10.1038/nature08455. [DOI] [PubMed] [Google Scholar]
- 21.Cumming RC, Lightfoot J, Beard K, Youssoufian H, O'Brien PJ, Buchwald M. Fanconi anemia group C protein prevents apoptosis in hematopoietic cells through redox regulation of GSTP1. Nat Med. 2001;7(7):814–820. doi: 10.1038/89937. [DOI] [PubMed] [Google Scholar]
- 22.Ito K, Hirao A, Arai F, et al. Regulation of oxidative stress by ATM is required for self-renewal of haematopoietic stem cells. Nature. 2004;431(7011):997–1002. doi: 10.1038/nature02989. [DOI] [PubMed] [Google Scholar]
- 23.Tothova Z, Kollipara R, Huntly BJ, et al. FoxOs are critical mediators of hematopoietic stem cell resistance to physiologic oxidative stress. Cell. 2007;128(2):325–339. doi: 10.1016/j.cell.2007.01.003. [DOI] [PubMed] [Google Scholar]
- 24.Chen C, Liu Y, Liu R, et al. TSC-mTOR maintains quiescence and function of hematopoietic stem cells by repressing mitochondrial biogenesis and reactive oxygen species. J Exp Med. 2008;205(10):2397–2408. doi: 10.1084/jem.20081297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Ito K, Hirao A, Arai F, et al. Reactive oxygen species act through p38 MAPK to limit the lifespan of hematopoietic stem cells. Nat Med. 2006;12(4):446–451. doi: 10.1038/nm1388. [DOI] [PubMed] [Google Scholar]
- 26.Ficara F, Murphy MJ, Lin M, Cleary ML. Pbx1 regulates self-renewal of long-term hematopoietic stem cells by maintaining their quiescence. Cell Stem Cell. 2008;2(5):484–496. doi: 10.1016/j.stem.2008.03.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Galan-Caridad JM, Harel S, Arenzana TL, et al. Zfx controls the self-renewal of embryonic and hematopoietic stem cells. Cell. 2007;129(2):345–357. doi: 10.1016/j.cell.2007.03.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Jude CD, Climer L, Xu D, Artinger E, Fisher JK, Ernst P. Unique and independent roles for MLL in adult hematopoietic stem cells and progenitors. Cell Stem Cell. 2007;1(3):324–337. doi: 10.1016/j.stem.2007.05.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Kranc KR, Schepers H, Rodrigues NP, et al. Cited2 is an essential regulator of adult hematopoietic stem cells. Cell Stem Cell. 2009;5(6):659–665. doi: 10.1016/j.stem.2009.11.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Matsuoka S, Oike Y, Onoyama I, et al. Fbxw7 acts as a critical fail-safe against premature loss of hematopoietic stem cells and development of T-ALL. Genes Dev. 2008;22(8):986–991. doi: 10.1101/gad.1621808. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Renstrom J, Istvanffy R, Gauthier K, et al. Secreted frizzled-related protein 1 extrinsically regulates cycling activity and maintenance of hematopoietic stem cells. Cell Stem Cell. 2009;5(2):157–167. doi: 10.1016/j.stem.2009.05.020. [DOI] [PubMed] [Google Scholar]
- 32.Trowbridge JJ, Snow JW, Kim J, Orkin SH. DNA methyltransferase 1 is essential for and uniquely regulates hematopoietic stem and progenitor cells. Cell Stem Cell. 2009;5(4):442–449. doi: 10.1016/j.stem.2009.08.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Walkley CR, Shea JM, Sims NA, Purton LE, Orkin SH. Rb regulates interactions between hematopoietic stem cells and their bone marrow microenvironment. Cell. 2007;129(6):1081–1095. doi: 10.1016/j.cell.2007.03.055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Liu Y, Elf SE, Miyata Y, et al. p53 regulates hematopoietic stem cell quiescence. Cell Stem Cell. 2009;4(1):37–48. doi: 10.1016/j.stem.2008.11.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Zhang J, Grindley JC, Yin T, et al. PTEN maintains haematopoietic stem cells and acts in lineage choice and leukaemia prevention. Nature. 2006;441(7092):518–522. doi: 10.1038/nature04747. [DOI] [PubMed] [Google Scholar]
- 36.Yang L, Wang L, Geiger H, Cancelas JA, Mo J, Zheng Y. Rho GTPase Cdc42 coordinates hematopoietic stem cell quiescence and niche interaction in the bone marrow. Proc Natl Acad Sci U S A. 2007;104(12):5091–5096. doi: 10.1073/pnas.0610819104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Thompson BJ, Jankovic V, Gao J, et al. Control of hematopoietic stem cell quiescence by the E3 ubiquitin ligase Fbw7. J Exp Med. 2008;205(6):1395–1408. doi: 10.1084/jem.20080277. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Hock H, Hamblen MJ, Rooke HM, et al. Gfi-1 restricts proliferation and preserves functional integrity of haematopoietic stem cells. Nature. 2004;431(7011):1002–1007. doi: 10.1038/nature02994. [DOI] [PubMed] [Google Scholar]
- 39.Kim I, Rodriguez-Enriquez S, Lemasters JJ. Selective degradation of mitochondria by mitophagy. Arch Biochem Biophys. 2007;462(2):245–253. doi: 10.1016/j.abb.2007.03.034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Mathew R, Karp CM, Beaudoin B, et al. Autophagy suppresses tumorigenesis through elimination of p62. Cell. 2009;137(6):1062–1075. doi: 10.1016/j.cell.2009.03.048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Sandoval H, Thiagarajan P, Dasgupta SK, et al. Essential role for Nix in autophagic maturation of erythroid cells. Nature. 2008;454(7201):232–235. doi: 10.1038/nature07006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Tal MC, Sasai M, Lee HK, Yordy B, Shadel GS, Iwasaki A. Absence of autophagy results in reactive oxygen species-dependent amplification of RLR signaling. Proc Natl Acad Sci U S A. 2009;106(8):2770–2775. doi: 10.1073/pnas.0807694106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Owusu-Ansah E, Banerjee U. Reactive oxygen species prime Drosophila haematopoietic progenitors for differentiation. Nature. 2009;461(7263):537–541. doi: 10.1038/nature08313. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Kundu M, Lindsten T, Yang CY, et al. Ulk1 plays a critical role in the autophagic clearance of mitochondria and ribosomes during reticulocyte maturation. Blood. 2008;112(4):1493–1502. doi: 10.1182/blood-2008-02-137398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Mortensen M, Ferguson DJ, Edelmann M, et al. Loss of autophagy in erythroid cells leads to defective removal of mitochondria and severe anemia in vivo. Proc Natl Acad Sci U S A. 2010;107(2):832–837. doi: 10.1073/pnas.0913170107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Zhang J, Ney PA. NIX induces mitochondrial autophagy in reticulocytes. Autophagy. 2008;4(3):354–356. doi: 10.4161/auto.5552. [DOI] [PubMed] [Google Scholar]
- 47.Zhang J, Randall MS, Loyd MR, et al. Mitochondrial clearance is regulated by Atg7-dependent and -independent mechanisms during reticulocyte maturation. Blood. 2009;114(1):157–164. doi: 10.1182/blood-2008-04-151639. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Tsantes AE, Bonovas S, Travlou A, Sitaras NM. Redox imbalance, macrocytosis, and RBC homeostasis. Antioxid Redox Signal. 2006;8(7-8):1205–1216. doi: 10.1089/ars.2006.8.1205. [DOI] [PubMed] [Google Scholar]
- 49.Marinkovic D, Zhang X, Yalcin S, et al. Foxo3 is required for the regulation of oxidative stress in erythropoiesis. J Clin Invest. 2007;117(8):2133–2144. doi: 10.1172/JCI31807. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Mortensen M, Simon AK. Nonredundant role of Atg7 in mitochondrial clearance during erythroid development. Autophagy. 2010;6(3):423–425. doi: 10.4161/auto.6.3.11528. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.